Insights in Post-Translational Modifications: Ubiquitin and SUMO
Abstract
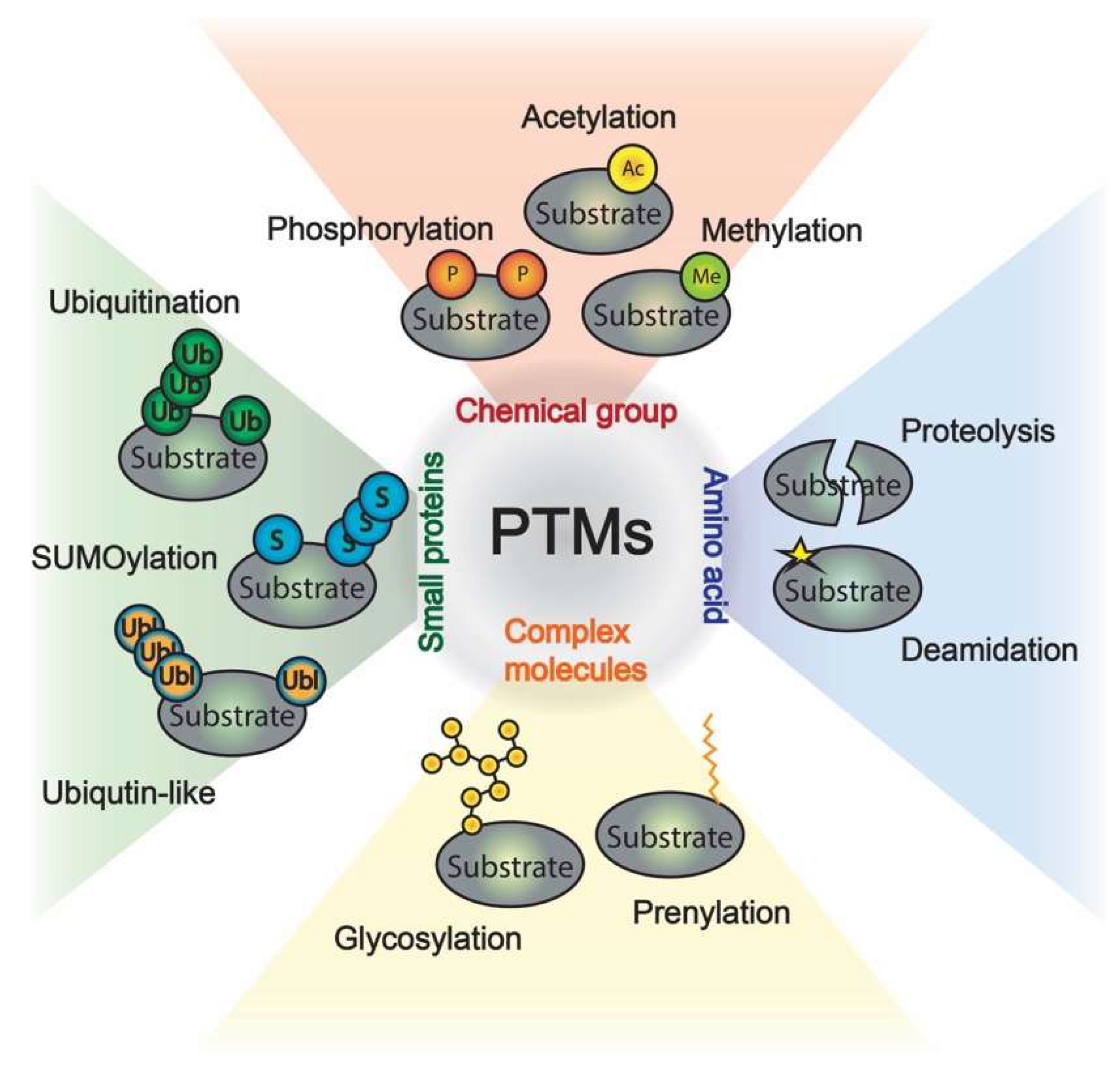
1. Introduction
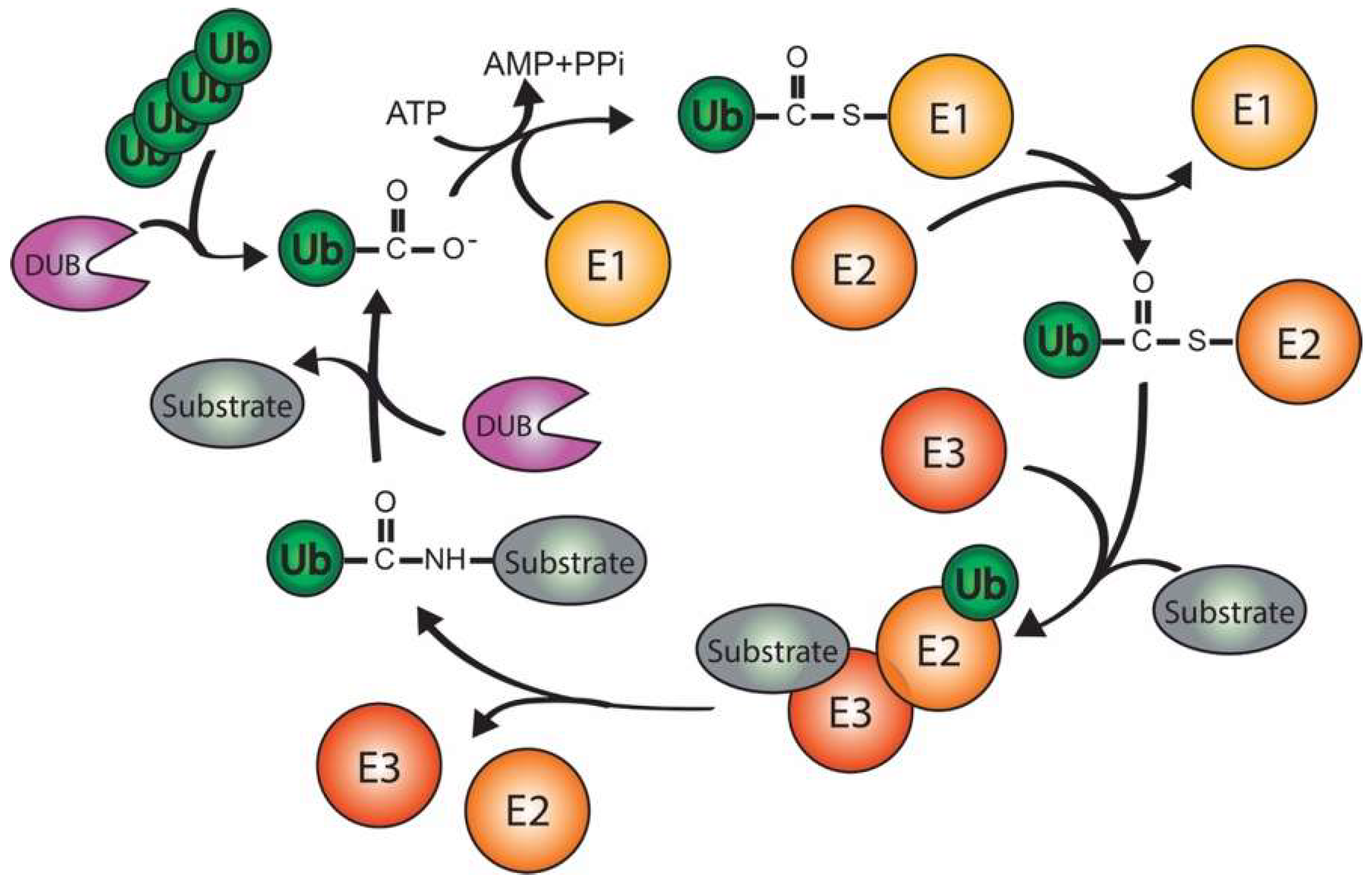
2. Ubiquitination
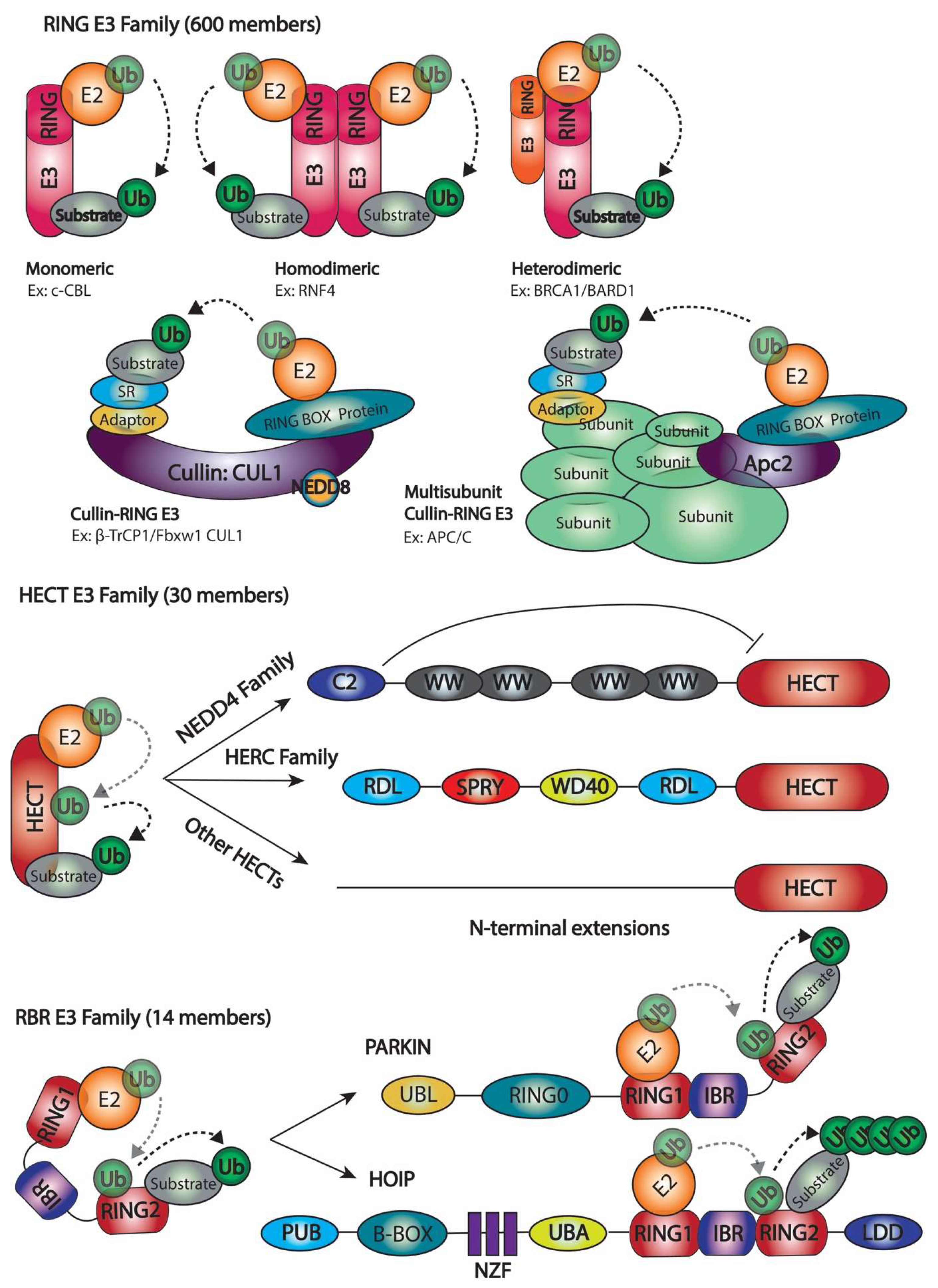
2.1. Number of Ubiquitination Enzymes
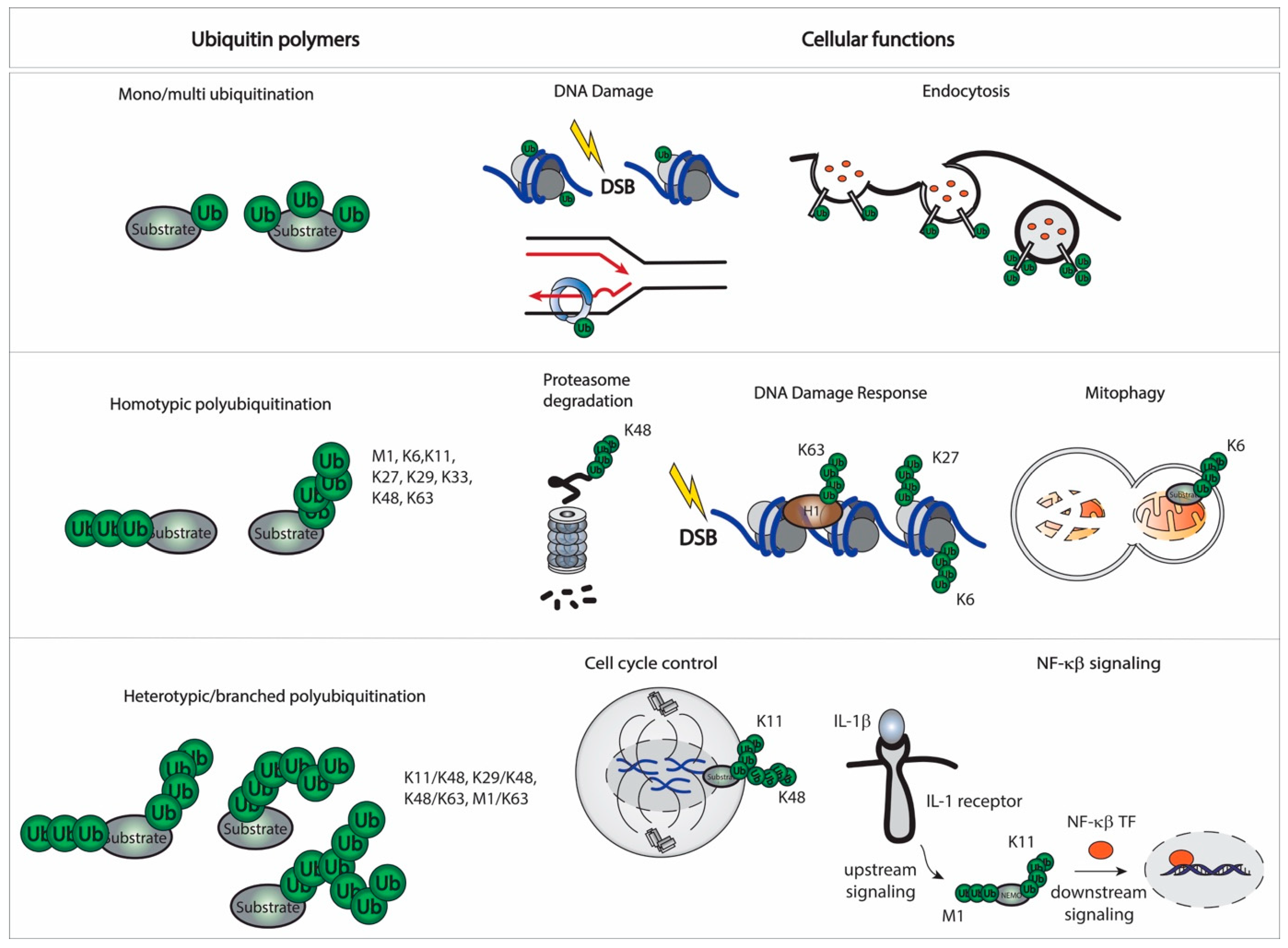
2.2. Linkages and Cellular Function
2.3. Crosstalk with Other Ubls
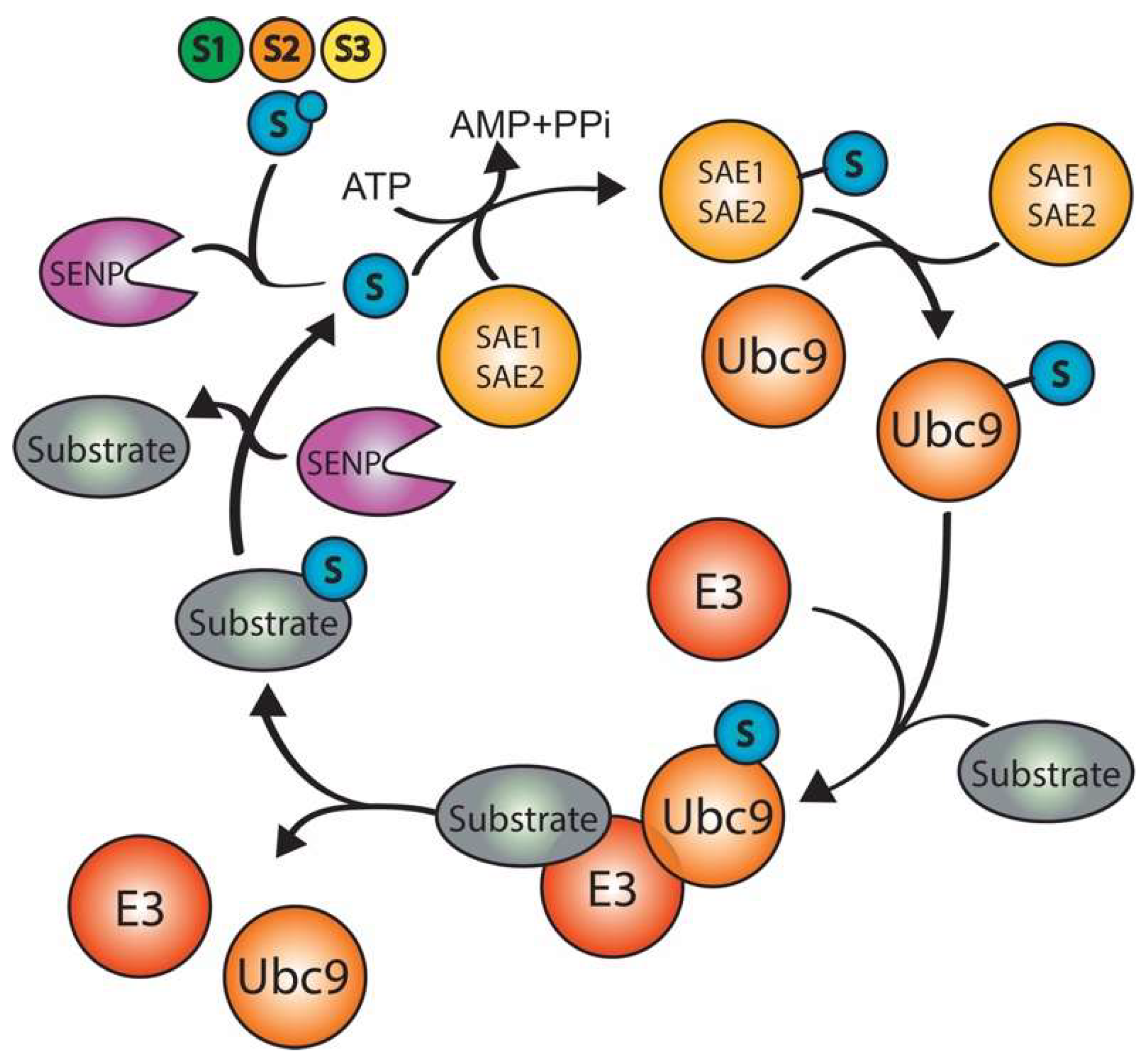
3. SUMOylation
3.1. SUMO E3 enzymes
3.2. SUMO Polymers
3.3. SUMO and Ubiquitin Crosstalk
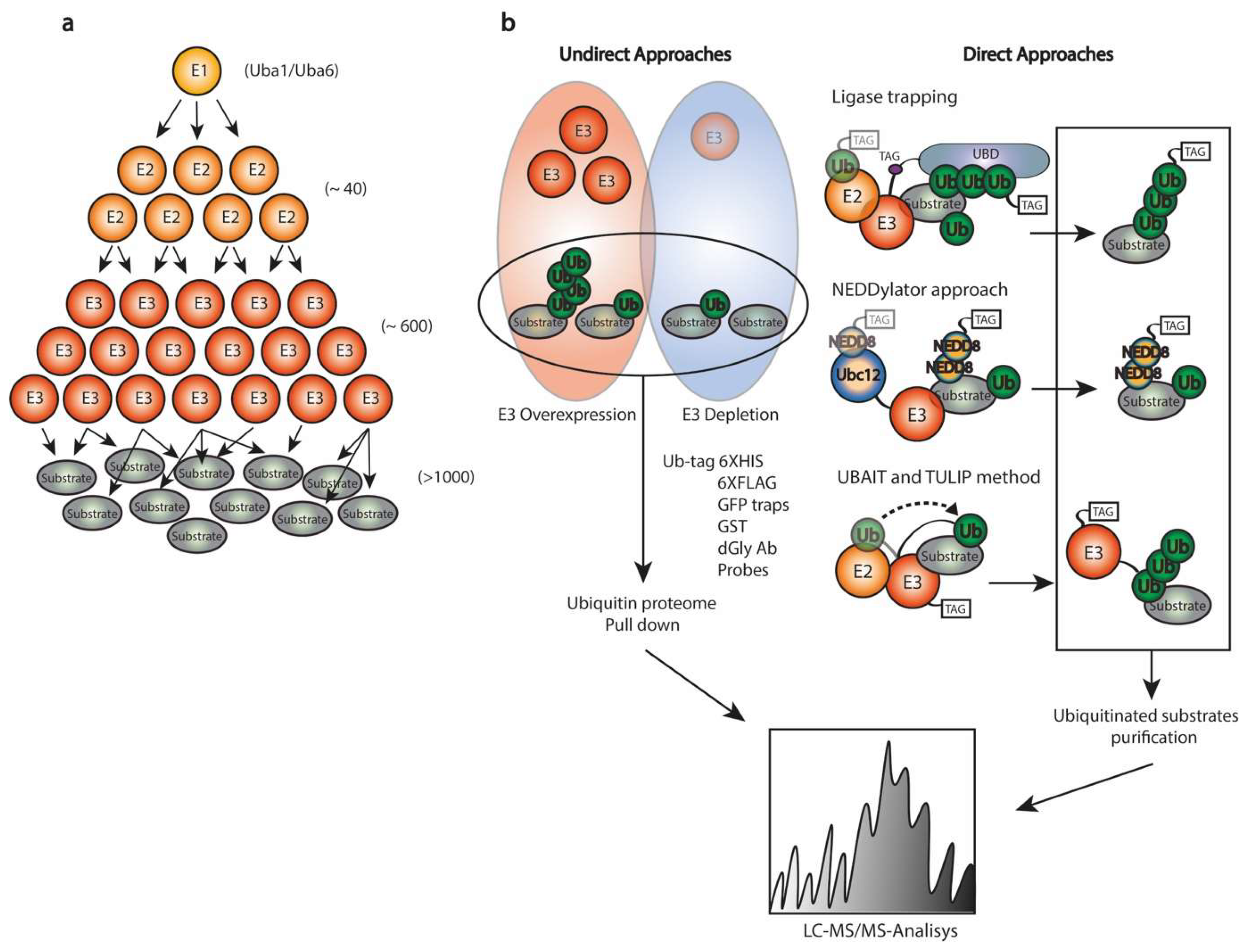
4. Proteomics for Substrate Identification
4.1. Mapping Sites
4.2. Identification of Novel Substrates
4.3. Other Ub/SUMO-Related MS-Based Proteomics Approaches
5. Conclusions and Future Perspectives
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
References
- Blazek, M.; Santisteban, T.S.; Zengerle, R.; Meier, M. Analysis of fast protein phosphorylation kinetics in single cells on a microfluidic chip. Lab Chip 2015, 15, 726–734. [Google Scholar] [CrossRef]
- Mohanty, P.; Chatterjee, K.S.; Das, R. NEDD8 Deamidation Inhibits Cullin RING Ligase Dynamics. Front. Immunol. 2021, 12, 695331. [Google Scholar] [CrossRef] [PubMed]
- Duan, G.; Walther, D. The roles of post-translational modifications in the context of protein interaction networks. PLoS Comput. Biol. 2015, 11, e1004049. [Google Scholar] [CrossRef] [PubMed]
- Lake, M.W.; Wuebbens, M.M.; Rajagopalan, K.V.; Schindelin, H. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature 2001, 414, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Xi, J.; Begley, T.P.; Nicholson, L.K. Solution structure of ThiS and implications for the evolutionary roots of ubiquitin. Nat. Struct. Biol. 2001, 8, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Pearce, M.J.; Mintseris, J.; Ferreyra, J.; Gygi, S.P.; Darwin, K.H. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science 2008, 322, 1104–1107. [Google Scholar] [CrossRef] [PubMed]
- Schulman, B.A.; Harper, J.W. Ubiquitin-like protein activation by E1 enzymes: The apex for downstream signalling pathways. Nat. Rev. Ther. Mol. Cell Biol. 2009, 10, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Aichem, A.; Sailer, C.; Ryu, S.; Catone, N.; Stankovic-Valentin, N.; Schmidtke, G.; Melchior, F.; Stengel, F.; Groettrup, M. The ubiquitin-like modifier FAT10 interferes with SUMO activation. Nat. Commun. 2019, 10, 4452. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Gonzalez-Prieto, R.; Xiao, Z.; Verlaan-de Vries, M.; Vertegaal, A.C.O. The STUbL RNF4 regulates protein group SUMOylation by targeting the SUMO conjugation machinery. Nat. Commun. 2017, 8, 1809. [Google Scholar] [CrossRef]
- Baek, K.; Krist, D.T.; Prabu, J.R.; Hill, S.; Klugel, M.; Neumaier, L.M.; von Gronau, S.; Kleiger, G.; Schulman, B.A. NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly. Nature 2020, 578, 461–466. [Google Scholar] [CrossRef]
- Fan, J.B.; Arimoto, K.; Motamedchaboki, K.; Yan, M.; Wolf, D.A.; Zhang, D.E. Identification and characterization of a novel ISG15-ubiquitin mixed chain and its role in regulating protein homeostasis. Sci. Rep. 2015, 5, 12704. [Google Scholar] [CrossRef] [PubMed]
- Grou, C.P.; Pinto, M.P.; Mendes, A.V.; Domingues, P.; Azevedo, J.E. The de novo synthesis of ubiquitin: Identification of deubiquitinases acting on ubiquitin precursors. Sci. Rep. 2015, 5, 12836. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, D.H.; Goldstein, G.; Niall, H.D. The complete amino acid sequence of ubiquitin, an adenylate cyclase stimulating polypeptide probably universal in living cells. Biochemistry 1975, 14, 2214–2218. [Google Scholar] [CrossRef]
- Scheffner, M.; Nuber, U.; Huibregtse, J.M. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 1995, 373, 81–83. [Google Scholar] [CrossRef] [PubMed]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Kirisako, T.; Kamei, K.; Murata, S.; Kato, M.; Fukumoto, H.; Kanie, M.; Sano, S.; Tokunaga, F.; Tanaka, K.; Iwai, K. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006, 25, 4877–4887. [Google Scholar] [CrossRef]
- Hershko, A.; Heller, H.; Elias, S.; Ciechanover, A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol. Chem. 1983, 258, 8206–8214. [Google Scholar] [CrossRef]
- Jin, J.; Li, X.; Gygi, S.P.; Harper, J.W. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature 2007, 447, 1135–1138. [Google Scholar] [CrossRef]
- Pelzer, C.; Kassner, I.; Matentzoglu, K.; Singh, R.K.; Wollscheid, H.P.; Scheffner, M.; Schmidtke, G.; Groettrup, M. UBE1L2, a novel E1 enzyme specific for ubiquitin. J. Biol. Chem. 2007, 282, 23010–23014. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.H.; Sun, Q.; Chen, Z.J. E1-L2 activates both ubiquitin and FAT10. Mol. Cell 2007, 27, 1014–1023. [Google Scholar] [CrossRef]
- Handley-Gearhart, P.M.; Stephen, A.G.; Trausch-Azar, J.S.; Ciechanover, A.; Schwartz, A.L. Human ubiquitin-activating enzyme, E1. Indication of potential nuclear and cytoplasmic subpopulations using epitope-tagged cDNA constructs. J. Biol. Chem. 1994, 269, 33171–33178. [Google Scholar] [CrossRef]
- Sugaya, K.; Ishihara, Y.; Inoue, S.; Tsuji, H. Characterization of ubiquitin-activating enzyme Uba1 in the nucleus by its mammalian temperature-sensitive mutant. PLoS ONE 2014, 9, e96666. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.D.; Ritterhoff, T.; Klevit, R.E.; Brzovic, P.S. E2 enzymes: More than just middle men. Cell Res. 2016, 26, 423–440. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Bengtson, M.H.; Ulbrich, A.; Matsuda, A.; Reddy, V.A.; Orth, A.; Chanda, S.K.; Batalov, S.; Joazeiro, C.A. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS ONE 2008, 3, e1487. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Beaudenon, S.L.; Kelley, M.L.; Waddell, M.B.; Yuan, W.; Schulman, B.A.; Huibregtse, J.M.; Krug, R.M. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-alpha/beta-induced ubiquitin-like protein. Proc. Natl. Acad. Sci. USA 2004, 101, 7578–7582. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.W.; Melchior, F.; Matunis, M.J.; Mahajan, R.; Tian, Q.; Anderson, P. Modification of Ran GTPase-activating protein by the small ubiquitin-related modifier SUMO-1 requires Ubc9, an E2-type ubiquitin-conjugating enzyme homologue. J. Biol. Chem. 1998, 273, 6503–6507. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.T.; Ayrault, O.; Hunt, H.W.; Taherbhoy, A.M.; Duda, D.M.; Scott, D.C.; Borg, L.A.; Neale, G.; Murray, P.J.; Roussel, M.F.; et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol. Cell 2009, 33, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Judes, A.; Bruch, A.; Klassen, R.; Helm, M.; Schaffrath, R. Sulfur transfer and activation by ubiquitin-like modifier system Uba4*Urm1 link protein urmylation and tRNA thiolation in yeast. Microb. Cell 2016, 3, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Tatsumi, K.; Sou, Y.S.; Tada, N.; Nakamura, E.; Iemura, S.; Natsume, T.; Kang, S.H.; Chung, C.H.; Kasahara, M.; Kominami, E.; et al. A novel type of E3 ligase for the Ufm1 conjugation system. J. Biol. Chem. 2010, 285, 5417–5427. [Google Scholar] [CrossRef] [PubMed]
- Shintani, T.; Mizushima, N.; Ogawa, Y.; Matsuura, A.; Noda, T.; Ohsumi, Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 1999, 18, 5234–5241. [Google Scholar] [CrossRef]
- Ichimura, Y.; Kirisako, T.; Takao, T.; Satomi, Y.; Shimonishi, Y.; Ishihara, N.; Mizushima, N.; Tanida, I.; Kominami, E.; Ohsumi, M.; et al. A ubiquitin-like system mediates protein lipidation. Nature 2000, 408, 488–492. [Google Scholar] [CrossRef]
- Ohi, M.D.; Vander Kooi, C.W.; Rosenberg, J.A.; Chazin, W.J.; Gould, K.L. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat. Struct. Biol. 2003, 10, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Lorick, K.L.; Jensen, J.P.; Fang, S.; Ong, A.M.; Hatakeyama, S.; Weissman, A.M. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 1999, 96, 11364–11369. [Google Scholar] [CrossRef]
- Mace, P.D.; Linke, K.; Feltham, R.; Schumacher, F.R.; Smith, C.A.; Vaux, D.L.; Silke, J.; Day, C.L. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J. Biol. Chem. 2008, 283, 31633–31640. [Google Scholar] [CrossRef]
- Bentley, M.L.; Corn, J.E.; Dong, K.C.; Phung, Q.; Cheung, T.K.; Cochran, A.G. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. EMBO J. 2011, 30, 3285–3297. [Google Scholar] [CrossRef]
- Brzovic, P.S.; Rajagopal, P.; Hoyt, D.W.; King, M.C.; Klevit, R.E. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol. 2001, 8, 833–837. [Google Scholar] [CrossRef]
- Liew, C.W.; Sun, H.; Hunter, T.; Day, C.L. RING domain dimerization is essential for RNF4 function. Biochem. J. 2010, 431, 23–29. [Google Scholar] [CrossRef]
- Metzger, M.B.; Pruneda, J.N.; Klevit, R.E.; Weissman, A.M. RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta 2014, 1843, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Lydeard, J.R.; Schulman, B.A.; Harper, J.W. Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep. 2013, 14, 1050–1061. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, A.; Stengel, F.; Zhang, Z.; Enchev, R.I.; Kong, E.H.; Morris, E.P.; Robinson, C.V.; da Fonseca, P.C.; Barford, D. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature 2011, 470, 227–232. [Google Scholar] [CrossRef]
- Huang, L.; Kinnucan, E.; Wang, G.; Beaudenon, S.; Howley, P.M.; Huibregtse, J.M.; Pavletich, N.P. Structure of an E6AP-UbcH7 complex: Insights into ubiquitination by the E2-E3 enzyme cascade. Science 1999, 286, 1321–1326. [Google Scholar] [CrossRef]
- Verdecia, M.A.; Joazeiro, C.A.; Wells, N.J.; Ferrer, J.L.; Bowman, M.E.; Hunter, T.; Noel, J.P. Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol. Cell 2003, 11, 249–259. [Google Scholar] [CrossRef]
- Wiesner, S.; Ogunjimi, A.A.; Wang, H.R.; Rotin, D.; Sicheri, F.; Wrana, J.L.; Forman-Kay, J.D. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell 2007, 130, 651–662. [Google Scholar] [CrossRef]
- Wenzel, D.M.; Lissounov, A.; Brzovic, P.S.; Klevit, R.E. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 2011, 474, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Reiter, K.H.; Klevit, R.E. Characterization of RING-Between-RING E3 Ubiquitin Transfer Mechanisms. Methods Mol. Biol. 2018, 1844, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Lechtenberg, B.C.; Rajput, A.; Sanishvili, R.; Dobaczewska, M.K.; Ware, C.F.; Mace, P.D.; Riedl, S.J. Structure of a HOIP/E2~ubiquitin complex reveals RBR E3 ligase mechanism and regulation. Nature 2016, 529, 546–550. [Google Scholar] [CrossRef] [PubMed]
- Kitada, T.; Asakawa, S.; Hattori, N.; Matsumine, H.; Yamamura, Y.; Minoshima, S.; Yokochi, M.; Mizuno, Y.; Shimizu, N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 1998, 392, 605–608. [Google Scholar] [CrossRef] [PubMed]
- Metzger, M.B.; Hristova, V.A.; Weissman, A.M. HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 2012, 125, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Sluimer, J.; Distel, B. Regulating the human HECT E3 ligases. Cell Mol. Life Sci. 2018, 75, 3121–3141. [Google Scholar] [CrossRef] [PubMed]
- Meyer, H.J.; Rape, M. Enhanced protein degradation by branched ubiquitin chains. Cell 2014, 157, 910–921. [Google Scholar] [CrossRef] [PubMed]
- Ziv, I.; Matiuhin, Y.; Kirkpatrick, D.S.; Erpapazoglou, Z.; Leon, S.; Pantazopoulou, M.; Kim, W.; Gygi, S.P.; Haguenauer-Tsapis, R.; Reis, N.; et al. A perturbed ubiquitin landscape distinguishes between ubiquitin in trafficking and in proteolysis. Mol. Cell Proteom. 2011, 10, M111.009753. [Google Scholar] [CrossRef]
- Passmore, L.A.; Barford, D. Getting into position: The catalytic mechanisms of protein ubiquitylation. Biochem. J. 2004, 379, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Alcon, P.; Shakeel, S.; Chen, Z.A.; Rappsilber, J.; Patel, K.J.; Passmore, L.A. FANCD2-FANCI is a clamp stabilized on DNA by monoubiquitination of FANCD2 during DNA repair. Nat. Struct. Mol. Biol. 2020, 27, 240–248. [Google Scholar] [CrossRef]
- Hedglin, M.; Aitha, M.; Pedley, A.; Benkovic, S.J. Replication protein A dynamically regulates monoubiquitination of proliferating cell nuclear antigen. J. Biol. Chem. 2019, 294, 5157–5168. [Google Scholar] [CrossRef] [PubMed]
- Thakar, T.; Leung, W.; Nicolae, C.M.; Clements, K.E.; Shen, B.; Bielinsky, A.K.; Moldovan, G.L. Ubiquitinated-PCNA protects replication forks from DNA2-mediated degradation by regulating Okazaki fragment maturation and chromatin assembly. Nat. Commun. 2020, 11, 2147. [Google Scholar] [CrossRef] [PubMed]
- Marsh, D.J.; Dickson, K.A. Writing Histone Monoubiquitination in Human Malignancy-The Role of RING Finger E3 Ubiquitin Ligases. Genes 2019, 10, 67. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; McGrath, B.; Bielas, S.L. Histone H2A Monoubiquitination in Neurodevelopmental Disorders. Trends Genet. 2017, 33, 566–578. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.R.; Clifford, G.; Bonnet, C.; Groth, A.; Wilson, M.D.; Chapman, J.R. BARD1 reads H2A lysine 15 ubiquitination to direct homologous recombination. Nature 2021, 596, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Haglund, K.; Sigismund, S.; Polo, S.; Szymkiewicz, I.; Di Fiore, P.P.; Dikic, I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 2003, 5, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Chau, V.; Tobias, J.W.; Bachmair, A.; Marriott, D.; Ecker, D.J.; Gonda, D.K.; Varshavsky, A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 1989, 243, 1576–1583. [Google Scholar] [CrossRef]
- Jin, L.; Williamson, A.; Banerjee, S.; Philipp, I.; Rape, M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 2008, 133, 653–665. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Wang, C.; Spencer, E.; Yang, L.; Braun, A.; You, J.; Slaughter, C.; Pickart, C.; Chen, Z.J. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 2000, 103, 351–361. [Google Scholar] [CrossRef]
- Plans, V.; Scheper, J.; Soler, M.; Loukili, N.; Okano, Y.; Thomson, T.M. The RING finger protein RNF8 recruits UBC13 for lysine 63-based self polyubiquitylation. J. Cell Biochem. 2006, 97, 572–582. [Google Scholar] [CrossRef]
- Shembade, N.; Harhaj, N.S.; Yamamoto, M.; Akira, S.; Harhaj, E.W. The human T-cell leukemia virus type 1 Tax oncoprotein requires the ubiquitin-conjugating enzyme Ubc13 for NF-kappaB activation. J. Virol. 2007, 81, 13735–13742. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, H.D.; Walden, H. Ubiquitin signalling in DNA replication and repair. Nat. Rev. Ther. Mol. Cell Biol. 2010, 11, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Bremm, A.; Komander, D. Emerging roles for Lys11-linked polyubiquitin in cellular regulation. Trends Biochem. Sci. 2011, 36, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Duong, D.M.; Seyfried, N.T.; Cheng, D.; Xie, Y.; Robert, J.; Rush, J.; Hochstrasser, M.; Finley, D.; Peng, J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 2009, 137, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Phu, L.; Izrael-Tomasevic, A.; Matsumoto, M.L.; Bustos, D.; Dynek, J.N.; Fedorova, A.V.; Bakalarski, C.E.; Arnott, D.; Deshayes, K.; Dixit, V.M.; et al. Improved quantitative mass spectrometry methods for characterizing complex ubiquitin signals. Mol. Cell Proteom. 2011, 10, M110.003756. [Google Scholar] [CrossRef]
- Durcan, T.M.; Tang, M.Y.; Perusse, J.R.; Dashti, E.A.; Aguileta, M.A.; McLelland, G.L.; Gros, P.; Shaler, T.A.; Faubert, D.; Coulombe, B.; et al. USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. EMBO J. 2014, 33, 2473–2491. [Google Scholar] [CrossRef]
- Ordureau, A.; Sarraf, S.A.; Duda, D.M.; Heo, J.M.; Jedrychowski, M.P.; Sviderskiy, V.O.; Olszewski, J.L.; Koerber, J.T.; Xie, T.; Beausoleil, S.A.; et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell 2014, 56, 360–375. [Google Scholar] [CrossRef] [PubMed]
- Wu-Baer, F.; Lagrazon, K.; Yuan, W.; Baer, R. The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J. Biol. Chem. 2003, 278, 34743–34746. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.R.; Solomon, E. BRCA1: BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum. Mol. Genet. 2004, 13, 807–817. [Google Scholar] [CrossRef] [PubMed]
- Michel, M.A.; Swatek, K.N.; Hospenthal, M.K.; Komander, D. Ubiquitin Linkage-Specific Affimers Reveal Insights into K6-Linked Ubiquitin Signaling. Mol. Cell 2017, 68, 233–246.e235. [Google Scholar] [CrossRef] [PubMed]
- Gatti, M.; Pinato, S.; Maiolica, A.; Rocchio, F.; Prato, M.G.; Aebersold, R.; Penengo, L. RNF168 promotes noncanonical K27 ubiquitination to signal DNA damage. Cell Rep. 2015, 10, 226–238. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Smits, A.H.; van Tilburg, G.B.; Jansen, P.W.; Makowski, M.M.; Ovaa, H.; Vermeulen, M. An Interaction Landscape of Ubiquitin Signaling. Mol. Cell 2017, 65, 941–955.e948. [Google Scholar] [CrossRef] [PubMed]
- Fei, C.; Li, Z.; Li, C.; Chen, Y.; Chen, Z.; He, X.; Mao, L.; Wang, X.; Zeng, R.; Li, L. Smurf1-mediated Lys29-linked nonproteolytic polyubiquitination of axin negatively regulates Wnt/beta-catenin signaling. Mol. Cell Biol. 2013, 33, 4095–4105. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Jeon, M.S.; Liao, L.; Yang, C.; Elly, C.; Yates, J.R., 3rd; Liu, Y.C. K33-linked polyubiquitination of T cell receptor-zeta regulates proteolysis-independent T cell signaling. Immunity 2010, 33, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Dittmar, G.; Winklhofer, K.F. Linear Ubiquitin Chains: Cellular Functions and Strategies for Detection and Quantification. Front. Chem. 2019, 7, 915. [Google Scholar] [CrossRef] [PubMed]
- Spit, M.; Rieser, E.; Walczak, H. Linear ubiquitination at a glance. J. Cell Sci. 2019, 132, jcs208512. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Liu, W.; Ye, Y.; Li, W. Ufd2p synthesizes branched ubiquitin chains to promote the degradation of substrates modified with atypical chains. Nat. Commun. 2017, 8, 14274. [Google Scholar] [CrossRef]
- Ohtake, F.; Saeki, Y.; Ishido, S.; Kanno, J.; Tanaka, K. The K48-K63 Branched Ubiquitin Chain Regulates NF-kappaB Signaling. Mol. Cell 2016, 64, 251–266. [Google Scholar] [CrossRef] [PubMed]
- Ohtake, F.; Tsuchiya, H.; Saeki, Y.; Tanaka, K. K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains. Proc. Natl. Acad. Sci. USA 2018, 115, E1401–E1408. [Google Scholar] [CrossRef] [PubMed]
- Wertz, I.E.; Newton, K.; Seshasayee, D.; Kusam, S.; Lam, C.; Zhang, J.; Popovych, N.; Helgason, E.; Schoeffler, A.; Jeet, S.; et al. Erratum: Phosphorylation and linear ubiquitin direct A20 inhibition of inflammation. Nature 2016, 532, 402. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.T.; Kim, K.P.; Lledias, F.; Kisselev, A.F.; Scaglione, K.M.; Skowyra, D.; Gygi, S.P.; Goldberg, A.L. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 2007, 282, 17375–17386. [Google Scholar] [CrossRef] [PubMed]
- Michel, M.A.; Elliott, P.R.; Swatek, K.N.; Simicek, M.; Pruneda, J.N.; Wagstaff, J.L.; Freund, S.M.; Komander, D. Assembly and specific recognition of k29- and k33-linked polyubiquitin. Mol. Cell 2015, 58, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Schwartz, D.; Elias, J.E.; Thoreen, C.C.; Cheng, D.; Marsischky, G.; Roelofs, J.; Finley, D.; Gygi, S.P. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003, 21, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Wegmann, S.; Meister, C.; Renz, C.; Yakoub, G.; Wollscheid, H.P.; Takahashi, D.T.; Mikicic, I.; Beli, P.; Ulrich, H.D. Linkage reprogramming by tailor-made E3s reveals polyubiquitin chain requirements in DNA-damage bypass. Mol. Cell 2022. [Google Scholar] [CrossRef]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef]
- Koyano, F.; Okatsu, K.; Kosako, H.; Tamura, Y.; Go, E.; Kimura, M.; Kimura, Y.; Tsuchiya, H.; Yoshihara, H.; Hirokawa, T.; et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 2014, 510, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Lamoliatte, F.; McManus, F.P.; Maarifi, G.; Chelbi-Alix, M.K.; Thibault, P. Uncovering the SUMOylation and ubiquitylation crosstalk in human cells using sequential peptide immunopurification. Nat. Commun. 2017, 8, 14109. [Google Scholar] [CrossRef]
- Liang, Y.C.; Lee, C.C.; Yao, Y.L.; Lai, C.C.; Schmitz, M.L.; Yang, W.M. SUMO5, a Novel Poly-SUMO Isoform, Regulates PML Nuclear Bodies. Sci. Rep. 2016, 6, 26509. [Google Scholar] [CrossRef] [PubMed]
- Evdokimov, E.; Sharma, P.; Lockett, S.J.; Lualdi, M.; Kuehn, M.R. Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J. Cell Sci. 2008, 121, 4106–4113. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wansleeben, C.; Zhao, S.; Miao, P.; Paschen, W.; Yang, W. SUMO2 is essential while SUMO3 is dispensable for mouse embryonic development. EMBO Rep. 2014, 15, 878–885. [Google Scholar] [CrossRef] [PubMed]
- Khoo, K.H.; Verma, C.S.; Lane, D.P. Drugging the p53 pathway: Understanding the route to clinical efficacy. Nat. Rev. Drug Discov. 2014, 13, 217–236. [Google Scholar] [CrossRef] [PubMed]
- Kessler, J.D.; Kahle, K.T.; Sun, T.; Meerbrey, K.L.; Schlabach, M.R.; Schmitt, E.M.; Skinner, S.O.; Xu, Q.; Li, M.Z.; Hartman, Z.C.; et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science 2012, 335, 348–353. [Google Scholar] [CrossRef]
- Su, H.L.; Li, S.S. Molecular features of human ubiquitin-like SUMO genes and their encoded proteins. Gene 2002, 296, 65–73. [Google Scholar] [CrossRef]
- Nayak, A.; Muller, S. SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol. 2014, 15, 422. [Google Scholar] [CrossRef]
- Bernier-Villamor, V.; Sampson, D.A.; Matunis, M.J.; Lima, C.D. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 2002, 108, 345–356. [Google Scholar] [CrossRef]
- Pichler, A.; Gast, A.; Seeler, J.S.; Dejean, A.; Melchior, F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 2002, 108, 109–120. [Google Scholar] [CrossRef]
- Reverter, D.; Lima, C.D. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 2005, 435, 687–692. [Google Scholar] [CrossRef]
- Johnson, E.S.; Gupta, A.A. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 2001, 106, 735–744. [Google Scholar] [CrossRef]
- Kahyo, T.; Nishida, T.; Yasuda, H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 2001, 8, 713–718. [Google Scholar] [CrossRef]
- Yunus, A.A.; Lima, C.D. Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol. Cell 2009, 35, 669–682. [Google Scholar] [CrossRef]
- Li, C.; McManus, F.P.; Plutoni, C.; Pascariu, C.M.; Nelson, T.; Alberici Delsin, L.E.; Emery, G.; Thibault, P. Quantitative SUMO proteomics identifies PIAS1 substrates involved in cell migration and motility. Nat. Commun. 2020, 11, 834. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, D.; Muller, S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl. Acad. Sci. USA 2002, 99, 2872–2877. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.S.; Zou, L. The SUMO (Small Ubiquitin-like Modifier) Ligase PIAS3 Primes ATR for Checkpoint Activation. J. Biol. Chem. 2016, 291, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Sachdev, S.; Bruhn, L.; Sieber, H.; Pichler, A.; Melchior, F.; Grosschedl, R. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 2001, 15, 3088–3103. [Google Scholar] [CrossRef]
- Potts, P.R.; Yu, H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol. Cell Biol. 2005, 25, 7021–7032. [Google Scholar] [CrossRef]
- Andrews, E.A.; Palecek, J.; Sergeant, J.; Taylor, E.; Lehmann, A.R.; Watts, F.Z. Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage. Mol. Cell Biol. 2005, 25, 185–196. [Google Scholar] [CrossRef]
- Moreno-Onate, M.; Herrero-Ruiz, A.M.; Garcia-Dominguez, M.; Cortes-Ledesma, F.; Ruiz, J.F. RanBP2-Mediated SUMOylation Promotes Human DNA Polymerase Lambda Nuclear Localization and DNA Repair. J. Mol. Biol. 2020, 423, 3965–3979. [Google Scholar] [CrossRef]
- Dawlaty, M.M.; Malureanu, L.; Jeganathan, K.B.; Kao, E.; Sustmann, C.; Tahk, S.; Shuai, K.; Grosschedl, R.; van Deursen, J.M. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell 2008, 133, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Eisenhardt, N.; Chaugule, V.K.; Koidl, S.; Droescher, M.; Dogan, E.; Rettich, J.; Sutinen, P.; Imanishi, S.Y.; Hofmann, K.; Palvimo, J.J.; et al. A new vertebrate SUMO enzyme family reveals insights into SUMO-chain assembly. Nat. Struct. Mol. Biol. 2015, 22, 959–967. [Google Scholar] [CrossRef] [PubMed]
- Koidl, S.; Eisenhardt, N.; Fatouros, C.; Droescher, M.; Chaugule, V.K.; Pichler, A. The SUMO2/3 specific E3 ligase ZNF451-1 regulates PML stability. Int. J. Biochem. Cell Biol. 2016, 79, 478–487. [Google Scholar] [CrossRef]
- Guervilly, J.H.; Takedachi, A.; Naim, V.; Scaglione, S.; Chawhan, C.; Lovera, Y.; Despras, E.; Kuraoka, I.; Kannouche, P.; Rosselli, F.; et al. The SLX4 complex is a SUMO E3 ligase that impacts on replication stress outcome and genome stability. Mol. Cell 2015, 57, 123–137. [Google Scholar] [CrossRef]
- Kagey, M.H.; Melhuish, T.A.; Wotton, D. The polycomb protein Pc2 is a SUMO E3. Cell 2003, 113, 127–137. [Google Scholar] [CrossRef]
- Weger, S.; Hammer, E.; Heilbronn, R. Topors acts as a SUMO-1 E3 ligase for p53 in vitro and in vivo. FEBS Lett. 2005, 579, 5007–5012. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gutierrez, P.; Juarez-Vicente, F.; Gallardo-Chamizo, F.; Charnay, P.; Garcia-Dominguez, M. The transcription factor Krox20 is an E3 ligase that sumoylates its Nab coregulators. EMBO Rep. 2011, 12, 1018–1023. [Google Scholar] [CrossRef] [PubMed]
- Tago, K.; Chiocca, S.; Sherr, C.J. Sumoylation induced by the Arf tumor suppressor: A p53-independent function. Proc. Natl. Acad. Sci. USA 2005, 102, 7689–7694. [Google Scholar] [CrossRef]
- Zhao, X.; Sternsdorf, T.; Bolger, T.A.; Evans, R.M.; Yao, T.P. Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol. Cell Biol. 2005, 25, 8456–8464. [Google Scholar] [CrossRef]
- Subramaniam, S.; Mealer, R.G.; Sixt, K.M.; Barrow, R.K.; Usiello, A.; Snyder, S.H. Rhes, a physiologic regulator of sumoylation, enhances cross-sumoylation between the basic sumoylation enzymes E1 and Ubc9. J. Biol. Chem. 2010, 285, 20428–20432. [Google Scholar] [CrossRef]
- Roth, W.; Sustmann, C.; Kieslinger, M.; Gilmozzi, A.; Irmer, D.; Kremmer, E.; Turck, C.; Grosschedl, R. PIASy-deficient mice display modest defects in IFN and Wnt signaling. J. Immunol. 2004, 173, 6189–6199. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.A.; Kim, R.; Christofk, H.; Gao, J.; Lawson, G.; Wu, H. Protein inhibitor of activated STAT Y (PIASy) and a splice variant lacking exon 6 enhance sumoylation but are not essential for embryogenesis and adult life. Mol. Cell Biol. 2004, 24, 5577–5586. [Google Scholar] [CrossRef] [PubMed]
- Werner, A.; Flotho, A.; Melchior, F. The RanBP2/RanGAP1*SUMO1/Ubc9 complex is a multisubunit SUMO E3 ligase. Mol. Cell 2012, 46, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.; Tan, S.H.; Karpova, T.S.; McNally, J.G.; Dasso, M. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 2002, 156, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Hamada, M.; Haeger, A.; Jeganathan, K.B.; van Ree, J.H.; Malureanu, L.; Walde, S.; Joseph, J.; Kehlenbach, R.H.; van Deursen, J.M. Ran-dependent docking of importin-beta to RanBP2/Nup358 filaments is essential for protein import and cell viability. J. Cell Biol. 2011, 194, 597–612. [Google Scholar] [CrossRef] [PubMed]
- Aslanukov, A.; Bhowmick, R.; Guruju, M.; Oswald, J.; Raz, D.; Bush, R.A.; Sieving, P.A.; Lu, X.; Bock, C.B.; Ferreira, P.A. RanBP2 modulates Cox11 and hexokinase I activities and haploinsufficiency of RanBP2 causes deficits in glucose metabolism. PLoS Genet. 2006, 2, e177. [Google Scholar] [CrossRef] [PubMed]
- Abascal, F.; Tress, M.L.; Valencia, A. Alternative splicing and co-option of transposable elements: The case of TMPO/LAP2alpha and ZNF451 in mammals. Bioinformatics 2015, 31, 2257–2261. [Google Scholar] [CrossRef]
- Karvonen, U.; Jaaskelainen, T.; Rytinki, M.; Kaikkonen, S.; Palvimo, J.J. ZNF451 is a novel PML body- and SUMO-associated transcriptional coregulator. J. Mol. Biol. 2008, 382, 585–600. [Google Scholar] [CrossRef]
- Schellenberg, M.J.; Lieberman, J.A.; Herrero-Ruiz, A.; Butler, L.R.; Williams, J.G.; Munoz-Cabello, A.M.; Mueller, G.A.; London, R.E.; Cortes-Ledesma, F.; Williams, R.S. ZATT (ZNF451)-mediated resolution of topoisomerase 2 DNA-protein cross-links. Science 2017, 357, 1412–1416. [Google Scholar] [CrossRef]
- De La Cruz-Herrera, C.F.; Shire, K.; Siddiqi, U.Z.; Frappier, L. A genome-wide screen of Epstein-Barr virus proteins that modulate host SUMOylation identifies a SUMO E3 ligase conserved in herpesviruses. PLoS Pathog. 2018, 14, e1007176. [Google Scholar] [CrossRef]
- Rodriguez, M.S.; Dargemont, C.; Hay, R.T. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 2001, 276, 12654–12659. [Google Scholar] [CrossRef] [PubMed]
- Tatham, M.H.; Jaffray, E.; Vaughan, O.A.; Desterro, J.M.; Botting, C.H.; Naismith, J.H.; Hay, R.T. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 2001, 276, 35368–35374. [Google Scholar] [CrossRef]
- Hendriks, I.A.; Vertegaal, A.C. A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 2016, 17, 581–595. [Google Scholar] [CrossRef] [PubMed]
- Matic, I.; Schimmel, J.; Hendriks, I.A.; van Santen, M.A.; van de Rijke, F.; van Dam, H.; Gnad, F.; Mann, M.; Vertegaal, A.C. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol. Cell 2010, 39, 641–652. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Hsu, C.T.; Ting, C.Y.; Liu, L.F.; Hwang, J. Assembly of a polymeric chain of SUMO1 on human topoisomerase I in vitro. J. Biol. Chem. 2006, 281, 8264–8274. [Google Scholar] [CrossRef] [PubMed]
- Vertegaal, A.C.; Ogg, S.C.; Jaffray, E.; Rodriguez, M.S.; Hay, R.T.; Andersen, J.S.; Mann, M.; Lamond, A.I. A proteomic study of SUMO-2 target proteins. J. Biol. Chem. 2004, 279, 33791–33798. [Google Scholar] [CrossRef]
- Vogt, B.; Hofmann, K. Bioinformatical detection of recognition factors for ubiquitin and SUMO. Methods Mol. Biol. 2012, 832, 249–261. [Google Scholar] [CrossRef] [PubMed]
- González-Prieto, R.; Eifler-Olivi, K.; Claessens, L.A.; Willemstein, E.; Xiao, Z.; Ormeno, C.M.T.; Ovaa, H.; Ulrich, H.D.; Vertegaal, A.C.O. Global non-covalent SUMO interaction networks reveal SUMO-dependent stabilization of the non-homologous end joining complex. Cell Rep. 2021, 34, 108691. [Google Scholar] [CrossRef]
- Hendriks, I.A.; Lyon, D.; Su, D.; Skotte, N.H.; Daniel, J.A.; Jensen, L.J.; Nielsen, M.L. Site-specific characterization of endogenous SUMOylation across species and organs. Nat. Commun. 2018, 9, 2456. [Google Scholar] [CrossRef]
- Jansen, N.S.; Vertegaal, A.C.O. A Chain of Events: Regulating Target Proteins by SUMO Polymers. Trends Biochem. Sci. 2021, 46, 113–123. [Google Scholar] [CrossRef]
- Vertegaal, A.C. SUMO chains: Polymeric signals. Biochem. Soc. Trans. 2010, 38, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Skilton, A.; Ho, J.C.; Mercer, B.; Outwin, E.; Watts, F.Z. SUMO chain formation is required for response to replication arrest in S. pombe. PLoS ONE 2009, 4, e6750. [Google Scholar] [CrossRef] [PubMed]
- Tatham, M.H.; Geoffroy, M.C.; Shen, L.; Plechanovova, A.; Hattersley, N.; Jaffray, E.G.; Palvimo, J.J.; Hay, R.T. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 2008, 10, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Lallemand-Breitenbach, V.; Jeanne, M.; Benhenda, S.; Nasr, R.; Lei, M.; Peres, L.; Zhou, J.; Zhu, J.; Raught, B.; de The, H. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 2008, 10, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Seifert, A.; Chua, J.S.; Maure, J.F.; Golebiowski, F.; Hay, R.T. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 2012, 26, 1196–1208. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, D.; Arnaoutov, A.; Dasso, M. The SUMO protease SENP6 is essential for inner kinetochore assembly. J. Cell Biol. 2010, 188, 681–692. [Google Scholar] [CrossRef]
- Galanty, Y.; Belotserkovskaya, R.; Coates, J.; Jackson, S.P. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 2012, 26, 1179–1195. [Google Scholar] [CrossRef]
- Salas-Lloret, D.; Agabitini, G.; Gonzalez-Prieto, R. TULIP2: An Improved Method for the Identification of Ubiquitin E3-Specific Targets. Front. Chem. 2019, 7, 802. [Google Scholar] [CrossRef]
- Lecona, E.; Rodriguez-Acebes, S.; Specks, J.; Lopez-Contreras, A.J.; Ruppen, I.; Murga, M.; Munoz, J.; Mendez, J.; Fernandez-Capetillo, O. USP7 is a SUMO deubiquitinase essential for DNA replication. Nat. Struct. Mol. Biol. 2016, 23, 270–277. [Google Scholar] [CrossRef]
- Hendriks, I.A.; Schimmel, J.; Eifler, K.; Olsen, J.V.; Vertegaal, A.C.O. Ubiquitin-specific Protease 11 (USP11) Deubiquitinates Hybrid Small Ubiquitin-like Modifier (SUMO)-Ubiquitin Chains to Counteract RING Finger Protein 4 (RNF4). J. Biol. Chem. 2015, 290, 15526–15537. [Google Scholar] [CrossRef]
- Pfeiffer, A.; Luijsterburg, M.S.; Acs, K.; Wiegant, W.W.; Helfricht, A.; Herzog, L.K.; Minoia, M.; Bottcher, C.; Salomons, F.A.; van Attikum, H.; et al. Ataxin-3 consolidates the MDC1-dependent DNA double-strand break response by counteracting the SUMO-targeted ubiquitin ligase RNF4. EMBO J. 2017, 36, 1066–1083. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, I.A.; Lyon, D.; Young, C.; Jensen, L.J.; Vertegaal, A.C.; Nielsen, M.L. Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat. Struct. Mol. Biol. 2017, 24, 325–336. [Google Scholar] [CrossRef]
- Iconomou, M.; Saunders, D.N. Systematic approaches to identify E3 ligase substrates. Biochem. J. 2016, 473, 4083–4101. [Google Scholar] [CrossRef] [PubMed]
- Akimov, V.; Barrio-Hernandez, I.; Hansen, S.V.F.; Hallenborg, P.; Pedersen, A.K.; Bekker-Jensen, D.B.; Puglia, M.; Christensen, S.D.K.; Vanselow, J.T.; Nielsen, M.M.; et al. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 2018, 25, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Bennett, E.J.; Huttlin, E.L.; Guo, A.; Li, J.; Possemato, A.; Sowa, M.E.; Rad, R.; Rush, J.; Comb, M.J.; et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 2011, 44, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.A.; Beli, P.; Weinert, B.T.; Nielsen, M.L.; Cox, J.; Mann, M.; Choudhary, C. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell Proteom. 2011, 10, M111.013284. [Google Scholar] [CrossRef] [PubMed]
- Swatek, K.N.; Usher, J.L.; Kueck, A.F.; Gladkova, C.; Mevissen, T.E.T.; Pruneda, J.N.; Skern, T.; Komander, D. Insights into ubiquitin chain architecture using Ub-clipping. Nature 2019, 572, 533–537. [Google Scholar] [CrossRef]
- Matsumoto, M.L.; Dong, K.C.; Yu, C.; Phu, L.; Gao, X.; Hannoush, R.N.; Hymowitz, S.G.; Kirkpatrick, D.S.; Dixit, V.M.; Kelley, R.F. Engineering and structural characterization of a linear polyubiquitin-specific antibody. J. Mol. Biol. 2012, 418, 134–144. [Google Scholar] [CrossRef]
- Newton, K.; Matsumoto, M.L.; Ferrando, R.E.; Wickliffe, K.E.; Rape, M.; Kelley, R.F.; Dixit, V.M. Using linkage-specific monoclonal antibodies to analyze cellular ubiquitylation. Methods Mol. Biol. 2012, 832, 185–196. [Google Scholar] [CrossRef]
- Mattern, M.; Sutherland, J.; Kadimisetty, K.; Barrio, R.; Rodriguez, M.S. Using Ubiquitin Binders to Decipher the Ubiquitin Code. Trends Biochem. Sci. 2019, 44, 599–615. [Google Scholar] [CrossRef]
- Kliza, K.; Husnjak, K. Resolving the Complexity of Ubiquitin Networks. Front. Mol. Biosci. 2020, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Hakala, K.; Weintraub, S.T.; Shiio, Y. Quantitative proteomic identification of the BRCA1 ubiquitination substrates. J. Proteome Res. 2011, 10, 5191–5198. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sarraf, S.A.; Raman, M.; Guarani-Pereira, V.; Sowa, M.E.; Huttlin, E.L.; Gygi, S.P.; Harper, J.W. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 2013, 496, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.W.; Nagel, J.; Hoving, S.; Gerrits, B.; Bauer, A.; Thomas, J.R.; Kirschner, M.W.; Schirle, M.; Luchansky, S.J. Quantitative Lys--Gly-Gly (diGly) proteomics coupled with inducible RNAi reveals ubiquitin-mediated proteolysis of DNA damage-inducible transcript 4 (DDIT4) by the E3 ligase HUWE1. J. Biol. Chem. 2014, 289, 28942–28955. [Google Scholar] [CrossRef]
- Kim, B.J.; Chan, D.W.; Jung, S.Y.; Chen, Y.; Qin, J.; Wang, Y. The Histone Variant MacroH2A1 Is a BRCA1 Ubiquitin Ligase Substrate. Cell Rep. 2017, 19, 1758–1766. [Google Scholar] [CrossRef]
- Mark, K.G.; Simonetta, M.; Maiolica, A.; Seller, C.A.; Toczyski, D.P. Ubiquitin ligase trapping identifies an SCF(Saf1) pathway targeting unprocessed vacuolar/lysosomal proteins. Mol. Cell 2014, 53, 148–161. [Google Scholar] [CrossRef]
- Loveless, T.B.; Topacio, B.R.; Vashisht, A.A.; Galaang, S.; Ulrich, K.M.; Young, B.D.; Wohlschlegel, J.A.; Toczyski, D.P. DNA Damage Regulates Translation through beta-TRCP Targeting of CReP. PLoS Genet. 2015, 11, e1005292. [Google Scholar] [CrossRef]
- Streich, F.C., Jr.; Lima, C.D. Strategies to Trap Enzyme-Substrate Complexes that Mimic Michaelis Intermediates During E3-Mediated Ubiquitin-Like Protein Ligation. Methods Mol. Biol. 2018, 1844, 169–196. [Google Scholar] [CrossRef]
- Zhuang, M.; Guan, S.; Wang, H.; Burlingame, A.L.; Wells, J.A. Substrates of IAP ubiquitin ligases identified with a designed orthogonal E3 ligase, the NEDDylator. Mol. Cell 2013, 49, 273–282. [Google Scholar] [CrossRef]
- O’Connor, H.F.; Lyon, N.; Leung, J.W.; Agarwal, P.; Swaim, C.D.; Miller, K.M.; Huibregtse, J.M. Ubiquitin-Activated Interaction Traps (UBAITs) identify E3 ligase binding partners. EMBO Rep. 2015, 16, 1699–1712. [Google Scholar] [CrossRef]
- Coyaud, E.; Mis, M.; Laurent, E.M.; Dunham, W.H.; Couzens, A.L.; Robitaille, M.; Gingras, A.C.; Angers, S.; Raught, B. BioID-based Identification of Skp Cullin F-box (SCF)beta-TrCP1/2 E3 Ligase Substrates. Mol. Cell Proteom. 2015, 14, 1781–1795. [Google Scholar] [CrossRef]
- Barroso-Gomila, O.; Trulsson, F.; Muratore, V.; Canosa, I.; Merino-Cacho, L.; Cortazar, A.R.; Perez, C.; Azkargorta, M.; Iloro, I.; Carracedo, A.; et al. Identification of proximal SUMO-dependent interactors using SUMO-ID. Nat. Commun. 2021, 12, 6671. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.C.; Rhee, D.Y.; Duda, D.M.; Kelsall, I.R.; Olszewski, J.L.; Paulo, J.A.; de Jong, A.; Ovaa, H.; Alpi, A.F.; Harper, J.W.; et al. Two Distinct Types of E3 Ligases Work in Unison to Regulate Substrate Ubiquitylation. Cell 2016, 166, 1198–1214.e1124. [Google Scholar] [CrossRef] [PubMed]
- Horn-Ghetko, D.; Krist, D.T.; Prabu, J.R.; Baek, K.; Mulder, M.P.C.; Klugel, M.; Scott, D.C.; Ovaa, H.; Kleiger, G.; Schulman, B.A. Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly. Nature 2021, 590, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Senft, D.; Qi, J.; Ronai, Z.A. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat. Rev. Ther. Cancer 2018, 18, 69–88. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Meng, T.; Chen, L.; Wei, W.; Wang, P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal. Transduct. Target 2020, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Rabellino, A.; Melegari, M.; Tompkins, V.S.; Chen, W.; Van Ness, B.G.; Teruya-Feldstein, J.; Conacci-Sorrell, M.; Janz, S.; Scaglioni, P.P. PIAS1 Promotes Lymphomagenesis through MYC Upregulation. Cell Rep. 2016, 15, 2266–2278. [Google Scholar] [CrossRef] [PubMed]
- Rayner, S.L.; Morsch, M.; Molloy, M.P.; Shi, B.; Chung, R.; Lee, A. Using proteomics to identify ubiquitin ligase-substrate pairs: How novel methods may unveil therapeutic targets for neurodegenerative diseases. Cell Mol. Life Sci. 2019, 76, 2499–2510. [Google Scholar] [CrossRef]
- Bachmair, A.; Finley, D.; Varshavsky, A. In vivo half-life of a protein is a function of its amino-terminal residue. Science 1986, 234, 179–186. [Google Scholar] [CrossRef]
- Liu, P.; Cong, X.; Liao, S.; Jia, X.; Wang, X.; Dai, W.; Zhai, L.; Zhao, L.; Ji, J.; Ni, D.; et al. Global identification of phospho-dependent SCF substrates reveals a FBXO22 phosphodegron and an ERK-FBXO22-BAG3 axis in tumorigenesis. Cell Death Differ. 2022, 29, 1–13. [Google Scholar] [CrossRef]
- Langston, S.P.; Grossman, S.; England, D.; Afroze, R.; Bence, N.; Bowman, D.; Bump, N.; Chau, R.; Chuang, B.C.; Claiborne, C.; et al. Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer. J. Med. Chem. 2021, 64, 2501–2520. [Google Scholar] [CrossRef] [PubMed]
- Kroonen, J.S.; Vertegaal, A.C.O. Targeting SUMO Signaling to Wrestle Cancer. Trends Cancer 2021, 7, 496–510. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Schoonderwoerd, M.J.A.; Kroonen, J.S.; de Graaf, I.J.; Sluijter, M.; Ruano, D.; Gonzalez-Prieto, R.; Verlaan-de Vries, M.; Rip, J.; Arens, R.; et al. Targeting pancreatic cancer by TAK-981: A SUMOylation inhibitor that activates the immune system and blocks cancer cell cycle progression in a preclinical model. Gut 2022, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Sha, Z.; Blyszcz, T.; Gonzalez-Prieto, R.; Vertegaal, A.C.O.; Goldberg, A.L. Inhibiting ubiquitination causes an accumulation of SUMOylated newly synthesized nuclear proteins at PML bodies. J. Biol. Chem. 2019, 294, 15218–15234. [Google Scholar] [CrossRef]
- Mojsa, B.; Tatham, M.H.; Davidson, L.; Liczmanska, M.; Branigan, E.; Hay, R.T. Identification of SUMO Targets Associated With the Pluripotent State in Human Stem Cells. Mol. Cell Proteom. 2021, 20, 100164. [Google Scholar] [CrossRef]
- Joung, J.; Konermann, S.; Gootenberg, J.S.; Abudayyeh, O.O.; Platt, R.J.; Brigham, M.D.; Sanjana, N.E.; Zhang, F. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 2017, 12, 828–863. [Google Scholar] [CrossRef]
- Tang, C.P.; Clark, O.; Ferrarone, J.R.; Campos, C.; Lalani, A.S.; Chodera, J.D.; Intlekofer, A.M.; Elemento, O.; Mellinghoff, I.K. GCN2 kinase activation by ATP-competitive kinase inhibitors. Nat. Chem. Biol. 2022, 18, 207–215. [Google Scholar] [CrossRef]
- Weller, C.E.; Dhall, A.; Ding, F.; Linares, E.; Whedon, S.D.; Senger, N.A.; Tyson, E.L.; Bagert, J.D.; Li, X.; Augusto, O.; et al. Aromatic thiol-mediated cleavage of N-O bonds enables chemical ubiquitylation of folded proteins. Nat. Commun. 2016, 7, 12979. [Google Scholar] [CrossRef]
- Bondalapati, S.; Eid, E.; Mali, S.M.; Wolberger, C.; Brik, A. Total chemical synthesis of SUMO-2-Lys63-linked diubiquitin hybrid chains assisted by removable solubilizing tags. Chem. Sci. 2017, 8, 4027–4034. [Google Scholar] [CrossRef]
- Mulder, M.P.C.; Merkx, R.; Witting, K.F.; Hameed, D.S.; El Atmioui, D.; Lelieveld, L.; Liebelt, F.; Neefjes, J.; Berlin, I.; Vertegaal, A.C.O.; et al. Total Chemical Synthesis of SUMO and SUMO-Based Probes for Profiling the Activity of SUMO-Specific Proteases. Angew. Chem. Int. Ed. Engl. 2018, 57, 8958–8962. [Google Scholar] [CrossRef]
- Wang, Y.S.; Wu, K.P.; Jiang, H.K.; Kurkute, P.; Chen, R.H. Branched Ubiquitination: Detection Methods, Biological Functions and Chemical Synthesis. Molecules 2020, 25, 5200. [Google Scholar] [CrossRef] [PubMed]
| E1 | Complex Formation | UBL | Cellular Process | E2 | Reference |
|---|---|---|---|---|---|
| Uba1 Uba6 | Monomer Monomer | Ub | Ubiquitination | 38 | [23] |
| Uba7 | Monomer | ISG15 | ISGylation | Ubch8 | [25] |
| Sae1/Uba2 | Heterodimer | SUMO | SUMOylation | Ubc9 | [26] |
| Nae1/Uba3 | Heterodimer | NEDD8 | NEDDylation | Uba12 | [27] |
| Uba4/Mocs3 | Homodimer | URM1 | URMylation | Unknown | [28] |
| Uba5 | Homodimer | UFM1 | UFMylation | Ufc1 | [29] |
| Atg7 | Homodimer | ATG8/ATG12 | Autophagy | ATG3/ATG10 | [30,31] |
| Class | E3 Ligase | SUMO | Substrates | Chains | Functions | Reference |
|---|---|---|---|---|---|---|
| PIAS | PIAS1 | SUMO 1, SUMO2/3 | p53, PCNA, Vimentin, etc. | K? | Check points regulation, DNA damage, cell migration | [102,103,104] |
| PIAS2 | SUMO1 | p53 | K7? | Check points | [105] | |
| PIAS3 | SUMO2/3 | ATR | K11? | DNA damage | [106] | |
| PIAS4 | SUMO1, SUMO2/3 | LEF1 | K11? | Wnt-signaling, DNA damage | [107] | |
| NSMCE2/NSE2 | SUMO1 | Rad18, TRAX, MMS21… | K7? | DNA damage | [108,109] | |
| NCP | RanBP2/Nup358 | SUMO 1 | Sp100, Top2, Borealin, DNApolα | K7? | Nuclear import, mitosis, DNA repair | [99,110,111] |
| ZNF451 | ZNF451-1/2 | SUMO2/3 | MCM4, PML | K11 | PML stability | [112,113] |
| ZNF451-3 | SUMO2/3 | MCM4 | K11 | [112] | ||
| KIAA 1586 | SUMO1, SUMO2/3 | MCM4 | K7? K11 | [112] | ||
| SLX4 | SLX4 Complex | SUMO1, SUMO2/3 | SLX4, XPF | K? | Genome maintenance, cell division | [114] |
| Other SUMO E3 enzymes | Pc2/CBX4 | SUMO1 | CtBP | - | Polycomb bodies | [115] |
| TOPORS | SUMO1 | p53 | - | Check points regulation | [116] | |
| Krox20 | SUMO1 | Nab | - | Krox20 autoregulation | [117] | |
| p14/Arf | SUMO1, SUMO2/3 | MDM2, NPM/B23 | K? | Tumor suppression | [118] | |
| HDAC4 | SUMO1 | MEF2 | Mono | Muscle cell differentiation | [119] | |
| Rhes | SUMO1, SUMO2/3 | Ubc9 | Multimono? | SUMOylation | [120] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salas-Lloret, D.; González-Prieto, R. Insights in Post-Translational Modifications: Ubiquitin and SUMO. Int. J. Mol. Sci. 2022, 23, 3281. https://doi.org/10.3390/ijms23063281
Salas-Lloret D, González-Prieto R. Insights in Post-Translational Modifications: Ubiquitin and SUMO. International Journal of Molecular Sciences. 2022; 23(6):3281. https://doi.org/10.3390/ijms23063281
Chicago/Turabian StyleSalas-Lloret, Daniel, and Román González-Prieto. 2022. "Insights in Post-Translational Modifications: Ubiquitin and SUMO" International Journal of Molecular Sciences 23, no. 6: 3281. https://doi.org/10.3390/ijms23063281
APA StyleSalas-Lloret, D., & González-Prieto, R. (2022). Insights in Post-Translational Modifications: Ubiquitin and SUMO. International Journal of Molecular Sciences, 23(6), 3281. https://doi.org/10.3390/ijms23063281







