Abstract
In a wide range of lymphoid neoplasms, the process of malignant transformation is associated with somatic mutations in B cells that affect the epigenetic machinery. Consequential alterations in histone modifications contribute to disease-specific changes in the transcriptional program. Affected genes commonly play important roles in cell cycle regulation, apoptosis-inducing signal transduction, and DNA damage response, thus facilitating the emergence of malignant traits that impair immune surveillance and favor the emergence of different B-cell lymphoma subtypes. In the last two decades, the field has made a major effort to develop therapies that target these epigenetic alterations. In this review, we discuss which epigenetic alterations occur in B-cell non-Hodgkin lymphoma. Furthermore, we aim to present in a close to comprehensive manner the current state-of-the-art in the preclinical and clinical development of epigenetic drugs. We focus on therapeutic strategies interfering with histone methylation and acetylation as these are most advanced in being deployed from the bench-to-bedside and have the greatest potential to improve the prognosis of lymphoma patients.
1. Introduction
The observation that the epigenetic mechanisms controlling the transcription of a wide range of genes involved in B-cell development are frequently dysregulated in lymphoid neoplasms, has recently centered the efforts of hematological cancer researchers worldwide. These alterations can affect DNA methylation, covalent histone modifications, protein recognition modules, and different chromatin remodelers. Among the latter, epigenetic modifications of histones are versatile marks that are intimately connected to lymphomagenesis. The term B-cell non-Hodgkin lymphoma (B-NHL) encompasses different neoplasms characterized by an abnormal proliferation of lymphoid B cells. Here, we provide an update on current efforts to develop therapeutic strategies to interfere with two major post-translational histone modifications altered in B-NHL: histone acetylation and methylation.
1.1. Characteristics of the Main B-NHL Subtypes
B-NHL are sub-divided into distinct categories based on the differentiation stage of the aberrant B cell and the presence of specific genetic alterations.
Diffuse large B-cell lymphoma (DLBCL) is the most common subtype of B-NHL, accounting for 25–35% of all cases. Although different genetic subsets with distinct genotypic, epigenetic, and clinical characteristics have been recently identified by high-throughput sequencing [1,2], the three molecular subtypes defined in the early 2000s by gene expression profiling, namely germinal center B-cell (GCB)-like, activated B-cell (ABC)-like, and unclassifiable [3], are still widely used in clinics. Patients with ABC-DLBCL or genetic alterations in MYC and BCL2 and/or BCL6, called double hit (DHL) or triple hit lymphomas (THL), generally have a poor survival prognosis. The standard of care for DLBCL is an immunochemotherapeutic regimen combining the chemotherapeutic drugs cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP) with the anti-CD20 monoclonal antibody rituximab (R-CHOP) [4,5].
Follicular lymphoma (FL) is a neoplasm originating from germinal center (GC) cells with a follicular pattern. It is the second most common B-NHL, accounting for 20% of all B-NHL cases. FL is characterized by the t(14;18)(q32;q21) translocation involving the BCL2 gene, present in 90% of grade 1–2 patients [6,7]. However, the clinical course is mostly indolent; about 20% of patients, despite treatment, relapse or progress to transformed-FL (t-FL), a more aggressive subtype. Treatment usually involves localized radiotherapy for early stages and rituximab combined with chemotherapy regimens like CHOP for advanced stages [7,8].
Burkitt lymphoma (BL) is another GC-derived lymphoma characterized by the deregulation of MYC due to translocations such as t(8;14)(q23;q32). Three subtypes have been described, namely endemic, sporadic, and immunodeficiency-associated form, which is mostly found in patients infected with the human immunodeficiency virus (HIV). Although BL is an aggressive neoplasm, most patients respond to intensive chemotherapeutic regimens [9,10].
Marginal zone lymphomas (MZL), accounting for 5–15% of B-NHL, originate from memory B cells. The three described clinical entities are splenic (SMZL), nodal (NMZL), and extra-nodal MZL (EMZL), arising from the marginal zone of the spleen, the lymph nodes, and the mucosa-associated lymphoid tissue (MALT), respectively. EMZL, the most common subtype, is associated with chronic inflammation, such as that derived from Helicobacter pylori infections [11,12,13]. Clinical evolution is mostly slow. Treatment usually involves antibiotic treatment for H. pylori-positive gastric EMZL, splenectomy for SMZL, radiotherapy for localized disease, and chemotherapy regimens combined with rituximab for advanced stages [14].
Mantle cell lymphoma (MCL) originates from mature B cells in the mantle zone of lymph nodes and accounts for 3–10% of B-NHL. Its molecular hallmark is the t(11;14)(q13;q32) translocation, which leads to the overexpression of cyclin D1 (CCND1). MCL has a poor prognosis due to diagnosis often at a disseminated stage and an aggressive clinical evolution. Treatment usually involves immunochemotherapy regimens such as R-CHOP, followed by rituximab maintenance, and autologous stem cell transplantation (ASCT) in fit cases [15,16] (Figure 1).
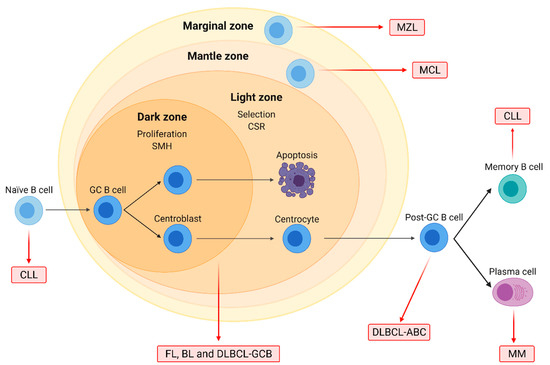
Figure 1.
Origin of the major B-cell non-Hodgkin lymphoma (B-NHL) subtypes. Naïve B-cells form germinal centers (GC) after interacting with antigens. In the dark zone, centroblasts proliferate and undergo somatic hypermutation (SMH), while in the light zone, centrocytes are selected based on BCR affinity and undergo class-switch recombination (CSR). GC cells are the normal counterparts of follicular lymphoma (FL), Burkitt lymphoma (BL), and diffuse large B-cell lymphoma (DLBCL) of the GC subtype (GCB). DLBCL of the activated B-cell (ABC) subtype originates from post-GC cells, and multiple myeloma (MM) arises from differentiated plasma cells. Chronic lymphocytic leukemia (CLL) may originate from either naïve or differentiated memory B cells. Mantle cell (MCL) and marginal zone lymphoma (MZL) arise from B cells located on the mantle and the marginal zone of lymphoid follicles, respectively.
In conclusion, B-NHL subtypes range in their severity from well-controlled indolent diseases to extremely aggressive forms that have an unmet need for the development of novel therapeutic options. Related B-cell-derived neoplasms further include multiple myeloma (MM) and chronic lymphocytic leukemia (CLL).
1.2. Epigenetic Modification of Histone Proteins in B-NHL
Epigenetics was defined in 1942 by Conrad H. Waddington as “the branch of biology which studies the causal interactions between genes and their products, which bring the phenotype into being” [17]. The more scientific knowledge evolved, the more epigenetics became understood as a group of molecular mechanisms constituting a level of memory of previous signals by marking genomic loci and determining the accessibility of embedded genes [18]. Thus, epigenetics is the bridge between the genotype and the phenotype, anchored in the structure and packaging of the genome into chromatin.
The chromatin inside the nucleic compartment is highly compacted and formed by RNA, DNA, and proteins. Importantly, chromatin has a three-dimensional structure that is dynamic and varies, not only between cells of the same or different cell types, but also during the lifespan of a cell itself [19]. Within the interphase nucleus, the chromatin is present as chromosomes, which occupy separate and distinct spaces denominated chromosome territories [20]. Differences in the state of compaction are visible when staining chromatin; less dense regions indicate more loosely packed euchromatin enriched in active transcription, while the strongly stained regions indicate denser heterochromatin harboring repressed genes and tandem repeats such as microsatellites, minisatellites, and transposons [21]. Heterochromatin is frequently enriched at the periphery of the nucleus and on the surface of the nucleolus. Molecular mechanisms exist to transform euchromatin into heterochromatin and vice versa, which allows genes to be expressed differently based on cell type and differentiation state.
The nucleosome is the structural unit of chromatin. The positioning of nucleosomes and their density is the first level of chromatin compaction [22]. In more recent years, attention has been drawn to chromatin motion as a separate phenomenon from compaction status [23]. Nucleosomes consist of 146 pairs of nucleotides wrapped in two loops around an octamer of 8 core histone proteins [24]. More specifically, each nucleosome contains two H2A histones, two H2B histones, two H3 histones, and two H4 histones. Histone H1 is not part of the nucleosome but stabilizes chromatin between nucleosomes to achieve a higher level of structure [25].
Several epigenetic mechanisms operate on the level of the nucleosome. Histone variants can replace replication-coupled histones and provide the nucleosome with different biochemical and biophysical properties [26]. The N-terminal and also some C-terminal tails of histones protrude out of the compact structure of the nucleosome [24] and serve as a platform for many post-translational modifications (PTMs) [27]. PTMs can also occur on the core histone fold, where they directly affect the histone-DNA interaction [28]. Many types of histones PTMs exist that include methylation, acetylation, ubiquitination, SUMOylation, citrullination, glycosylation, ADP-ribosylation, and phosphorylation [27]. The combinatorial nature of PTMs at histone residues led to the controversial hypothesis from Strahl and Allis of a “histone code” on top of the genetic code [29]. Most histone PTMs are catalyzed by enzymes. In the jargon of the chromatin community, we talk about these enzymes as “writers” of PTMs that are recognized by “readers” and removed by “erasers” [27]. The dynamic nature of these mechanisms and their interactions with the transcriptional machinery provide robustness to gene expression programs and a memory of extrinsic or intrinsic stimuli, thereby contributing to the identity and fate of a cell. This precise regulation is perturbed in cancer [30]. The relation with the transcriptional regulation is best understood for the mutual exclusive acetylation and methylation of lysine residues [31].
The writers and erasers of acetylation are histone acetyltransferases (HATs) and histone deacetylase (HDACs), respectively. HATs can be divided into several families: the GNAT family with GCN5; the MYST family including MOZ and Tip60; and the p300/CBP family, among others [32]. Similarly, HDACs are separated into three classes that have zinc-dependent enzymatic activity [33] and the Sirtuins that are NAD+-dependent [34]. The transfer of a methyl group from S-adenosyl methionine (SAM) to a lysine or arginine residue is facilitated by histone methyl transferases (HMTs) such as G9a, EZH2, or protein arginine methyltransferases (PRMTs) [35]. The corresponding erasers are histone demethylases (HDMs), of which LSD1 is an important example [35].
Independent from histones, cytosine methylation is a repressive mark occurring on the DNA molecule. DNA methyltransferases (DNMTs) add methyl groups to cytosine bases [36]. Importantly, DNA methylation can only be indirectly removed by TET enzymes and subsequent DNA repair pathways or by dilution through cell divisions [37].
Genes encoding chromatin-modifying enzymes are frequently mutated in B-cell lymphomas. In DLBCL patients, recurrent mutations affect the writers CBP (also named CREBBP), EP300, EZH2, DNMT3A, or KMT2D/MLL4, among others [38]. Chromatin-modifying enzymes can also be strongly overexpressed in B-NHL subtypes, for instance, the arginine methyltransferase PRMT5 in DLBCL and MCL [39]. Furthermore, oncogenic drivers of B-NHL, such as MYC and BCL-6, act as recruitment platforms for chromatin-modifying enzymes resulting in an altered epigenetic landscape. MYC interacts with the acetylation writers p300, GCN5, and Tip60; the erasers HDAC1 and HDAC3; the histone demethylases LSD1 and KDM4B; and the DNA methyltransferase DNMT3A [40]. BCL-6 can recruit CBP, class I, and II HDACs as well as components of the nucleosome remodeling NuRD complex or polycomb proteins [41,42,43,44] (Figure 2).
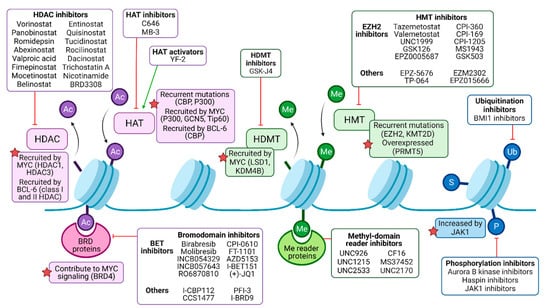
Figure 2.
Pharmacological modulation of deregulated histone modifiers in B-NHL. Histone acetylation (Ac) is catalyzed by histone acetyltransferases (HATs), frequently recruited by oncogenic drivers MYC and/or BCL-6 in malignant B cells, and may be targeted by either activators or inhibitors. Histone deacetylases (HDACs) mediate deacetylation and are the target of numerous inhibitory drugs, as some of them present recurrent mutations and B-NHL and may be recruited by MYC and/or BCL-6 as well. Bromodomain (BRD)-containing proteins can bind to acetylated residues, enhancing oncogenic signaling (such as MYC program, in the case of BRD4), and can be targeted by pan or isoform-specific inhibitors. Methylation (Me) is regulated by histone methyltransferases (HMTs) and histone demethylases (HDMTs). HMTs are common targets of epigenetic drugs, especially the EZH2 subunit of the polycomb repressor complex 2 (PRC2), since it is recurrently mutated in B-NHL. HDMT inhibitors can partially counteract MYC signaling, among other effects. Me domain reader proteins, such as those containing chromodomains, may be targeted by inhibitors as well. Other histone modifications include ubiquitination (Ub), sumoylation (S), and phosphorylation (P), but their role in B-NHL pathogenesis and their targeting require further studies.
In conclusion, mechanisms of epigenetic regulation are disrupted in B-NHL through mutations, overexpression, and false recruitment of chromatin-modifying enzymes. By targeting chromatin-modifying enzymes with epidrugs, the field aims at reverting epigenetic changes on chromatin for therapy [45]. The oldest epidrugs are azanucleosides that inhibit DNMT enzymes [46]. In this review, we focus on the current toolkit available to target PTMs on histone residues, providing rationales for the use of epidrugs in B-cell neoplasms and revisiting ongoing clinical trials.
2. The Pharmacological Targeting of Histone Acetylation
B-cell lymphoma show reduced levels of histone acetylation compared to untransformed B-cells, which is likely driving the malignant program [47]. The oncogene MYC, for example, silences loci encoding tumor suppressors involving HDAC3 [48]. Thus, targeting HDACs is a promising strategy to re-establish physiological histone acetylation levels in cancer cells and reactivate the expression of tumor suppressor genes. In addition, increasing histone acetylation in B-NHL cells can cause apoptosis induction, cell cycle arrest, DNA damage induction, or reduced proliferation [49]. HDAC inhibition also alters the acetylation status of non-histone substrates like p53, MYC, or NF-κB [50], which can indirectly affect gene expression through altered activity of the transcriptional machinery. Several compounds interfering with histone acetylation have entered the preclinical and clinical stages of drug development. In addition to inhibitors of HDACs, these also include inhibitors of HATs and compounds interfering with readers of the family of bromodomain and extra-terminal domain (BET)-containing proteins (Figure 2). We focus on B-NHL but on occasions extend our discussion to other hematopoietic malignancies.
2.1. Extensive Preclinical Development of HDAC Inhibitors
HDAC inhibitors (HDACi) against class I, II, and IV HDACs are grouped based on their chemical properties into hydroxamic acids (TSA, SAHA, panobinostat, rocilinostat), short-chain fatty acids (valproic acid), benzamides (entinostat, mocetinostat), and cyclic tetrapeptides (romidepsin) [49]. Although structurally different, the common mode of action of HDACi against class I, II, and IV lies in chelating the central zinc ion in the catalytic center [51]. On the contrary, sirtuin inhibitors (SIRTi) act as non-competitive inhibitors for NAD+, for example, nicotinamide (NAM) [52], or occupy the catalytic center, for example, Ex-527 [53], to prevent enzymatic activity.
HDACi were originally based on the structure of DMSO, which, at certain concentrations, can induce growth arrest in transformed cells [54]. This eventually led to the development of vorinostat, a potent inhibitor of class I and II HDACs [54]. Also, naturally occurring microbial metabolites, such as trichostatin A (TSA), can act as pan-HDACi [55]. In 2015, a database listed a total of 1445 natural or synthetic compounds with HDACi properties [56]. This number is probably surpassed by now as the design and synthesis of HDACi are ongoing by refining side chains of available inhibitors [57]. While most first-generation HDACi showed activity against several classes of HDAC family members (pan-HDACi), novel HDACi are aimed at targeting only one member or class (selective HDACi) to reduce off-target and, eventually, side effects in patients [57]. Novel highly-specific HDACi are, for example, directed against HDAC3 [58] or HDAC6 [59]. However, until now, there has been a huge discrepancy between the number of HDACi on hand and the number of Food and Drug Administration (FDA)-approved HDACi that include only the pan-HDACi vorinostat, romidepsin, panobinostat, and belinostat.
The first requisite before becoming available for clinical trials is to successfully pass preclinical development using a variety of mouse models. Eµ-Myc transgenic mice, for example, recapitulate MYC-induced B-cell lymphomagenesis by restricted Myc overexpression in the B-cell compartment using the strong endogenous Eµ enhancer [60]. Phenotypically, Eµ-Myc mice develop immature or mature B-cell lymphomas [60,61] with dramatic tumor heterogeneity between individual mice [62]. However, this should not be interpreted as a weakness of the model, as a recent study utilized Eµ-Myc mice for predicting chemotherapy treatment outcomes of human DLBCL patients [63].
Pan-HDACi such as dacinostat showed strong anti-proliferative effects in panels of B-NHL and MM cell lines [64]. Moreover, apoptosis induction upon pan-HDACi challenge required the pro-apoptotic proteins BID and BIM in primary lymphoma cells from Eµ-Myc mice [65]. Similarly, direct application of vorinostat or panobinostat in vivo in Eµ-Myc mice or lymphoma xenografts extended the median survival significantly via increased apoptosis and upregulated autophagic processes [65,66]. To identify singular HDAC family members mediating pro-tumoral functions, systemic depletions were conducted in Eµ-Myc cells revealing that knock-down of HDAC3 strongly reduced lymphoma cell proliferation and tumor mass in xenografted mice [67]. Besides, only the combinatorial knock-down of HDAC1 and HDAC2 led to a huge increase in apoptosis of MYC-driven lymphoma cells [67]. However, in contrast to B-ALL lines, pharmacological targeting of HDAC1/HDAC2 was less effective in reducing the viability of B-NHL cell lines as no upregulation of H2A.X was achieved [64].
HDAC6 has emerged as an interesting target for the development of specific HDAC inhibitors. In MYC-induced lymphoma, HDAC6 has a different function than HDAC1 [67]. Indeed, HDAC6 is unique with two catalytic domains, a mainly cytoplasmic location, and substrates such as tubulin and heat-shock proteins [68]. Current research on HDAC6 is fueled by specific inhibitors like rocilinostat, which induces an unfolded protein response (UPR) in DLBCL cells concomitant with overloading of the proteasome [69]. Interestingly, HDAC6 inhibitor treatment of BL and DLBCL cell lines induced MYC degradation, which was accompanied by apoptosis and even prevented lymphomagenesis in Eµ-Myc mice [70]. Of note, HDAC6 seems to have cancer-specific functions as Hdac6-knock-out mice showed no obvious phenotype under non-inflammatory conditions [71,72]. This explains the relatively mild side effect profile observed after clinical administration of HDAC6 inhibitors [70], providing an argument for single HDAC member inhibition instead of using pan-HDACi.
Many of the above-made observations for HDACi can be transferred to other lymphoid neoplasms, such as MM [73]. The preclinical model of Vk*MYC mice recapitulates the pathogenesis of MM, including typical signs of disease such as increased antibody production, splenomegaly, osteolytic lesions, and extensively increased numbers of CD138-positive plasma cells [74]. Interestingly, the now FDA-approved HDACi panobinostat prolonged survival of Vk*MYC mice by reducing CD138-positive cells and the M-spike caused by antibody secretion [75]. Besides, rocilinostat, in combination with bortezomib, induced apoptosis in vitro by activation of endoplasmic reticulum (ER) stress in MM cells and significantly increased survival of MM xenografts [76]. A recent study showed that rocilinostat treatment upregulated CD38 surface expression on MM cells which improved targeting by the anti-CD38 monoclonal antibody daratumumab [77]. Rocilinostat also increased CD20 surface expression on B-NHL cells [78], which could be combined with anti-CD20 monoclonal antibody treatment, pointing towards the combination of HDACi with immunotherapy.
The recognition of malignant cells by the immune system is generally known as immune surveillance, and many cancer cells prevent this interaction by blocking immune checkpoints. Importantly, HDAC3 inhibition using the novel molecule BRD3308 upregulated p21 resulting in a cell-intrinsic arrest of proliferation [79]. However, this was only effective in CBP/p300-mutated DLBCL, where BCL6-HDAC3 complexes can be found that repress p21 (CDKN1A) transcription [79]. BRD3308 also increased PDL1 and HLADR gene expression in DLBCL, which promoted CD4 and CD8 T-cell recruitment to the lymphoma site in a mouse model [79]. Similar effects on the tumor microenvironment were obtained for HDACi against class I or HDAC6 [80,81].
An important but less studied class of HDACs is class III (Sirtuins). In 2013, Amengual and colleagues combined the general SIRTi NAM with all four FDA-approved pan-HDACi and discovered strong synergies, especially in GC-DLBCL cell lines, by inducing Bcl-6 and p53 acetylation [82]. In addition, λ-MYC transgenic mice—another model for studying the development of mature B-cell lymphomas [83]—were treated with a combination of NAM and romidepsin with synergistic results in survival [82]. A comparison of isoform-specific functions in B-NHL gives a much more detailed view: SIRT1 expression was highly correlated with a worse survival prognosis in DLBCL [84]. In addition, SIRT1 was shown to activate AMPK in primary effusion lymphoma, and SIRT1 inhibition improved the survival of derived xenografts [85]. SIRT3, however, acts as a tumor suppressor in MCL [86] but as an oncogene in DLBCL by its influence on the cancer metabolism [87] and the positive regulation of IDH2 [88]. SIRT6 might also act as an oncogene as its knockdown reduced tumor volume in lymphoma models and increased expression of the negative cell cycle regulator p27 [89]. The clinical use of SIRTi will rely on the development of isoform-specific inhibitors, as currently performed against SIRT2 in B-NHL [90], and will finally shed more light on the impact on specific histone residues and affected target genes.
2.2. Preclinical Evaluation of HATs as Targets for B-NHL Therapy
Mutations in genes encoding the HATs, p300 and CBP, can be found in almost 40% of DLBCL and FL, resulting in impaired acetylation of histone residues but also of non-histone proteins, such as BCL-6 and p53 [44]. As p300 and CBP regulate super-enhancer networks in stimulated B-cells, inactivating mutations block terminal differentiation and increase responsiveness towards mitogenic stimuli [91]. Thus, the development of inhibitors against CBP/p300 seems contradictory at first [92]. However, CBP/p300 inhibitors such as C646 were found to reduce MYC expression as H3K18ac and H3K27ac marks were absent at the transcriptional start site, and no RNA polymerase II recruitment was present [93]. This, in turn, forced apoptosis induction of lymphoma cells and was associated with a strong tumor reduction in lymphoma xenografts [93].
Evidence for an oncogenic function of the HAT MOZ comes from Eµ-Myc mice where heterozygous knock-out increased median survival almost four-fold [94]. Thus, it is not surprising that MOZ inhibitors also caused cellular senescence and arrested lymphoma growth in murine transplant models [95]. In a similar way, homozygous loss of GCN5 led to a lifespan extension of Eµ-Myc mice by downregulating the expression of cell cycle-related genes such as E2f and Ccnd1 [96]. This can be pharmacologically explored in BL by drugging GCN5 with the compound MB-3 that induced a G2/M arrest in the cell cycle [97]. Conversely, heterozygous knock-out of the HAT Tip60 was shown to significantly diminish the lifespan of Eµ-Myc mice [98]. Although Tip60 inhibitors targeting the binding site of acetyl-CoA were developed [99], the preclinical data do not favor the use in B-NHL. It should be noted that also HAT activators exist, like YF-2, and are currently explored for the treatment of neurodegenerative diseases. As histone residues are commonly “under”-acetylated in B-NHL, a rationale for the testing of HAT activators is provided.
2.3. Experimental Insight in the Utility BET Inhibitors
Bromodomain-containing proteins can recognize acetylated lysine residues and bind to them via the bromodomain. Naturally, proteins harboring bromodomains include chromatin modifiers, such as HATs (CBP/p300), transcription factors (MLL), or transcription-associated proteins (TAF1) [100]. Importantly, the biological effects of inhibiting either the bromodomain or the catalytically active domain of HATs might be very distinct [101]. Among the bromodomain-containing proteins is the subfamily II of special interest. The members of this so-called BET family, i.e., mBRDT, BRD2, BRD3, and BRD4, act as adaptors for transcription factors and contribute to the precise regulation of gene expression [102]. The feasibility of inhibiting bromodomains was first shown for BRD4 [103,104].
BRD4 stimulates the kinase activity of positive transcription elongation factor b (P-TEFb), which in turn promotes Ser2 phosphorylation of RNA polymerase II, and therefore ongoing transcription [105]. Otherwise, RNA polymerase II would pause after transcribing a short strand of DNA, making successful elongation impossible [106]. MYC can recruit P-TEFb at active promoters and enhancers, causing transcriptional amplification [107]. Specifically in MM, BRD4 associates with super-enhancers related to key MM genes, such as CCDN2, PRDM1, XBP1, or MCL1 [108]. Moreover, BRD4 recruits enzymes involved in histone methylation, including lysine methyltransferases and arginine demethylases, thus, depleting BRD4 reduced H3K36 methylation [109].
BRD4 is druggable by two diazepine-derived compounds; the enantiomer-specific compound (+)-JQ1 and the synthetic histone mimetic I-BET762 (molibresib) [103,104]. (+)-JQ1 accomplished astonishing results in inducing apoptosis, cell cycle arrest, and senescence in BL, DLBCL, and MM cells as well as in derived mouse models [110,111,112]. Mechanistically, (+)-JQ1 blocks BRD4 recruitment by MYC for transcriptional activation and simultaneously directly downregulates expression of MYC, resulting in decreased MYC protein levels [110,111,113]. (+)-JQ1 also displaced BRD4 from super-enhancers impacting key oncogenic drivers [108]. Besides, BETi were shown to downregulate the expression of PD-L1 in an MYC-independent manner on lymphoma cells which engaged the immune response in vivo [114].
Subsequently, novel BETi were designed to achieve better suitability for clinical trials by increasing bioavailability or stability and reducing dose-limiting toxicities (DLT). For example, I-BET151 is a novel dimethylisoxazole BETi that is analog to I-BET762 with an enhanced half-life in vivo [115]. I-BET151 induced G1 phase arrest and apoptosis by reducing BCL-2 levels in leukemic cells [116]. Similarly, OTX015 (birabresib) had strong antiproliferative effects in a large panel of B-NHL cell lines from DLBCL, MCL, MZL, and MM subtypes by decreasing expression of E2F3 target genes and leading to a downregulated inflammatory signature, resulting in significant tumor volume reduction in xenografts [116]. The benzoisoxazoloazepine CPI-0610 decreased MYC transcripts in vivo and reduced leukemia xenograft tumor growth, which was synergistic with doxorubicin treatment [117]. However, toxicity studies showed that CPI-0610 treatment resulted in lymphoid depletion, hypocellularity of the bone marrow with associated anemia, and thrombocytopenia, among other side effects in animals [117].
Preclinical models also showed that combining BETi with the BH3 mimetic, BCL-2 antagonist venetoclax was beneficial in MYC-overexpressing lymphoma cells [118]. Mechanistically, the BCL-2 inhibitor was able to counteract the protective effect of the anti-apoptotic protein, frequently overexpressed in B-NHL, towards BETi-mediated upregulation of the pro-apoptotic BIM and subsequent triggering of apoptosis [118]. This combination significantly reduced the tumor burden of lymphoma xenografts and tremendously increased survival [118]. Combination of (+)-JQ1 and panobinostat synergistically induced apoptosis in MCL cells resistant to the Bruton’s tyrosine kinase (BTK) inhibitor ibrutinib [119]. Also, simultaneous inhibition of BET proteins and blockade of the chemokine receptor CXCR4 potentiated MYC reduction in DLBCL cells and reduced tumor volume of transplanted xenografts [120].
Finally, it should be noticed that non-BET bromodomain inhibitors exist that can be used to target chromatin-modifying enzymes or remodeling complexes. Inhibiting p300 or CBP is possible with bromodomain inhibitors like I-CBP112 or CCS1477 that do not target the catalytic site containing the acetyl transferase activity [121,122]. The bromodomain inhibitor PFI-3 targets SMARCA2, an ATP subunit of the SWI/SNF chromatin remodeling complex, and its association with the HMT NSD2 was found in a specific subset of MM [123]. PFI-3 induced apoptosis in these MM cells, probably by perturbing the expression of myeloma-relevant genes and without effects on BRD4 [123]. BRD9 is another component of the SWI/SNF complex that can be targeted by the non-BETi I-BRD9 [124]. Until today, the Structure Genomics Consortium, a joint venture of industry and academia, has generated high-affinity binders of many human bromodomains by combining different methodologies [125]. The possible usage of such non-BETi should be further explored in specific malignant entities for which rationales exist. Another recent development is dual inhibitors, in which BETi are fused with other inhibitory compounds which generated BET-CBP/p300 inhibitors, BET-BRD7/9 inhibitors, or BET/HDAC inhibitors [126,127,128]. Although strong and consistent anti-tumor activity was already shown for BET-CBP/p300-inhibitors in MM [126], general applications of dual inhibitors in B-NHL remain to be tested.
Taken together, distinct strategies are employed to target the imbalance between HATs and HDACs in the acetylation landscape of lymphoid neoplasms (Table 1). Depending on the disease context, inhibiting readers, writers, and readers of histone acetylation have been shown to be beneficial (Figure 2).

Table 1.
Drugs approved as second-line therapy for relapsed/refractory B-NHL and other lymphomas.
2.4. Ongoing Clinical Development of HDACi, BETi and HATi
2.4.1. HDACi
Vorinostat is the most extensively tested HDACi in clinical trials for lymphoma patients, both as a single agent and in combination. Reported overall response rates (ORR) for vorinostat monotherapy in cohorts with different subtypes of B-NHL range from 29% to 40% (NCT00253630, NCT00127140) [129,130]. Specifically, the best response was observed in FL patients, with an ORR of 49%, followed by 27% in MCL and only 5.5% in another trial for DLBCL (NCT00875056, NCT00097929) [131]. Although toxicity was generally manageable, thrombocytopenia was a common adverse event (AE), affecting up to 90% of participants in some of the trials. The combination of vorinostat with rituximab or rituximab-based regimens improved outcomes in some clinical trials. In a cohort of mostly relapsed FL, MCL, and MZL patients, vorinostat and rituximab achieved an ORR of 46% (NCT00720876) [132], increasing up to 65% in a similar cohort treated with rituximab, ifosfamide, carboplatin, and etoposide (R-ICE) (NCT00601718). In MCL, the combination with rituximab and cladribine resulted in an ORR of 39% in relapsed patients and an impressive 97% when used at the frontline (NCT00764517) [133]. In DLBCL, the combination with R-CHOP also achieved remarkable results, with an overall survival (OS) of 86% and progression-free survival (PFS) of 73% at 2 years (NCT00972478). All-grade AEs and serious AEs (SAEs) occurred in more than 70% and 30% of participants, respectively, in the four clinical trials mentioned, being especially relevant the presence of serious febrile neutropenia in 35% of DLBCL participants treated with vorinostat and R-CHOP. Other drug combinations with vorinostat demonstrated promising results as well. When vorinostat was combined with azacitidine, rituximab, and other chemotherapeutic drugs as preconditioning therapy before ASCT, event-free survival (EFS) at 100 days post-transplant was 66% in DLBCL and 100% in FL and MCL. Although some all-grade AEs, such as neutropenia and mucositis, affected over 90% of participants, no SAEs were reported (NCT01983969). The combination with the proteasome inhibitor bortezomib resulted in a modest ORR of 27% in MCL and 8% in DLBCL (NCT00703664), but results were more remarkable as maintenance therapy after ASCT, as B- and T-NHL patients presented 84% OS and 74% EFS at 6.5 years post-ASCT (NCT00992446). The combination of vorinostat, etoposide, and niacinamide was evaluated in a cohort with only four B-NHL patients, but one of them had a complete response (CR) (NCT00691210) [82], while the combination of vorinostat with the aurora A inhibitor alisertib led to 2 CR in a cohort of 12 relapsed and refractory (R/R) DLBCL patients (NCT01567709) [134]. Vorinostat combined with the immunomodulatory drug (IMiD) lenalidomide presented manageable toxicity; however, the trial was terminated early due to low recruitment. The mTOR inhibitor tacrolimus (NCT04220008, NCT03842696), the PARP inhibitor olaparib (NCT03259503), and the PD-1 immune checkpoint inhibitor pembrolizumab (NCT03150329), among others, are also currently evaluated in combination with vorinostat (Table 2).

Table 2.
Current clinical trials evaluating epi-drugs as single agents and/or in combination regimens in B-NHL patients.
The rest of HDACi evaluated in B-NHL patients have shown modest results so far. Panobinostat monotherapy, evaluated in a cohort of B- and T-NHL patients, led to median OS and PFS of 15 and 3 months, respectively, and an ORR of 21% (NCT01261247), and similar results were reported in a cohort of DLBCL patients refractory to R-CHOP treatment (NCT01523834). Two separate DLBCL clinical trials (NCT01238692, NCT01282476) reported no improvement of the outcome when combined with rituximab [135]. Panobinostat was also evaluated in combination with the mTOR inhibitor everolimus in 3 clinical trials in a cohort of B-, T-NHL and Hodgkin lymphoma (HL) patients (NCT00918333, NCT00967044, NCT00978432), in which the best response was an ORR of 33%, and OS and PFS of 35 and 4.2 months, respectively. Regarding toxicity, thrombocytopenia was the most notable AE, both in monotherapy and combination trials, reaching a frequency over 90% in some of them.
Romidepsin was evaluated as a single agent in a small cohort of 9 relapsed DLBCL and MCL patients, of which only 1 had a partial response (PR) (NCT00383565). Combinations have been evaluated in lymphoma cohorts formed mainly by patients with T-NHL, who achieved better outcomes than those with B-NHL. The best result was an ORR of 75% in four FL patients treated with romidepsin and pralatrexate (NCT01947140) [136], but much lower ORRs were reported in trials combining romidepsin with azacitidine (NCT01998035) [137] or gemcitabine, dexamethasone, and cisplatin (NCT01846390) [138]. An ongoing clinical trial is testing the combination with lenalidomide (NCT01755975).
Abexinostat monotherapy has shown promising results in FL patients in two clinical trials (NCT00724984, EudraCT-2009-013691-47), leading to an ORR of 56% and a PFS of 10.2 months, but achieving more modest outcomes in MCL and DLBCL [139]. Several ongoing trials are testing abexinostat as a single agent in different subtypes of NHL (NCT03600441, NCT04014696, NCT03936153, NCT03934567) and in combination with ibrutinib (NCT03939182).
Valproic acid has shown potent activity in DLBCL in combination with R-CHOP, achieving OS and PFS at 2 years of 97% and 85%, respectively, and an ORR of 90%. However, toxicity was prominent, as 81% of participants experienced grade 3 or 4 neutropenia, and notable auditory AE was common (NCT01622439) [140].
Fimepinostat monotherapy in DLBCL achieved an ORR of 47%, and median PFS was 3 months, but the combination with rituximab did not improve these outcomes (NCT01742988) [141].
Mocetinostat monotherapy has shown modest activity in DLBCL and FL patients so far, with an ORR below 20% and a PFS below 4 months (NCT00359086) [142].
Belinostat has shown no clinical benefit for lymphoma patients, as no response has been reported either as a monotherapy (NCT00303953, NCT01273155) or in combination with ibritumomab tiuxetan, an yttrium-90-labelled anti-CD20 monoclonal antibody (NCT01686165).
Entinostat (NCT02780804, NCT03179930), quisinostat (NCT00677105), tucidinostat (NCT04661943, NCT04337606, NCT04231448, NCT04025593, NCT04022005, NCT03974243), and rocilinostat (NCT02091063) have been or are currently being evaluated for B-NHL patients as well, either as single agents or in combination, but no results are available at the moment.
2.4.2. HATi
As for HAT inhibitors, only CCS1477-02 has entered clinical development for B-NHL, with an ongoing phase 1 trial for patients with hematological malignancies that is estimated to end in December 2021 (NCT04068597).
2.4.3. BETi
Regarding BET inhibitors, birabresib monotherapy was evaluated in 22 DLBCL patients, which showed an ORR of 10% and a high incidence of AEs, including thrombocytopenia and anemia affecting more than 90% (NCT01713582) [143]. A second trial testing birabresib discontinued enrollment due to the same lack of efficacy (NCT02698189).
RO6870810 as a single agent has demonstrated limited efficacy in DLBCL, with an ORR of 10.5%, median PFS of only 29 days, and prominent toxicity (NCT01987362) [144]. The combination with rituximab and venetoclax has been recently evaluated in lymphoma patients, with no results available yet (NCT03255096).
INCB054329 (NCT02431260) and INCB057643 (NCT02711137) monotherapies were evaluated in cohorts of cancer patients that included 4 and 16 cases of lymphoma, respectively. Both trials were terminated early as all patients discontinued due to lack of responses and notable toxicity [145].
CPI-0610 and FT-1101 have been evaluated in B-NHL patients, both with minimal efficacy: CPI-0610 led to a response in four DLBCL and one FL patient out of 64 cases with B-NHL (NCT01949883), and FT-1101 did not achieve a response in any of the ten B-NHL patients included in the trial (NCT02543879). Lastly, molibresib was recently evaluated as a single agent in a phase 2 trial (NCT01943851), and AZD5153 is being studied in combination with the PARP inhibitor (NCT03205176), but no efficacy results are available yet.
The appearance of DLTs after BETi treatment in clinical studies (Table 2) [146] provides an argument for combining BETi with other compounds to achieve lower effective dosing of BET antagonists. As Basheer and Huntly concluded for BETi, “monotherapy will not suffice to effectively treat complicated hematologic malignancies” [147]. Another argument for combination treatments with BETi is the notion of (+)-JQ1-insensitive genes in B-cell lymphoma likely due to transcriptional rearrangements, which are typical for post-GC lymphomas [148].
3. Targeting Histone Methylation and Some Other PTMs
The field of histone methylation has a complex nature due to the large number of players and different configurations of methylation modifications. Lysine residues can be mono-, di-, and tri-methylated, while arginine residues can be mono- or di-methylated, with di-methylation being symmetric or asymmetric [156]. If carefully interpreted, methylation marks are useful for the functional annotation of the genome, in particular, non-coding regulatory regions [157]. Mono-methylation of H3K4 marks active promoter sites and enhancers. Tri-methylation of H3K4 is indicative of a promoter driving the active transcription of its gene, but its co-occurrence with tri-methylation of H3K27 correlates with a poised transcriptional state. Larger regions of tri-methylation of H3K27 alone are indicative of a repressed heterochromatin state (Figure 2).
In DLBCL and FL, a large sequencing study revealed frequent somatic mutations in the PRC2 subunit EZH2 [158]. This is mirrored in the recent re-classification of DLBCL subtypes which now contains a cluster of EZH2MUT lymphomas [1,2]. The frequent tyrosine 641 (Y641) mutation of EZH2 in lymphomas is a gain-of-function mutation that enhances tri-methylation of H3K27 in concert with EZH2WT that performs the H3K27 mono-methylation, eventually shutting down key B-cell genes [157,158]. Moreover, EZH2MUT also reprograms interactions of lymphoma cells with other immune cells in the niche [159]. Besides, mutations in EZH2 can also be found in BL, and its expression is upregulated in pre-plasmablasts, implicating additional roles in B-cell development and associated pathophysiological conditions [160,161].
3.1. Preclinical Development of Histone Methyl Transferase and Demethylase Inhibitors
The overactivity of HMTs in disease provides a rationale for the development of enzymatic inhibitors. Indeed, most drugs against an HMT are available for EZH2, starting from unspecific drugs like DZNep to specific dual EZH1/2 inhibitors, like UNC1999 that is used in B-NHL treatment, and finally, the very selective EZH2i valemetostat and tazemetostat [162]. EZH2 inhibitors specifically target EZH2MUT lymphoma cells, restoring physiological H3K27me3 levels [163,164]. This reactivates expression of genes like TXNIP and TNFRSF21, resulting in cell cycle arrest, cell death, and reduced tumor load of xenografts [163,164]. However, also non-malignant cells are affected by reduced tri-methylation [164].
The HMT G9a mediates mono- and di-methylation of H3K9, which is followed by tri-methylation via SUV39H1 that is sensitive to chaetocin [165]. Inhibition of SUV39H1 enabled differentiation of leukemic cells and was combined with other epi-drugs [165].
The drug EPZ-5676 (pinomestat) targets specifically the H3K79 methyltransferase DOTL1 [166]. The H3K79 histone residue is located in the core histone fold [28], and inhibition of DOTL1 might be favorable for a specific leukemia subtype [166].
Not only lysine methyltransferases were identified as driving forces of B-NHL but also arginine methyl transferases such as CARM1 and PRMT5. CARM1 inhibition using TP-064 reduced the proliferation of DLBCL cells which was most effective when these cells carried inactivating mutations in genes encoding the HATs CBP or p300 [167]. CARM1 inhibition reduced CBP/p300 target gene expression, as these loci exhibited a loss of H3K4me3 and H3K27ac signals [167]. Besides, treatment with the more bioavailable CARM1 inhibitor EZM2302 reduced tumor volume of p300MUT DLBCL cells in xenograft models, which was highly synergetic with p300/CBP inhibitors [167].
The other example, PRMT5, is overexpressed in some B-cell lymphoma lines in an MYC-dependent manner [168,169]. PRMT5 directly methylates H3R8 and H4R3, which results in hypermethylation and transcriptional repression at target genes, such as RB1 [168]. Importantly, knock-down of PRMT5 upregulated pRB protein levels, impaired proliferation of lymphoma cells, and induced cell death as detected by caspase-3 cleavage [39]. PRMT5 also methylates non-histone proteins, which are crucial for ribosome biogenesis [169]. As proliferating lymphoma cells rely on increased translation, knock-out of PRMT5 doubled the lifespan of Eµ-Myc mice [169]. The PRMT5 inhibitor EPZ015666 (GSK3235025) showed striking anti-proliferative properties in vitro and in vivo against MCL, with an IC50 in the low nanomolar range [170]. As PRMT5 also directly interacts with BCL-6, it is not surprising that PRMT5 inhibition slowed down the proliferation of DLBCL cells [171].
In addition to the development of HMT inhibitors, efforts are being made to target erasers and readers of histone methylation. A major impact on lymphoma formation has the lysine demethylase LSD1 as it interacts with BCL-6 to repress B-cell termination [172]. Cell type-specific knock-out of LSD1 prolonged survival of lymphoma-prone mice [172], which supports the use of LSD1 inhibitors in B-NHL. The inhibitor GSK-J4 targets the lysine demethylase KDM6B, which is partially overexpressed in B-NHL [173]. Although GSK-J4 did not show strong effects on B-NHL cell proliferation, the combination with other chemotherapeutic agents was highly synergistic in inducing apoptosis [173].
In contrast to acetylation marks that are only read by bromodomain-containing proteins, the diversity of methylation marks is also reflected in a number of different modules able to bind methylated lysine or arginine residues [155]. Interestingly, these methyl-“reader” domains are found in many proteins with different functions. Some examples include chromodomains that can attach to H3K9me2 or H3K9me3, among others, like the heterochromatin factor HP1 [174]. However, the chromodomain of the HAT Tip60 was thought to attach to H3K9me3 or H3K4me1, but this was not confirmed in vitro [175]. The tudor domain found, for example, in JMJD2A or 53BP1, binds methylated histone H3K4 and H4K20 residues [176]. The PHD finger, which is found in several chromatin-modifying enzymes, recognizes H3K4me2 and H3K4me3 but is, surprisingly, also able to bind methylated DNMT enzymes [177,178]. This recalls that there are different ways to inhibit the same enzyme, representing a magnitude of options if appropriate inhibitors are developed. To summarize it in other words: “Only six of the hundreds of known methyl-lysine reader proteins have been targeted” [155].
An interesting example for a bromodomain-containing protein to target is CBX7, a polycomb protein, which is highly expressed in FL [179]. In mouse models, the combination of MYC with CBX7 overexpression resulted in a faster lymphoma development [180]. CBX7 binds via its chromodomain trimethylated H3K9 and H3K27 [180], occupies the INK4A/ARF locus, and silences expression of the corresponding tumor suppressors [181]. Recently developed inhibitors against CBX7 [182] might restore the INK4A/ARF-axis in malignant cells and thus, prevent lymphoma cell proliferation.
3.2. EZH2 Inhibitors in Clinical Development
Almost the entire clinical development of HMT inhibitors is focused on targeting EZH2. Among the SAM-competitive inhibitors developed to interfere with EZH2, tazemetostat (E7438/EPZ6438) [183] has shown improved potency and pharmacokinetic properties compared to other agents and can be administered orally in animals [184]. This compound is under evaluation in a series of clinical trials in both solid and hematological tumors. In a phase 1 trial (NCT01897571), it showed anticancer activity and a favorable safety profile in patients with R/R B-NHL, as durable objective responses (DOR) were observed in 38% of the cases, including some CRs, with only grade 1 and 2 AEs [185]. In a subsequent open-label, single-arm, multicenter phase 2 trial with n = 99 evaluable R/R FL patients receiving 800 mg tazemetostat twice a day until progressive disease or withdrawal, the ORR was 69% in the EZH2MUT cohort (n = 45 patients) and 35% in the EZH2WT cohort (n = 54 patients). Median DOR was 10.9 months in the EZH2MUT cohort and 13.0 months in the EZH2WT cohort; median PFS was 13.8 months and 11.1 months, respectively. Treatment with tazemetostat was generally well-tolerated, and no treatment-related deaths were observed. Serious treatment-related AEs were reported only in 4% of the 99 patients [186]. Following this study, which confirmed tazemetostat to be a safe and effective new therapeutic drug for patients with R/R FL, the FDA granted accelerated approval for tazemetostat in June 2020. Eligible adult patients either have FL positive for an EZH2 mutation as detected by an FDA-approved test and received at least two prior systemic therapies or have R/R FL with no satisfactory alternative treatment options [187].
In parallel, tazemetostat has also been evaluated in combination studies, firstly in a phase 1b clinical trial with R-CHOP in DLBCL patients. This study determined that a twice-daily administration of 800 mg tazemetostat was the optimum dose for phase 2 evaluation and reported grade 3–4 hematologic AEs in 8/17 patients [152]. Another phase 1b/3 randomized study (NCT04224493) is currently recruiting patients with histologically confirmed FL (grades 1–3A; EZH2WT or EZH2MUT status), treated previously with ≥1 line of chemotherapy, immunotherapy, or chemoimmunotherapy, to determine and assess the recommended phase 3 dose, efficacy, and safety of lenalidomide and rituximab (R2) + tazemetostat vs. R2 + placebo in patients with R/R FL [153].
GSK2816126 (thereafter GSK126) is another methyltransferase inhibitor capable of inhibiting both EZH2WT and EZH2MUT with similar potency in the low nanomolar range of concentrations and with high selectivity for EZH2 over EZH1 (>100-fold increased potency) and 20 other methyltransferases [187]. This compound has been shown to display significant anti-tumor activity in xenograft models of EZH2MUT DLBCL [188]. Based on these preclinical results, a multicenter, open-label, dose-escalation study (NCT02082977) was carried out in patients with advanced cancers, including 20 B-NHL cases, to evaluate the safety and to determine the maximum-tolerated dose (MTD), the pharmacokinetics, and pharmacodynamics of GSK126. All patients had at least one AE, with the most common being nausea and vomiting in about half of the patients. The results showed insufficient evidence of clinical activity, with only one patient showing a partial radiologic response and 34% of patients having achieved an SD while 51% of them underwent disease progression [188]. Therefore, the compound was not further investigated in clinical settings. One explanation for these discrepancies between preclinical and clinical studies was the presence of myeloid-derived suppressor cells, which may be increased in immunocompetent hosts after treatment, thus impairing the antitumoral activity of the compound [189].
EI1 is a cell-permeable indolocarboxamide compound that acts as a selective SAM-competitive inhibitor of the EZH2 methyltransferase activity with an IC50 of 13–15 nM against H3K27me marks, showing activity against EZH2WT or EZH2MUT-containing PRC2. This compound has been shown to suppress cellular H3K27 di-methylation and tri-methylation in the low micromolar ranges of concentrations in DLBCL cultures and to exert anti-proliferative activity in DLBCL cell lines with either EZH2WT or EZH2MUT [190]. EPZ011989 is another selective and orally bioavailable EZH2 inhibitor that equipotently inhibits EZH2WT or EZH2MUT with an inhibition constant (Ki) of <3 nM and with the capacity to significantly inhibit DLBCL tumor growth in mouse xenografts [191].
CPI-169, an indole-based selective EZH2 inhibitor, first showed significant antitumor activity and pharmacodynamic (PD) target engagement in a DLBCL mouse xenograft model of EZH2MUT, but with limited oral bioavailability [192]. Nonetheless, this compound was able to exert significant antitumor activity in this model when administered intraperitoneally, together with a synergistic tumor growth inhibition when combined with the BRD4 inhibitor, CPI-203 [193]. CPI-1205 (lirametostat), an orally available, subsequently developed compound by the same company, harbored IC50 values of 2 nM and 52 nM for EZH2 and EZH1, respectively. This compound exerted antitumoral activity in in vitro and in vivo models of EZH2MUT DLBCL by selectively binding to the EZH2 catalytic pocket [194]. A phase 1 sequential dose-escalation and expansion study (NCT02395601) was launched in patients with progressive B-cell lymphomas to define MTD and DLT of CPI-1205. Preliminary results from this trial revealed that most treatment-related AES were grade 2 or lower, with no DLTs reached. Authors reported one CR, five SD (three remained on SD ≥ 6 months) with target engagement confirmed by immunohistochemical detection of H3K27me3, and concluded that the drug was well tolerated with manageable toxicities and evidence of antitumor activity [154]. CPI-0209 is a second-generation EZH2 inhibitor that showed robust PD effects in a recent phase 1 trial in different subtypes of solid tumors, making this agent well-tolerated with manageable treatment-related AEs [195].
Activated IGF-1R, PI3K, and MAPK pathways confer resistance to EZH2 inhibition in DLBCL [196]. To improve antitumor efficacy of targeting methyltransferase activity, simultaneous targeting of both EZH2 and its homolog EZH1 [197] has been considered. UNC1999 was the first orally bioavailable SAM-competitive inhibitor of both EZH2WT and EZH2MUT and EZH1, having the ability to selectively kill EZH2MUT DLBCL cell lines [198]. More recently, two orally bioavailable EZH1/2 dual inhibitors, (R)-OR-S1 and (R)-OR-S2, demonstrated improved antitumor activity in in vitro and in vivo models of EZH2MUT DLBCL with no significant toxicity [199]. An ongoing phase 1 study (NCT02732275) is showing that the clinical lead, DS-3201b, exerts early clinical activity and has the potential to be an orally available, novel therapeutic option for B-NHL [200]. Among the latest molecules, the lactam-derived and orally bioavailable EZH2 antagonist PF-06821497 [201] and SHR2554 are potent and orally available EZH2 antagonists, being evaluated in open-label, multi center, phase 1 dose-escalation trials with R/R FL and DLBCL patients (NCT03460977) and R/R mature lymphoid neoplasms (NCT03603951), respectively. Apart from EZH2 inhibitors, the PRMT5 inhibitor EPZ015938 (GSK3326595) with improved activity, when compared to EPZ015666, is currently being used in a dose-escalation study (NCT02783300) with B-NHL patients.
EZH2 has become a hotspot of research in the field of GC-derived B-cell lymphoma, and this interest rapidly led to the emergence of several new drugs that made a quick transfer from bench-to-bedside. These advances also led to the discovery of more functions and roles of EZH2 in tumorigenesis. However, tazemetostat is the only molecule approved to date to treat a subgroup of patients. Continuous efforts are still to be made in the development of (1) EZH2 inhibitors with higher efficiency, lower toxicity, and higher selectivity and (2) dual EZH1/EZH2 antagonists able to counteract EZH1 compensatory effects in EZH2-compromised cells.
3.3. Targeting Other Posttranslational Modifications of Histones
The distinct patterns of epigenetic aberrations may provide novel therapeutic approaches targeting histone modifications. Besides histone methylation and acetylation, the N-termini of histone tails are modified by many other processes, such as phosphorylation, glycosylation, ubiquitination, and sumoylation [202,203,204,205]. However, these modifications are much less explored in the B-NHL field. Here, we discuss the first observations suggesting that interfering with histone phosphorylation, ubiquitination and sumoylation could have therapeutic potential.
Histone H3 has a crucial role in chromatin compaction and chromosome formation [206]. Phosphorylation of H3 at serine 10 (phospho-H3) results in immediate condensation of the chromatin in the late G2 phase of the cell cycle, and thus, is a marker present in dividing cells [207]. Indeed, mitotic activity determined by phospho-H3 counting has been experimentally and clinically used to measure cell proliferation in different types of cancer, including lymphoma [207,208,209,210]. Thus, the discovery of compounds that target histone phosphorylation is an interesting approach for the treatment of oncological patients. In their study, Méhes et al. [211] showed an enhanced G2 phase transition due to the abrupt chromatin phosphorylation in neoplastic B cells. Moreover, during the G2 phase, phospho-H3 co-localizes with the Aurora B kinase, which is known to be frequently overrepresented in NHL patients [211,212]. Interestingly, both preclinical [213] and clinical [214] data have supported the potential for the Aurora kinase inhibition as a new therapy for DLBCL patients. Similarly, small molecule inhibitors of another enzyme that phosphorylates histone H3, Haspin, have been developed as efficient anti-mitotic cancer therapeutics [215]. In B-cell lymphoma xenograft models, the oral administration of the Haspin inhibitor SEL120 revealed encouraging results towards this potential treatment [216].
Several novel therapeutic approaches are being evaluated for the management of R/R B-cell lymphomas [217]. It has been shown that Janus kinase (JAK) signaling is deregulated in several hematologic malignancies [218]. Interestingly, Rui et al. [219] demonstrated that JAK1 contributes to tumor growth and survival of ABC-DLBCL cells through a regulatory mechanism dependent on phosphorylation of histone H3 at tyrosine 41, which supports the development of JAK1 inhibitors for ABC-DLBCL therapy. However, some contradictory data showed an increased risk of B-cell lymphoma development in patients treated with JAK inhibitors by the impairment of immune surveillance [220,221,222], highlighting the importance of assessing the clinical impact of these drugs. Although an increase in phospho-H3 has been associated with poor prognosis in several cancers, the exact mechanisms and the clinical relevance of the histone H3 phosphorylation pattern in B-cell lymphoma requires further clarification.
Recent studies have suggested that sumoylation, a PTM in which a small ubiquitin-like modifier (SUMO) is covalently conjugated to lysine residues, is involved in the regulation of several cellular key processes, such as chromatin occupancy, and is most often associated with reduced gene expression [223]. It has been shown that TRAF6, an E3 ubiquitin ligase correlated with poor prognosis of DLBCL patients [224], can be modified by sumoylation, which in turn may repress the gene transcription through recruitment of HDACs in B cells [225]. Although the role of TRAF6 regulation in lymphomagenesis is still unclear, its activity may have therapeutic implications in DLBCL.
Histone ubiquitination is another important PTM, as its deregulation drives oncogenesis by altering the transcription of key tumor suppressors and oncogenes, which may promote cell proliferation [226]. Indeed, genomic data showed that the genes encoding histone E3 ubiquitin ligases and deubiquitinating enzymes are frequently altered in cancers. The pharmacological inhibition of BMI1 (an activator of the RING1 E3 ubiquitin ligase) and H2AK119ub1 levels both induced apoptosis and prolonged survival in xenograft models of acute myeloid leukemia [227]. Moreover, histone H2B has been shown to be overexpressed and strongly ubiquitinated at K120, which is related to NKX2-1 expression and chromosomal rearrangements in DLBCL cells [228]. In this study, the authors found an aberrant activity of NKX2-1 by in silico expression analysis performed in 204 DLBCL patient samples and suggested that the permissive chromatin structure at the NKX2-1 gene may be associated with deregulation of histone ubiquitination enzymes in DLBCL. However, the therapeutic potentials of NKX2-1 through histone modification require further examinations.
In conclusion, further insights are needed before the development of epigenetic drugs targeting other histone modifications than acetylation and methylation would be warranted for B-NHL and especially its aggressive subtypes.
4. Conclusions and Future Directions
Epigenetic alterations have been shown to play a significant role in a wide range of lymphoid and myeloid neoplasms, mainly in regard to their impact on the regulation of gene expression. In the last 10 to 20 years, major advances have been made in the comprehension of the role of histone modifiers in normal and malignant hematopoiesis. This has spurred the simultaneous development of hundreds of small molecule inhibitors able to target dysregulated or mutated histone modifiers. Despite the remarkable preclinical activity of some of these molecules in selected cancer subtypes, and especially in the relapse setting, these new epigenetic therapies are generally associated with modest overall response rates. Thus, additional in vitro/in vivo studies are still required to develop more effective, more selective, and less toxic epigenetic drugs.
To bypass the current limitations in epidrug development, the field has started to implement high-throughput molecular tools for the identification of disease-specific epigenetic alterations. In parallel, genetic screenings have been demonstrated to be powerful tools able to identify novel drug targets for combination therapies. In particular, strategies aiming at synthetic lethality with disease-specific mutations and other drugs are expected to limit the off-target effects of monotherapies. Another interesting approach is the development of dual inhibitors simultaneously targeting unrelated classes of chromatin-modifying enzymes. Rational-based treatment choices, in particular for combination therapies, will require the development of response-predicting biomarkers. The use of epigenetic signatures has great potential to provide these biomarkers. In any case, future studies will need to consider cancer therapies in the context of their tumor microenvironment, including immune effector cells. The development of ex vivo organoid-like models for B-NHL is needed to accelerate preclinical drug testing.
Author Contributions
M.F.-S., R.W., J.C.S., M.-M.L.P., M.B. and G.R. made a substantial contribution to all aspects of the preparation of this manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
G.R. acknowledges supports from Fondo de Investigación Sanitaria PI18/01383, Spanish Ministry of Science and Innovation, European Regional Development Fund (ERDF) “Una manera de hacer Europa”. J.C.S. and M.F.-S. were recipients of a Sara Borrell research contract (CD19/00228) and a predoctoral fellowship (FI19/00338) from Instituto de Salud Carlos III, respectively. Research in the Buschbeck lab is supported by the following grants and contracts: RTI2018-094005-B-I00 from ERDF/Spanish Ministry of Science and Innovation —Agencia Estatal de Investigación (M.B.), the Marie Skłodowska Curie Training network ‘INTERCEPT-MDS’ H2020-MSCA-ITN-2020-953407, funded by the European Comission (M.B.), the Deutsche José Carreras Leukämie Stiftung (DJCLS) grant 14R/2018 (M.B.), the Catalan Agency for Management of University and Research Grants (AGAUR) grant 2017-SGR-305 (M.B.), and the “Fundació La Marató de TV3” grant 257/C/2019 (M.B.). Research at the IJC is supported by the ‘La Caixa’ Foundation, the Fundació Internacional Josep Carreras, Celgene Spain and the CERCA Programme/Generalitat de Catalunya.
Conflicts of Interest
G.R. received research support from TG Therapeutics, and M.B. from CellCentric Ltd. Remaining authors declare no conflict of interest.
Abbreviations
AE: adverse event; ASCT, autologous stem cell transplantation; B-NHL, B-cell non-Hodgkin lymphoma; BET, bromodomain and extra-terminal; BL, Burkitt lymphoma; CLL, chronic lymphocytic leukemia; CR, complete response; DLBCL, diffuse large B-cell lymphoma; DLT, dose-limiting toxicity; DNMT, DNA methyltransferase; DOR, durable objective response; EFS, event-free survival; FL, follicular lymphoma; GC, germinal center; GDP, Gemcitabine, Dexamethasone and Cisplatin; HAT, histone acetyltransferase; HDAC, histone deacetylase; HDM, histone demethylase; HL; Hodgkin lymphoma; HMT, histone methyl transferase; IMiD, immunomodulatory drug; iNHL, indolent non-Hodgkin lymphoma; LPL, lymphoplasmacytic lymphoma; MCL, mantle cell lymphoma; mCR, metabolic complete response; MM, multiple myeloma; MTD, maximum-tolerated dose; MZL, marginal zone lymphoma; NAM, nicotinamide; NHL, non-Hodgkin lymphoma; NP, neutropenia; N. R., not reached; ORR, overall response rate; OS, overall survival; PFS, progression-free survival; PMBCL, primary mediastinal B-cell lymphoma; PRC2, polycomb repressor complex 2; PRMT, protein arginine methyltransferase; PTM, post-translational modifications; R-CHOP, rituximab, cyclophosphamide, doxorubicin, vincristine and prednisone; R-ICE, rituximab, ifosfamide, carboplatin and etoposide; R/R, relapsed and refractory; SAE, serious adverse event; SD, stable disease; T-NHL, T-cell non-Hodgkin lymphoma; TP, thrombocytopenia; TRAE, treatment-related adverse event.
References
- Chapuy, B.; Stewart, C.; Dunford, A.J.; Kim, J.; Kamburov, A.; Redd, R.A.; Lawrence, M.S.; Roemer, M.G.M.; Li, A.J.; Ziepert, M.; et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med. 2018, 24, 679–690. [Google Scholar] [CrossRef]
- Schmitz, R.; Wright, G.W.; Huang, D.W.; Johnson, C.A.; Phelan, J.D.; Wang, J.Q.; Roulland, S.; Kasbekar, M.; Young, R.M.; Shaffer, A.L.; et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2018, 378, 1396–1407. [Google Scholar] [CrossRef]
- Alizadeh, A.A.; Elsen, M.B.; Davis, R.E.; Ma, C.L.; Lossos, I.S.; Rosenwald, A.; Boldrick, J.C.; Sabet, H.; Tran, T.; Yu, X.; et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000, 403, 503–511. [Google Scholar] [CrossRef]
- Gascoyne, R.D.; Campo, E.; Jaffe, E.S.; Chan, W.C. Diffuse Large B-Cell Lymphoma, NOS. In WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues; Swerdlow, S.H., Campo, E., Harris, N.L., Jaffe, E.S., Pileri, S.A., Stein, H., Thiele, J., Eds.; International Agency for Research on Cancer (IARC): Lyon, France, 2017; pp. 291–297. [Google Scholar]
- Liu, Y.; Barta, S.K. Diffuse large B-cell lymphoma: 2019 update on diagnosis, risk stratification, and treatment. Am. J. Hematol. 2019, 94, 604–616. [Google Scholar] [CrossRef]
- Jaffe, E.S.; Harris, N.L.; Swerdlow, S.H.; Ott, G.; Nathwani, B.N.; de Jong, D.; Yoshino, T.; Spagnolo, D.; Gascoyne, R.D. Follicular lymphoma. In WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues; Swerdlow, S.H., Campo, E., Harris, N.L., Jaffe, E.S., Pileri, S.A., Stein, H., Thiele, J., Eds.; International Agency for Research on Cancer (IARC): Lyon, France, 2017; pp. 266–277. [Google Scholar]
- Carbone, A.; Roulland, S.; Gloghini, A.; Younes, A.; von Keudell, G.; López-Guillermo, A.; Fitzgibbon, J. Follicular lymphoma. Nat. Rev. Dis. Prim. 2019, 5, 83. [Google Scholar] [CrossRef]
- Dreyling, M.; Ghielmini, M.; Rule, S.; Salles, G.A.; Vitolo, U.; Ladetto, M. Newly diagnosed and relapsed follicular lymphoma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2016, 27, v83–v90. [Google Scholar] [CrossRef] [PubMed]
- Leoncini, L.; Campo, E.; Stein, H.; Harris, N.L.; Jaffe, E.S.; Kluin, P.M. Burkitt Lymphoma. In WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues; Swerdlow, S.H., Campo, E., Harris, N.L., Jaffe, E.S., Pileri, S.A., Stein, H., Thiele, J., Eds.; International Agency for Research on Cancer (IARC): Lyon, France, 2017; pp. 330–334. [Google Scholar]
- Casulo, C.; Friedberg, J.W. Burkitt lymphoma- a rare but challenging lymphoma. Best Pract. Res. Clin. Haematol. 2018, 31, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Piris, M.A.; Isaacson, P.G.; Swerdlow, S.H.; Thieblemont, C.; Pittaluga, S.; Rossi, D.; Harris, N.L. Splenic Marginal Zone Lymphoma. In WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues; Swerdlow, S.H., Campo, E., Harris, N.L., Jaffe, E.S., Pileri, S.A., Stein, H., Thiele, J., Eds.; International Agency for Research on Cancer (IARC): Lyon, France, 2017; pp. 223–225. [Google Scholar]
- Campo, E.; Pileri, S.A.; Jaffe, E.S.; Nathwani, B.N.; Stein, H.; Müller-Hermelink, H.K. Nodal Marginal Zone Lymphoma. In WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues; Swerdlow, S.H., Campo, E., Harris, N.L., Jaffe, E.S., Pileri, S.A., Stein, H., Thiele, J., Eds.; International Agency for Research on Cancer (IARC): Lyon, France, 2017; pp. 263–265. [Google Scholar]
- Cook, J.R.; Isaacson, P.G.; Chott, A.; Nakamura, S.; Müller-Hermelink, H.K.; Harris, N.L.; Swerdlow, S.H. Extranodal Marginal Zone Lymphoma of Mucosa-Associated Lymphoid Tissue (MALT Lymphoma). In WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues; Swerdlow, S.H., Campo, E., Harris, N.L., Jaffe, E.S., Pileri, S.A., Stein, H., Thiele, J., Eds.; International Agency for Research on Cancer (IARC): Lyon, France, 2017; pp. 259–262. [Google Scholar]
- Sindel, A.; Al-Juhaishi, T.; Yazbeck, V. Marginal Zone Lymphoma: State-of-the-Art Treatment. Curr. Treat. Options Oncol. 2019, 20, 90. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Seto, M.; Müller-Hermelink, H.K. Mantle Cell Lymphoma. In WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues; Swerdlow, S.H., Campo, E., Harris, N.L., Jaffe, E.S., Pileri, S.A., Stein, H., Thiele, J., Eds.; International Agency for Research on Cancer (IARC): Lyon, France, 2017; pp. 285–290. [Google Scholar]
- Roué, G.; Sola, B. Management of drug resistance in mantle cell lymphoma. Cancers 2020, 12, 1565. [Google Scholar] [CrossRef]
- Waddington, C.H. The epigenotype. 1942. Int. J. Epidemiol. 2012, 41, 10–13. [Google Scholar] [CrossRef] [PubMed]
- Spitz, F.; Furlong, E.E.M. Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 2012, 13, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Hauer, M.H.; Gasser, S.M. Chromatin and nucleosome dynamics in DNA damage and repair. Genes Dev. 2017, 31, 2204–2221. [Google Scholar] [CrossRef]
- Misteli, T. The Self-Organizing Genome: Principles of Genome Architecture and Function. Cell 2020, 183, 28–45. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, E.M.; Dekker, J. Mechanisms and Functions of Chromosome Compartmentalization. Trends Biochem. Sci. 2020, 45, 385–396. [Google Scholar] [CrossRef]
- Jiang, C.; Pugh, B.F. Nucleosome positioning and gene regulation: Advances through genomics. Nat. Rev. Genet. 2009, 10, 161–172. [Google Scholar] [CrossRef]
- Shaban, H.A.; Barth, R.; Recoules, L.; Bystricky, K. Hi-D: Nanoscale mapping of nuclear dynamics in single living cells. Genome Biol. 2020, 21, 1–21. [Google Scholar] [CrossRef]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- Garcia-Saez, I.; Menoni, H.; Boopathi, R.; Shukla, M.S.; Soueidan, L.; Noirclerc-Savoye, M.; Le Roy, A.; Skoufias, D.A.; Bednar, J.; Hamiche, A.; et al. Structure of an H1-Bound 6-Nucleosome Array Reveals an Untwisted Two-Start Chromatin Fiber Conformation. Mol. Cell 2018, 72, 902–915.e7. [Google Scholar] [CrossRef] [PubMed]
- Buschbeck, M.; Hake, S.B. Variants of core histones and their roles in cell fate decisions, development and cancer. Nat. Rev. Mol. Cell Biol. 2017, 18, 299–314. [Google Scholar] [CrossRef]
- Tessarz, P.; Kouzarides, T. Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 2014, 15, 703–708. [Google Scholar] [CrossRef]
- Tropberger, P.; Schneider, R. Scratching the (lateral) surface of chromatin regulation by histone modifications. Nat. Struct. Mol. Biol. 2013, 20, 657–661. [Google Scholar] [CrossRef] [PubMed]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Audia, J.E.; Campbell, R.M. Histone modifications and cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.A.J.; Shilatifard, A. Reevaluating the roles of histone-modifying enzymes and their associated chromatin modifications in transcriptional regulation. Nat. Genet. 2020, 52, 1271–1281. [Google Scholar] [CrossRef]
- Roth, S.Y.; Denu, J.M.; Allis, C.D. Histone Acetyltransferases. Annu. Rev. Biochem. 2001, 70, 81–120. [Google Scholar] [CrossRef]
- Barneda-Zahonero, B.; Parra, M. Histone deacetylases and cancer. Mol. Oncol. 2012, 6, 579–589. [Google Scholar] [CrossRef]
- Bosch-Presegué, L.; Vaquero, A. Sirtuin-dependent epigenetic regulation in the maintenance of genome integrity. FEBS J. 2015, 282, 1745–1767. [Google Scholar] [CrossRef]
- Morera, L.; Lübbert, M.; Jung, M. Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin. Epigenetics 2016, 8, 16. [Google Scholar] [CrossRef]
- Blecua, P.; Martinez-Verbo, L.; Esteller, M. The DNA methylation landscape of hematological malignancies: An update. Mol. Oncol. 2020, 14, 1616–1639. [Google Scholar] [CrossRef]
- Rasmussen, K.D.; Helin, K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016, 30, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.; Zhang, J.; Davis, N.S.; Moffitt, A.B.; Love, C.L.; Waldrop, A.; Leppa, S.; Pasanen, A.; Meriranta, L.; Karjalainen-Lindsberg, M.L.; et al. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell 2017, 171, 481–494.e15. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.; Karkhanis, V.; Tae, S.; Yan, F.; Smith, P.; Ayers, L.W.; Agostinelli, C.; Pileri, S.; Denis, G.V.; Baiocchi, R.A.; et al. Protein arginine methyltransferase 5 (PRMT5) inhibition induces lymphoma cell death through reactivation of the retinoblastoma tumor suppressor pathway and polycomb repressor complex 2 (PRC2) Silencing. J. Biol. Chem. 2013, 288, 35534–35547. [Google Scholar] [CrossRef]
- Poole, C.J.; van Riggelen, J. MYC—master regulator of the cancer epigenome and transcriptome. Genes 2017, 8, 142. [Google Scholar] [CrossRef]
- Fujita, N.; Jaye, D.L.; Geigerman, C.; Akyildiz, A.; Mooney, M.R.; Boss, J.M.; Wade, P.A. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell 2004, 119, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Lemercier, C.; Brocard, M.P.; Puvion-Dutilleul, F.; Kao, H.Y.; Albagli, O.; Khochbin, S. Class II histone deacetylases are directly recruited by BCL6 transcriptional repressor. J. Biol. Chem. 2002, 277, 22045–22052. [Google Scholar] [CrossRef]
- Gearhart, M.D.; Corcoran, C.M.; Wamstad, J.A.; Bardwell, V.J. Polycomb Group and SCF Ubiquitin Ligases Are Found in a Novel BCOR Complex That Is Recruited to BCL6 Targets. Mol. Cell. Biol. 2006, 26, 6880–6889. [Google Scholar] [CrossRef]
- Pasqualucci, L.; Dominguez-Sola, D.; Chiarenza, A.; Fabbri, G.; Grunn, A.; Trifonov, V.; Kasper, L.H.; Lerach, S.; Tang, H.; Ma, J.; et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature 2011, 471, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Berdasco, M.; Esteller, M. Clinical epigenetics: Seizing opportunities for translation. Nat. Rev. Genet. 2019, 20, 109–127. [Google Scholar] [CrossRef]
- Diesch, J.; Zwick, A.; Garz, A.K.; Palau, A.; Buschbeck, M.; Götze, K.S. A clinical-molecular update on azanucleoside-based therapy for the treatment of hematologic cancers. Clin. Epigenetics 2016, 8, 1–11. [Google Scholar] [CrossRef]
- Morin, R.D.; Mendez-Lago, M.; Mungall, A.J.; Goya, R.; Mungall, K.L.; Corbett, R.D.; Johnson, N.A.; Severson, T.M.; Chiu, R.; Field, M.; et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 2011, 476, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, X.; Fiskus, W.; Lin, J.; Lwin, T.; Rao, R.; Zhang, Y.; Chan, J.C.; Fu, K.; Marquez, V.E.; et al. Coordinated Silencing of MYC-Mediated miR-29 by HDAC3 and EZH2 as a Therapeutic Target of Histone Modification in Aggressive B-Cell Lymphomas. Cancer Cell 2012, 22, 506–523. [Google Scholar] [CrossRef] [PubMed]
- New, M.; Olzscha, H.; La Thangue, N.B. HDAC inhibitor-based therapies: Can we interpret the code? Mol. Oncol. 2012, 6, 637–656. [Google Scholar] [CrossRef]
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009, 325, 834–840. [Google Scholar] [CrossRef] [PubMed]
- Finnin, M.S.; Donigian, J.R.; Cohen, A.; Richon, V.M.; Rifkind, R.A.; Marks, P.A.; Breslow, R.; Pavletich, N.P. Structures of TSA and SAHA. Nature 1999, 401, 188–193. [Google Scholar] [CrossRef]
- Guan, X.; Lin, P.; Knoll, E.; Chakrabarti, R. Mechanism of inhibition of the human sirtuin enzyme SIRT3 by nicotinamide: Computational and experimental studies. PLoS ONE 2014, 9, e107729. [Google Scholar]
- Gertz, M.; Fischer, F.; Nguyen, G.T.T.; Lakshminarasimhan, M.; Schutkowski, M.; Weyand, M.; Steegborn, C. Ex-527 inhibits Sirtuins by exploiting their unique NAD+-dependent deacetylation mechanism. Proc. Natl. Acad. Sci. USA 2013, 110, E2772–E2781. [Google Scholar] [CrossRef]
- Marks, P.A.; Breslow, R. Dimethyl sulfoxide to vorinostat: Development of this histone deacetylase inhibitor as an anticancer drug. Nat. Biotechnol. 2007, 25, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, M.; Horinouchi, S.; Beppu, T. Rn Trichostatin A and trapoxin: Novel. BioEssays 1995, 17, 423–430. [Google Scholar] [CrossRef]
- Murugan, K.; Sangeetha, S.; Ranjitha, S.; Vimala, A.; Al-Sohaibani, S.; Rameshkumar, G. HDACiDB: A database for histone deacetylase inhibitors. Drug Des. Devel. Ther. 2015, 9, 2257–2264. [Google Scholar] [CrossRef]
- Yang, F.; Zhao, N.; Ge, D.; Chen, Y. Next-generation of selective histone deacetylase inhibitors. RSC Adv. 2019, 9, 19571–19583. [Google Scholar] [CrossRef]
- Liu, J.; Yu, Y.; Kelly, J.; Sha, D.; Alhassan, A.B.; Yu, W.; Maletic, M.M.; Duffy, J.L.; Klein, D.J.; Holloway, M.K.; et al. Discovery of Highly Selective and Potent HDAC3 Inhibitors Based on a 2-Substituted Benzamide Zinc Binding Group. ACS Med. Chem. Lett. 2020, 11, 2476–2483. [Google Scholar] [CrossRef]
- Leonhardt, M.; Sellmer, A.; Krämer, O.H.; Dove, S.; Elz, S.; Kraus, B.; Beyer, M.; Mahboobi, S. Design and biological evaluation of tetrahydro-β-carboline derivatives as highly potent histone deacetylase 6 (HDAC6) inhibitors. Eur. J. Med. Chem. 2018, 152, 329–357. [Google Scholar] [CrossRef]
- Adams, J.M.; Harris, A.W.; Pinkert, C.A.; Corcoran, L.M.; Alexander, W.S.; Cory, S.; Palmiter, R.D.; Brinster, R.L. The c-myc oncogene driven by immunglubulin enhancers iduces lymphoid malignancy in transgenic mice. Nature 1985, 318, 533–538. [Google Scholar] [CrossRef]
- Sidman, C.L.; Denial, T.M.; Marshall, J.D.; Roths, J.B. Multiple mechanisms of tumorigenesis in E??-myc transgenic mice. Cancer Res. 1993, 53, 1665–1669. [Google Scholar]
- Sabò, A.; Kress, T.R.; Pelizzola, M.; de Pretis, S.; Gorski, M.M.; Tesi, A.; Morelli, M.J.; Bora, P.; Doni, M.; Verrecchia, A.; et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 2014, 511, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Schleich, K.; Kase, J.; Dörr, J.R.; Trescher, S.; Bhattacharya, A.; Yu, Y.; Wailes, E.M.; Fan, D.N.Y.; Lohneis, P.; Milanovic, M.; et al. H3K9me3-mediated epigenetic regulation of senescence in mice predicts outcome of lymphoma patients. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef]
- Stubbs, M.C.; Kim, W.; Bariteau, M.; Davis, T.; Vempati, S.; Minehart, J.; Witkin, M.; Qi, J.; Krivtsov, A.V.; Bradner, J.E.; et al. Selective inhibition of HDAC1 and HDAC2 as a potential therapeutic option for B-ALL. Clin. Cancer Res. 2015, 21, 2348–2358. [Google Scholar] [CrossRef] [PubMed]
- Lindemann, R.K.; Newbold, A.; Whitecross, K.F.; Cluse, L.A.; Frew, A.J.; Ellis, L.; Williams, S.; Wiegmans, A.P.; Dear, A.E.; Scott, C.L.; et al. Analysis of the apoptotic and therapeutic activities of histone deacetylase inhibitors by using a mouse model of B cell lymphoma. Proc. Natl. Acad. Sci. USA 2007, 104, 8071–8076. [Google Scholar] [CrossRef]
- Ellis, L.; Bots, M.; Lindemann, R.K.; Bolden, J.E.; Newbold, A.; Cluse, L.A.; Scott, C.L.; Strasser, A.; Atadja, P.; Lowe, S.W.; et al. The histone deacetylase inhibitors LAQ824 and LBH589 do not require death receptor signaling or a functional apoptosome to mediate tumor cell death or therapeutic efficacy. Blood 2009, 114, 380–393. [Google Scholar] [CrossRef] [PubMed]
- Matthews, G.M.; Mehdipour, P.; Cluse, L.A.; Falkenberg, K.J.; Wang, E.; Roth, M.; Santoro, F.; Vidacs, E.; Stanley, K.; House, C.M.; et al. Functional-genetic dissection of HDAC dependencies in mouse lymphoid and myeloid malignancies. Blood 2015, 126, 2392–2403. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, D.; Zhou, J. Histone Deacetylase 6 as a Therapeutic Target in B cell-associated Hematological Malignancies. Front. Pharmacol. 2020, 11, 1–8. [Google Scholar] [CrossRef]
- Amengual, J.E.; Johannet, P.; Lombardo, M.; Zullo, K.; Hoehn, D.; Bhagat, G.; Scotto, L.; Jirau-Serrano, X.; Radeski, D.; Heinen, J.; et al. Dual targeting of protein degradation pathways with the selective HDAC6 inhibitor ACY-1215 and bortezomib is synergistic in lymphoma. Clin. Cancer Res. 2015, 21, 4663–4675. [Google Scholar] [CrossRef] [PubMed]
- Winkler, R.; Mägdefrau, A.-S.; Kleemann, M.; Beyer, M.; Linke, K.; Hansen, L.; Schaffer, A.-M.; Hoffmann, M.E.; Poepsel, S.; Heyd, F.; et al. Targeting the MYC interaction network in B-cell lymphoma via histone deacetylase 6 inhibition. BioRxiv 2021. [Google Scholar] [CrossRef]
- Bobrowska, A.; Paganetti, P.; Matthias, P.; Bates, G.P. Hdac6 knock-out increases tubulin acetylation but does not modify disease progression in the R6/2 mouse model of Huntington’s disease. PLoS ONE 2011, 6, e20696. [Google Scholar] [CrossRef] [PubMed]
- Rosenjack, J.; Hodges, C.A.; Darrah, R.J.; Kelley, T.J. HDAC6 depletion improves cystic fibrosis mouse airway responses to bacterial challenge. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Imai, Y.; Hirano, M.; Kobayashi, M.; Futami, M.; Tojo, A. HDAC inhibitors exert anti-myeloma effects through multiple modes of action. Cancers 2019, 11, 475. [Google Scholar] [CrossRef]
- Chesi, M.; Robbiani, D.F.; Sebag, M.; Chng, W.J.; Affer, M.; Tiedemann, R.; Valdez, R.; Palmer, S.E.; Haas, S.S.; Stewart, A.K.; et al. AID-Dependent Activation of a MYC Transgene Induces Multiple Myeloma in a Conditional Mouse Model of Post-Germinal Center Malignancies. Cancer Cell 2008, 13, 167–180. [Google Scholar] [CrossRef]
- Chesi, M.; Matthews, G.M.; Garbitt, V.M.; Palmer, S.E.; Shortt, J.; Lefebure, M.; Stewart, A.K.; Johnstone, R.W.; Leif Bergsagel, P. Drug response in a genetically engineered mouse model of multiple myeloma is predictive of clinical efficacy. Blood 2012, 120, 376–385. [Google Scholar] [CrossRef]
- Santo, L.; Hideshima, T.; Kung, A.L.; Tseng, J.-C.; Tamang, D.; Yang, M.; Jarpe, M.; van Duzer, J.H.; Mazitschek, R.; Ogier, W.C.; et al. Preclinical activity, pharmacodynamic, and pharmacokinetic properties of a selective HDAC6 inhibitor, ACY-1215, in combination with bortezomib in multiple myeloma. Blood 2012, 119, 2579–2589. [Google Scholar] [CrossRef]
- García-Guerrero, E.; Götz, R.; Doose, S.; Sauer, M.; Rodríguez-Gil, A.; Nerreter, T.; Kortüm, K.M.; Pérez-Simón, J.A.; Einsele, H.; Hudecek, M.; et al. Upregulation of CD38 expression on multiple myeloma cells by novel HDAC6 inhibitors is a class effect and augments the efficacy of daratumumab. Leukemia 2021, 35, 201–214. [Google Scholar] [CrossRef]
- Bobrowicz, M.; Dwojak, M.; Pyrzynska, B.; Stachura, J.; Muchowicz, A.; Berthel, E.; Dalla-Venezia, N.; Kozikowski, M.; Siernicka, M.; Miazek, N.; et al. HDAC6 inhibition upregulates CD20 levels and increases the efficacy of anti-CD20 monoclonal antibodies. Blood 2017, 130, 1628–1638. [Google Scholar] [CrossRef] [PubMed]
- Mondello, P.; Tadros, S.; Teater, M.; Fontan, L.; Chang, A.Y.; Jain, N.; Yang, H.; Singh, S.; Ying, H.Y.; Chu, C.S.; et al. Selective inhibition of HDAC3 targets synthetic vulnerabilities and activates immune surveillance in lymphoma. Cancer Discov. 2020, 10, 440–459. [Google Scholar] [CrossRef]
- Knox, T.; Sahakian, E.; Banik, D.; Hadley, M.; Palmer, E.; Noonepalle, S.; Kim, J.; Powers, J.; Gracia-Hernandez, M.; Oliveira, V.; et al. Selective HDAC6 inhibitors improve anti-PD-1 immune checkpoint blockade therapy by decreasing the anti-inflammatory phenotype of macrophages and down-regulation of immunosuppressive proteins in tumor cells. Sci. Rep. 2019, 9, 1–17. [Google Scholar]
- Wang, X.; Waschke, B.C.; Woolaver, R.A.; Chen, Z.; Zhang, G.; Piscopio, A.D.; Liu, X.; Wang, J.H. Histone deacetylase inhibition sensitizes PD1 blockade-resistant b-cell lymphomas. Cancer Immunol. Res. 2019, 7, 1318–1331. [Google Scholar] [CrossRef]
- Amengual, J.E.; Clark-Garvey, S.; Kalac, M.; Scotto, L.; Marchi, E.; Neylon, E.; Johannet, P.; Wei, Y.; Zain, J.; O’Connor, O.A. Sirtuin and pan-class I/II deacetylase (DAC) inhibition is synergistic in preclinical models and clinical studies of lymphoma. Blood 2013, 122, 2104–2113. [Google Scholar] [CrossRef]
- Varano, G.; Raffel, S.; Sormani, M.; Zanardi, F.; Lonardi, S.; Zasada, C.; Perucho, L.; Petrocelli, V.; Haake, A.; Lee, A.K.; et al. The B-cell receptor controls fitness of MYC-driven lymphoma cells via GSK3ß inhibition. Nature 2017, 546, 302–306. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.Y.; Hwang, S.H.; Kwon, K.S.; Kim, K.R.; Choi, H.N.; Lee, N.R.; Kwak, J.Y.; Park, B.H.; Park, H.S.; Chung, M.J.; et al. SIRT1 expression is associated with poor prognosis of diffuse large B-cell lymphoma. Am. J. Surg. Pathol. 2008, 32, 1523–1531. [Google Scholar] [CrossRef]
- He, M.; Tan, B.; Vasan, K.; Yuan, H.; Cheng, F.; Ramos da Silva, S.; Lu, C.; Gao, S.J. SIRT1 and AMPK pathways are essential for the proliferation and survival of primary effusion lymphoma cells. J. Pathol. 2017, 242, 309–321. [Google Scholar] [CrossRef]
- Yu, W.; Denu, R.A.; Krautkramer, K.A.; Grindle, K.M.; Yang, D.T.; Asimakopoulos, F.; Hematti, P.; Denu, J.M. Loss of SIRT3 provides growth advantage for B cell malignancies. J. Biol. Chem. 2016, 291, 3268–3279. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chiang, Y.L.; Lyssiotis, C.A.; Teater, M.R.; Hong, J.Y.; Shen, H.; Wang, L.; Hu, J.; Jing, H.; Chen, Z.; et al. Non-oncogene Addiction to SIRT3 Plays a Critical Role in Lymphomagenesis. Cancer Cell 2019, 35, 916–931.e9. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Zhu, Y.; Park, S.H.; Liu, G.; O’Brien, J.; Jiang, H.; Gius, D. SIRT3-mediated dimerization of IDH2 directs cancer cell metabolism and tumor growth. Cancer Res. 2017, 77, 3990–3999. [Google Scholar] [CrossRef]
- Yang, J.; Li, Y.; Zhang, Y.; Fang, X.; Chen, N.; Zhou, X.; Wang, X. Sirt6 promotes tumorigenesis and drug resistance of diffuse large B-cell lymphoma by mediating PI3K/Akt signaling. J. Exp. Clin. Cancer Res. 2020, 39, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Sripathy, S.; Webster, A.; Park, A.; Lao, U.; Hsu, J.H.; Loe, T.; Bedalov, A.; Simon, J.A. Discovery of selective SIRT2 inhibitors as therapeutic agents in B-cell lymphoma and other malignancies. Molecules 2020, 25, 455. [Google Scholar] [CrossRef]
- Zhang, J.; Vlasevska, S.; Wells, V.A.; Nataraj, S.; Holmes, A.B.; Duval, R.; Meyer, S.N.; Mo, T.; Basso, K.; Brindle, P.K.; et al. The CREBBP acetyltransferase is a haploinsufficient tumor suppressor in B-cell lymphoma. Cancer Discov. 2017, 7, 323–337. [Google Scholar] [CrossRef] [PubMed]
- He, Z.X.; Wei, B.F.; Zhang, X.; Gong, Y.P.; Ma, L.Y.; Zhao, W. Current development of CBP/p300 inhibitors in the last decade. Eur. J. Med. Chem. 2021, 209, 112861. [Google Scholar] [CrossRef]
- Ogiwara, H.; Sasaki, M.; Mitachi, T.; Oike, T.; Higuchi, S.; Tominaga, Y.; Kohno, T. Targeting p300 addiction in CBP-deficient cancers causes synthetic lethality by apoptotic cell death due to abrogation of MYC expression. Cancer Discov. 2016, 6, 430–445. [Google Scholar] [CrossRef]
- Sheikh, B.N.; Lee, S.C.W.; El-Saafin, F.; Vanyai, H.K.; Hu, Y.; Pang, S.H.M.; Grabow, S.; Strasser, A.; Nutt, S.L.; Alexander, W.S.; et al. MOZ regulates B-cell progenitors and, consequently, Moz haploinsufficiency dramatically retards MYC-induced lymphoma development. Blood 2015, 125, 1910–1921. [Google Scholar] [CrossRef]
- Baell, J.B.; Leaver, D.J.; Hermans, S.J.; Kelly, G.L.; Brennan, M.S.; Downer, N.L.; Nguyen, N.; Wichmann, J.; McRae, H.M.; Yang, Y.; et al. Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth. Nature 2018, 560, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Farria, A.T.; Plummer, J.B.; Salinger, A.P.; Shen, J.; Lin, K.; Lu, Y.; McBride, K.M.; Koutelou, E.; Dent, S.Y.R. Transcriptional Activation of MYC-Induced Genes by GCN5 Promotes B-cell Lymphomagenesis. Cancer Res. 2020, 80, 5543–5553. [Google Scholar] [CrossRef]
- Farria, A.T.; Mustachio, L.M.; Coban Akdemir, Z.H.; Dent, S.Y.R. GCN5 HAT inhibition reduces human Burkitt lymphoma cell survival through reduction of MYC target gene expression and impeding BCR signaling pathways. Oncotarget 2019, 10, 5847–5858. [Google Scholar] [CrossRef]
- Gorrini, C.; Squatrito, M.; Luise, C.; Syed, N.; Perna, D.; Wark, L.; Martinato, F.; Sardella, D.; Verrecchia, A.; Bennett, S.; et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature 2007, 448, 1063–1067. [Google Scholar] [CrossRef]
- Ghizzoni, M.; Wu, J.; Gao, T.; Haisma, H.J.; Dekker, F.J.; George Zheng, Y. 6-alkylsalicylates are selective Tip60 inhibitors and target the acetyl-CoA binding site. Eur. J. Med. Chem. 2012, 47, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, T.; Filippakopoulos, P. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat. Rev. Mol. Cell Biol. 2017, 18, 246–262. [Google Scholar] [CrossRef]
- Ebrahimi, A.; Sevinç, K.; Gürhan Sevinç, G.; Cribbs, A.P.; Philpott, M.; Uyulur, F.; Morova, T.; Dunford, J.E.; Göklemez, S.; Arı, Ş.; et al. Bromodomain inhibition of the coactivators CBP/EP300 facilitate cellular reprogramming. Nat. Chem. Biol. 2019, 15, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Stathis, A.; Bertoni, F. BET proteins as targets for anticancer treatment. Cancer Discov. 2018, 8, 24–36. [Google Scholar] [CrossRef]
- Filippakopoulos, P.; Qi, J.; Picaud, S.; Shen, Y.; Smith, W.B.; Fedorov, O.; Morse, E.M.; Keates, T.; Hickman, T.T.; Felletar, I.; et al. Selective inhibition of BET bromodomains. Nature 2010, 468, 1067–1073. [Google Scholar] [CrossRef] [PubMed]
- Nicodeme, E.; Jeffrey, K.L.; Schaefer, U.; Beinke, S.; Dewell, S.; Chung, C.W.; Chandwani, R.; Marazzi, I.; Wilson, P.; Coste, H.; et al. Suppression of inflammation by a synthetic histone mimic. Nature 2010, 468, 1119–1123. [Google Scholar] [CrossRef]
- Itzen, F.; Greifenberg, A.K.; Bösken, C.A.; Geyer, M. Brd4 activates P-TEFb for RNA polymerase II CTD phosphorylation. Nucleic Acids Res. 2014, 42, 7577–7590. [Google Scholar] [CrossRef] [PubMed]
- Jonkers, I.; Lis, J.T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015, 16, 167–177. [Google Scholar] [CrossRef]
- Lin, C.Y.; Lovén, J.; Rahl, P.B.; Paranal, R.M.; Burge, C.B.; Bradner, J.E.; Lee, T.I.; Young, R.A. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 2012, 151, 56–67. [Google Scholar] [CrossRef]
- Lovén, J.; Hoke, H.A.; Lin, C.Y.; Lau, A.; Orlando, D.A.; Vakoc, C.R.; Bradner, J.E.; Lee, T.I.; Young, R.A. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013, 153, 320–334. [Google Scholar] [CrossRef]
- Rahman, S.; Sowa, M.E.; Ottinger, M.; Smith, J.A.; Shi, Y.; Harper, J.W.; Howley, P.M. The Brd4 Extraterminal Domain Confers Transcription Activation Independent of pTEFb by Recruiting Multiple Proteins, Including NSD3. Mol. Cell. Biol. 2011, 31, 2641–2652. [Google Scholar] [CrossRef] [PubMed]
- Mertz, J.A.; Conery, A.R.; Bryant, B.M.; Sandy, P.; Balasubramanian, S.; Mele, D.A.; Bergeron, L.; Sims, R.J. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc. Natl. Acad. Sci. USA 2011, 108, 16669–16674. [Google Scholar] [CrossRef]
- Delmore, J.E.; Issa, G.C.; Lemieux, M.E.; Rahl, P.B.; Shi, J.; Jacobs, H.M.; Kastritis, E.; Gilpatrick, T.; Paranal, R.M.; Qi, J.; et al. BET Bromodomain Inhibition as a Therapeutic Strategy to Target c-Myc. Cell 2011, 146, 904–917. [Google Scholar] [CrossRef]
- Trabucco, S.E.; Gerstein, R.M.; Evens, A.M.; Bradner, J.E.; Shultz, L.D.; Greiner, D.L.; Zhang, H. Inhibition of bromodomain proteins for the treatment of human diffuse large B-cell lymphoma. Clin. Cancer Res. 2015, 21, 113–122. [Google Scholar] [CrossRef]
- Aird, F.; Kandela, I.; Mantis, C. Replication Study: BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Elife 2017, 6, 1–15. [Google Scholar] [CrossRef]
- Hogg, S.J.; Vervoort, S.J.; Deswal, S.; Ott, C.J.; Li, J.; Cluse, L.A.; Beavis, P.A.; Darcy, P.K.; Martin, B.P.; Spencer, A.; et al. BET-Bromodomain Inhibitors Engage the Host Immune System and Regulate Expression of the Immune Checkpoint Ligand PD-L1. Cell Rep. 2017, 18, 2162–2174. [Google Scholar] [CrossRef]
- Dawson, M.A.; Prinjha, R.K.; Dittmann, A.; Giotopoulos, G.; Bantscheff, M.; Chan, W.-I.; Robson, S.C.; Chung, C.; Hopf, C.; Savitski, M.M.; et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 2011, 478, 529–533. [Google Scholar] [CrossRef]
- Boi, M.; Gaudio, E.; Bonetti, P.; Kwee, I.; Bernasconi, E.; Tarantelli, C.; Rinaldi, A.; Testoni, M.; Cascione, L.; Ponzoni, M.; et al. The BET Bromodomain Inhibitor OTX015 Affects Pathogenetic Pathways in Preclinical B-cell Tumor Models and Synergizes with Targeted Drugs. Clin. Cancer Res. 2015, 21, 1628–1639. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, B.K.; Gehling, V.S.; Hewitt, M.C.; Vaswani, R.G.; Côté, A.; Leblanc, Y.; Nasveschuk, C.G.; Bellon, S.; Bergeron, L.; Campbell, R.; et al. Identification of a Benzoisoxazoloazepine Inhibitor (CPI-0610) of the Bromodomain and Extra-Terminal (BET) Family as a Candidate for Human Clinical Trials. J. Med. Chem. 2016, 59, 1330–1339. [Google Scholar] [CrossRef] [PubMed]
- Cummin, T.E.C.; Cox, K.L.; Murray, T.D.; Turaj, A.H.; Dunning, L.; English, V.L.; Fell, R.; Packham, G.; Ma, Y.; Powell, B.; et al. BET inhibitors synergize with venetoclax to induce apoptosis in MYC-driven lymphomas with high BCL-2 expression. Blood Adv. 2020, 4, 3316–3328. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Shah, B.; Fiskus, W.; Qi, J.; Rajapakshe, K.; Coarfa, C.; Li, L.; Devaraj, S.G.T.; Sharma, S.; Zhang, L.; et al. Synergistic activity of BET protein antagonist-based combinations in mantle cell lymphoma cells sensitive or resistant to ibrutinib. Blood 2015, 126, 1565–1574. [Google Scholar] [CrossRef]
- Recasens-Zorzo, C.; Cardesa-Salzmann, T.; Petazzi, P.; Ros-Blanco, L.; Esteve-Arenys, A.; Clot, G.; Guerrero-Hernández, M.; Rodríguez, V.; Soldini, D.; Valera, A.; et al. Pharmacological modulation of CXCR4 cooperates with BET bromodomain inhibition in diffuse large B-cell lymphoma. Haematologica 2019, 104, 778–788. [Google Scholar] [CrossRef] [PubMed]
- Picaud, S.; Fedorov, O.; Thanasopoulou, A.; Leonards, K.; Jones, K.; Meier, J.; Olzscha, H.; Monteiro, O.; Martin, S.; Philpott, M.; et al. Generation of a selective small molecule inhibitor of the CBP/p300 bromodomain for Leukemia therapy. Cancer Res. 2015, 75, 5106–5119. [Google Scholar] [CrossRef]
- Welti, J.; Sharp, A.; Brooks, N.; Yuan, W.; McNair, C.; Chand, S.N.; Pal, A.; Figueiredo, I.; Riisnaes, R.; Gurel, B.; et al. Targeting p300/CBP axis in lethal prostate cancer. Cancer Discov. 2021, 44, CD-20-0751. [Google Scholar] [CrossRef] [PubMed]
- Chong, P.S.Y.; Chooi, J.Y.; Lim, J.S.L.; Toh, S.H.M.; Tan, T.Z.; Chng, W.-J. SMARCA2 is a novel interactor of NSD2 and regulates pro-metastatic PTP4A3 through chromatin remodeling in t(4;14) multiple myeloma. Cancer Res. 2021, 81, 2332–2344. [Google Scholar] [CrossRef] [PubMed]
- Theodoulou, N.H.; Bamborough, P.; Bannister, A.J.; Becher, I.; Bit, R.A.; Che, K.H.; Chung, C.W.; Dittmann, A.; Drewes, G.; Drewry, D.H.; et al. Discovery of I-BRD9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition. J. Med. Chem. 2016, 59, 1425–1439. [Google Scholar] [CrossRef] [PubMed]
- Kougnassoukou Tchara, P.E.; Filippakopoulos, P.; Lambert, J.P. Emerging tools to investigate bromodomain functions. Methods 2020, 184, 40–52. [Google Scholar] [CrossRef]
- Ryan, K.R.; Giles, F.; Morgan, G.J. Targeting both BET and CBP/EP300 proteins with the novel dual inhibitors NEO2734 and NEO1132 leads to anti-tumor activity in multiple myeloma. Eur. J. Haematol. 2021, 106, 90–99. [Google Scholar] [CrossRef]
- Hügle, M.; Regenass, P.; Warstat, R.; Hau, M.; Schmidtkunz, K.; Lucas, X.; Wohlwend, D.; Einsle, O.; Jung, M.; Breit, B.; et al. 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines. J. Med. Chem. 2020, 63, 15603–15620. [Google Scholar] [CrossRef] [PubMed]
- Ren, Q.; Gao, W. Current status in the discovery of dual BET/HDAC inhibitors. Bioorganic Med. Chem. Lett. 2021, 31, 127671. [Google Scholar] [CrossRef]
- Watanabe, T.; Kato, H.; Kobayashi, Y.; Yamasaki, S.; Morita-Hoshi, Y.; Yokoyama, H.; Morishima, Y.; Ricker, J.L.; Otsuki, T.; Miyagi-Maesima, A.; et al. Potential efficacy of the oral histone deacetylase inhibitor vorinostat in a phase I trial in follicular and mantle cell lymphoma. Cancer Sci. 2010, 101, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Kirschbaum, M.; Frankel, P.; Popplewell, L.; Zain, J.; Delioukina, M.; Pullarkat, V.; Matsuoka, D.; Pulone, B.; Rotter, A.J.; Espinoza-Delgado, I.; et al. Phase II Study of Vorinostat for Treatment of Relapsed or Refractory Indolent Non-Hodgkin’s Lymphoma and Mantle Cell Lymphoma. J. Clin. Oncol. 2011, 29, 1198–1203. [Google Scholar] [CrossRef]
- Crump, M.; Coiffier, B.; Jacobsen, E.D.; Sun, L.; Ricker, J.L.; Xie, H.; Frankel, S.R.; Randolph, S.S.; Cheson, B.D. Phase II trial of oral vorinostat (suberoylanilide hydroxamic acid) in relapsed diffuse large-B-cell lymphoma. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2008, 19, 964–969. [Google Scholar] [CrossRef]
- Chen, R.; Frankel, P.; Popplewell, L.; Siddiqi, T.; Ruel, N.; Rotter, A.; Thomas, S.H.; Mott, M.; Nathwani, N.; Htut, M.; et al. A phase II study of vorinostat and rituximab for treatment of newly diagnosed and relapsed/refractory indolent non-Hodgkin lymphoma. Haematologica 2015, 100, 357–362. [Google Scholar] [CrossRef]
- Spurgeon, S.E.; Sharma, K.; Claxton, D.F.; Ehmann, C.; Pu, J.; Shimko, S.; Stewart, A.; Subbiah, N.; Palmbach, G.; LeBlanc, F.; et al. Phase 1–2 study of vorinostat (SAHA), cladribine and rituximab (SCR) in relapsed B-cell non-Hodgkin lymphoma and previously untreated mantle cell lymphoma. Br. J. Haematol. 2019, 186, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, T.; Frankel, P.; Beumer, J.H.; Kiesel, B.F.; Christner, S.; Ruel, C.; Song, J.Y.; Chen, R.; Kelly, K.R.; Ailawadhi, S.; et al. Phase 1 study of the Aurora kinase A inhibitor alisertib (MLN8237) combined with the histone deacetylase inhibitor vorinostat in lymphoid malignancies. Leuk. Lymphoma 2020, 61, 309–317. [Google Scholar] [CrossRef]
- Assouline, S.E.; Nielsen, T.H.; Yu, S.; Alcaide, M.; Chong, L.; MacDonald, D.; Tosikyan, A.; Kukreti, V.; Kezouh, A.; Petrogiannis-Haliotis, T.; et al. Phase 2 study of panobinostat with or without rituximab in relapsed diffuse large B-cell lymphoma. Blood 2016, 128, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Amengual, J.E.; Lichtenstein, R.; Lue, J.; Sawas, A.; Deng, C.; Lichtenstein, E.; Khan, K.; Atkins, L.; Rada, A.; Kim, H.A.; et al. A phase 1 study of romidepsin and pralatrexate reveals marked activity in relapsed and refractory T-cell lymphoma. Blood 2018, 131, 397–407. [Google Scholar] [CrossRef]
- O’Connor, O.A.; Falchi, L.; Lue, J.K.; Marchi, E.; Kinahan, C.; Sawas, A.; Deng, C.; Montanari, F.; Amengual, J.E.; Kim, H.A.; et al. Oral 5-azacytidine and romidepsin exhibit marked activity in patients with PTCL: A multicenter phase 1 study. Blood 2019, 134, 1395–1405. [Google Scholar] [CrossRef]
- Reiman, T.; Savage, K.J.; Crump, M.; Cheung, M.C.; MacDonald, D.; Buckstein, R.; Couban, S.; Piliotis, E.; Imrie, K.; Spaner, D.; et al. A phase I study of romidepsin, gemcitabine, dexamethasone and cisplatin combination therapy in the treatment of peripheral T-cell and diffuse large B-cell lymphoma; the Canadian cancer trials group LY.15 study. Leuk. Lymphoma 2019, 60, 912–919. [Google Scholar] [CrossRef]
- Ribrag, V.; Kim, W.S.; Bouabdallah, R.; Lim, S.T.; Coiffier, B.; Illes, A.; Lemieux, B.; Dyer, M.J.S.; Offner, F.; Felloussi, Z.; et al. Safety and efficacy of abexinostat, a pan-histone deacetylase inhibitor, in non-Hodgkin lymphoma and chronic lymphocytic leukemia: Results of a phase II study. Haematologica 2017, 102, 903–909. [Google Scholar] [CrossRef]
- Drott, K.; Hagberg, H.; Papworth, K.; Relander, T.; Jerkeman, M. Valproate in combination with rituximab and CHOP as first-line therapy in diffuse large B-cell lymphoma (VALFRID). Blood Adv. 2018, 2, 1386–1392. [Google Scholar] [CrossRef] [PubMed]
- Oki, Y.; Kelly, K.R.; Flinn, I.; Patel, M.R.; Gharavi, R.; Ma, A.; Parker, J.; Hafeez, A.; Tuck, D.; Younes, A. CUDC-907 in relapsed/refractory diffuse large B-cell lymphoma, including patients with MYC-alterations: Results from an expanded phase I trial. Haematologica 2017, 102, 1923–1930. [Google Scholar] [CrossRef]
- Batlevi, C.L.; Crump, M.; Andreadis, C.; Rizzieri, D.; Assouline, S.E.; Fox, S.; Van Der Jagt, R.H.C.; Copeland, A.; Potvin, D.; Chao, R.; et al. A phase 2 study of mocetinostat, a histone deacetylase inhibitor, in relapsed or refractory lymphoma. Br. J. Haematol. 2017, 178, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Amorim, S.; Stathis, A.; Gleeson, M.; Iyengar, S.; Magarotto, V.; Leleu, X.; Morschhauser, F.; Karlin, L.; Broussais, F.; Rezai, K.; et al. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: A dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol. 2016, 3, e196–e204. [Google Scholar] [CrossRef]
- Shapiro, G.I.; LoRusso, P.; Dowlati, A.; T. Do, K.; Jacobson, C.A.; Vaishampayan, U.; Weise, A.; Caimi, P.F.; Eder, J.P.; French, C.A.; et al. A Phase 1 study of RO6870810, a novel bromodomain and extra-terminal protein inhibitor, in patients with NUT carcinoma, other solid tumours, or diffuse large B-cell lymphoma. Br. J. Cancer 2020, 124, 744–753. [Google Scholar] [CrossRef]
- Falchook, G.; Rosen, S.; LoRusso, P.; Watts, J.; Gupta, S.; Coombs, C.C.; Talpaz, M.; Kurzrock, R.; Mita, M.; Cassaday, R.; et al. Development of 2 bromodomain and extraterminal inhibitors with distinct pharmacokinetic and pharmacodynamic profiles for the treatment of advanced malignancies. Clin. Cancer Res. 2020, 26, 1247–1257. [Google Scholar] [CrossRef]
- Basheer, F.; Huntly, B.J.P. BET bromodomain inhibitors in leukemia. Exp. Hematol. 2015, 43, 718–731. [Google Scholar] [CrossRef] [PubMed]
- Donato, E.; Croci, O.; Sabò, A.; Muller, H.; Morelli, M.J.; Pelizzola, M.; Campaner, S. Compensatory RNA polymerase 2 loading determines the efficacy and transcriptional selectivity of JQ1 in Myc-driven tumors. Leukemia 2017, 31, 479–490. [Google Scholar] [CrossRef]
- ClinicalTrials.gov Database. Available online: https://clinicaltrials.gov/ (accessed on 16 November 2021).
- Puvvada, S.D.; Guillén-Rodríguez, J.M.; Rivera, X.I.; Heard, K.; Inclan, L.; Schmelz, M.; Schatz, J.H.; Persky, D.O. A Phase II Exploratory Study of PXD-101 (Belinostat) Followed by Zevalin in Patients with Relapsed Aggressive High-Risk Lymphoma. Oncology 2017, 93, 401–405. [Google Scholar] [CrossRef]
- Blum, K.A.; Abramson, J.; Maris, M.; Flinn, I.; Goy, A.; Mertz, J.; Sims, R.; Garner, F.; Senderowicz, A.; Younes, A. 41OA phase I study of CPI-0610, a bromodomain and extra terminal protein (BET) inhibitor in patients with relapsed or refractory lymphoma. Ann. Oncol. 2018, 29, iii7. [Google Scholar] [CrossRef]
- Patel, M.R.; Garcia-Manero, G.; Paquette, R.; Dinner, S.; Donnellan, W.B.; Grunwald, M.R.; Ribadeneira, M.D.; Schroeder, P.; Brevard, J.; Wilson, L.; et al. The BET Inhibitor FT-1101 As a Single Agent in Patients with Relapsed or Refractory Hematologic Malignancies. Blood 2019, 134, 3907. [Google Scholar] [CrossRef]
- Sarkozy, C.; Morschhauser, F.; Dubois, S.; Molina, T.; Michot, J.M.; Cullières-Dartigues, P.; Suttle, B.; Karlin, L.; Le Gouill, S.; Picquenot, J.M.; et al. A LYSA phase Ib study of tazemetostat (EPZ-6438) plus R-CHOP in patients with newly diagnosed diffuse large B-cell lymphoma (DLBCL) with poor prognosis features. Clin. Cancer Res. 2020, 26, 3145–3153. [Google Scholar] [CrossRef] [PubMed]
- Leonard, J.; Batlevi, C.L.; Gabrail, N.; Pagel, J.M.; Yang, J.; Whalen, J.; Adib, D. A Phase 1b/3 Randomized, Double-Blind, 3-Stage Study of Tazemetostat or Placebo Plus Lenalidomide and Rituximab in Patients with Relapsed/Refractory Follicular Lymphoma. Clin. Lymphoma Myeloma Leuk. 2020, 20, S279. [Google Scholar] [CrossRef]
- Harb, W.; Abramson, J.; Lunning, M.; Goy, A.; Maddocks, K.; Lebedinsky, C.; Senderowicz, A.; Trojer, P.; Bradley, W.D.; Flinn, I. A phase 1 study of CPI-1205, a small molecule inhibitor of EZH2, preliminary safety in patients with B-cell lymphomas. Ann. Oncol. 2018, 29, iii7. [Google Scholar] [CrossRef]
- Milosevich, N.; Hof, F. Chemical Inhibitors of Epigenetic Methyllysine Reader Proteins. Biochemistry 2016, 55, 1570–1583. [Google Scholar] [CrossRef]
- Roadmap Epigenomics Consortium; Kundaje, A.; Meuleman, W.; Ernst, J.; Bilenky, M.; Yen, A.; Heravi-Moussavi, A.; Kheradpour, P.; Zhang, Z.; Wang, J.; et al. Integrative analysis of 111 reference human epigenomes. Nature 2015, 518, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Morin, R.D.; Johnson, N.A.; Severson, T.M.; Mungall, A.J.; An, J.; Goya, R.; Paul, J.E.; Boyle, M.; Woolcock, B.W.; Kuchenbauer, F.; et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 2010, 42, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Sneeringer, C.J.; Scott, M.P.; Kuntz, K.W.; Knutson, S.K.; Pollock, R.M.; Richon, V.M.; Copeland, R.A. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc. Natl. Acad. Sci. USA 2010, 107, 20980–20985. [Google Scholar] [CrossRef] [PubMed]
- Béguelin, W.; Teater, M.; Meydan, C.; Hoehn, K.B.; Phillip, J.M.; Soshnev, A.A.; Venturutti, L.; Rivas, M.A.; Calvo-Fernández, M.T.; Gutierrez, J.; et al. Mutant EZH2 Induces a Pre-malignant Lymphoma Niche by Reprogramming the Immune Response. Cancer Cell 2020, 37, 655–673.e11. [Google Scholar] [CrossRef]
- Love, C.; Sun, Z.; Jima, D.; Li, G.; Zhang, J.; Miles, R.; Richards, K.L.; Dunphy, C.H.; Choi, W.L.; Srivastava, G.; et al. The genetic landscape of mutations in Burkitt lymphoma. Nat. Genet. 2012, 44, 1321–1325. [Google Scholar] [CrossRef]
- Herviou, L.; Jourdan, M.; Martinez, A.M.; Cavalli, G.; Moreaux, J. EZH2 is overexpressed in transitional preplasmablasts and is involved in human plasma cell differentiation. Leukemia 2019, 33, 2047–2060. [Google Scholar] [CrossRef]
- Eich, M.-L.; Athar, M.; Ferguson, J.E.; Varambally, S. EZH2-Targeted Therapies in Cancer: Hype or a Reality. Cancer Res. 2020, 80, 5449–5458. [Google Scholar] [CrossRef] [PubMed]
- McCabe, M.T.; Ott, H.M.; Ganji, G.; Korenchuk, S.; Thompson, C.; Van Aller, G.S.; Liu, Y.; Pietra, A.D.; LaFrance, L.V.; Mellinger, M.; et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 2012, 492, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Knutson, S.K.; Kawano, S.; Minoshima, Y.; Warholic, N.M.; Huang, K.C.; Xiao, Y.; Kadowaki, T.; Uesugi, M.; Kuznetsov, G.; Kumar, N.; et al. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol. Cancer Ther. 2014, 13, 842–854. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.S.; Chen, J.Y.; Tsai, H.J.; Chen, T.Y.; Hung, W.C. The SUV39H1 inhibitor chaetocin induces differentiation and shows synergistic cytotoxicity with other epigenetic drugs in acute myeloid leukemia cells. Blood Cancer J. 2015, 5, 1–6. [Google Scholar] [CrossRef]
- Stein, E.M.; Garcia-Manero, G.; Rizzieri, D.A.; Tibes, R.; Berdeja, J.G.; Savona, M.R.; Jongen-Lavrenic, M.; Altman, J.K.; Thomson, B.; Blakemore, S.J.; et al. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood 2018, 131, 2661–2669. [Google Scholar] [CrossRef]
- Veazey, K.J.; Cheng, D.; Lin, K.; Villarreal, O.D.; Gao, G.; Perez-Oquendo, M.; Van, H.T.; Stratton, S.A.; Green, M.; Xu, H.; et al. CARM1 inhibition reduces histone acetyltransferase activity causing synthetic lethality in CREBBP/EP300-mutated lymphomas. Leukemia 2020, 34, 3269–3285. [Google Scholar] [CrossRef]
- Wang, L.; Pal, S.; Sif, S. Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol. Cell. Biol. 2008, 28, 6262–6277. [Google Scholar] [CrossRef]
- Koh, C.M.; Bezzi, M.; Low, D.H.P.; Ang, W.X.; Teo, S.X.; Gay, F.P.H.; Al-Haddawi, M.; Tan, S.Y.; Osato, M.; Sabò, A.; et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature 2015, 523, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Chan-Penebre, E.; Kuplast, K.G.; Majer, C.R.; Boriack-Sjodin, P.A.; Wigle, T.J.; Johnston, L.D.; Rioux, N.; Munchhof, M.J.; Jin, L.; Jacques, S.L.; et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat. Chem. Biol. 2015, 11, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Fernando, T.M.; Lossos, C.; Yusufova, N.; Liu, F.; Fontán, L.; Durant, M.; Geng, H.; Melnick, J.; Luo, Y.; et al. PRMT5 interacts with the BCL6 oncoprotein and is required for germinal center formation and lymphoma cell survival. Blood 2018, 132, 2026–2039. [Google Scholar] [CrossRef] [PubMed]
- Hatzi, K.; Geng, H.; Doane, A.S.; Meydan, C.; LaRiviere, R.; Cardenas, M.; Duy, C.; Shen, H.; Vidal, M.N.C.; Baslan, T.; et al. Histone demethylase LSD1 is required for germinal center formation and BCL6-driven lymphomagenesis. Nat. Immunol. 2019, 20, 86–96. [Google Scholar] [CrossRef]
- Mathur, R.; Sehgal, L.; Havranek, O.; Köhrer, S.; Khashab, T.; Jain, N.; Burger, J.A.; Neelapu, S.S.; Davis, R.E.; Samaniego, F. Inhibition of demethylase KDM6B sensitizes diffuse large B-cell lymphoma to chemotherapeutic drugs. Haematologica 2017, 102, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Eissenberg, J.C. Structural biology of the chromodomain: Form and function. Gene 2012, 496, 69–78. [Google Scholar] [CrossRef]
- Zhang, Y.; Lei, M.; Yang, X.; Feng, Y.; Yang, Y.; Loppnau, P.; Li, Y.; Yang, Y.; Min, J.; Liu, Y. Structural and histone binding studies of the chromo barrel domain of TIP60. FEBS Lett. 2018, 592, 1221–1232. [Google Scholar] [CrossRef]
- Huang, Y.; Fang, J.; Bedford, M.T.; Zhang, Y.; Xu, R.M. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science 2006, 312, 748–751. [Google Scholar] [CrossRef]
- Shi, X.; Hong, T.; Walter, K.L.; Ewalt, M.; Michishita, E.; Hung, T.; Carney, D.; Peña, P.; Lan, F.; Kaadige, M.R.; et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 2006, 442, 96–99. [Google Scholar] [CrossRef]
- Estève, P.O.; Terragni, J.; Deepti, K.; Chin, H.G.; Dai, N.; Espejo, A.; Corrêa, I.R.; Bedford, M.T.; Pradhan, S. Methyllysine reader plant homeodomain (PHD) finger protein 20-like 1 (PHF20L1) antagonizes DNA (cytosine-5) methyltransferase 1 (DNMT1) proteasomal degradation. J. Biol. Chem. 2014, 289, 8277–8287. [Google Scholar] [CrossRef]
- Scott, C.L.; Gil, J.; Hernando, E.; Teruya-Feldstein, J.; Narita, M.; Martínez, D.; Visakorpi, T.; Mu, D.; Cordon-Cardo, C.; Peters, G.; et al. Role of the chromobox protein CBX7 in lymphomagenesis. Proc. Natl. Acad. Sci. USA 2007, 104, 5389–5394. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, E.; Duncan, E.M.; Masui, O.; Gil, J.; Heard, E.; Allis, C.D. Mouse Polycomb Proteins Bind Differentially to Methylated Histone H3 and RNA and Are Enriched in Facultative Heterochromatin. Mol. Cell. Biol. 2006, 26, 2560–2569. [Google Scholar] [CrossRef] [PubMed]
- Bracken, A.P.; Kleine-Kohlbrecher, D.; Dietrich, N.; Pasini, D.; Gargiulo, G.; Beekman, C.; Theilgaard-Mönch, K.; Minucci, S.; Porse, B.T.; Marine, J.C.; et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007, 21, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Simhadri, C.; Gignac, M.C.; Anderson, C.J.; Milosevich, N.; Dheri, A.; Prashar, N.; Flemmer, R.T.; Dev, A.; Henderson, T.G.; Douglas, S.F.; et al. Structure-Activity Relationships of Cbx7 Inhibitors, Including Selectivity Studies against Other Cbx Proteins. ACS Omega 2016, 1, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Knutson, S.K.; Warholic, N.M.; Wigle, T.J.; Klaus, C.R.; Allain, C.J.; Raimondi, A.; Scott, M.P.; Chesworth, R.; Moyer, M.P.; Copeland, R.A.; et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc. Natl. Acad. Sci. USA 2013, 110, 7922–7927. [Google Scholar] [CrossRef]
- Italiano, A.; Soria, J.C.; Toulmonde, M.; Michot, J.M.; Lucchesi, C.; Varga, A.; Coindre, J.M.; Blakemore, S.J.; Clawson, A.; Suttle, B.; et al. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: A first-in-human, open-label, phase 1 study. Lancet Oncol. 2018, 19, 649–659. [Google Scholar] [CrossRef]
- Morschhauser, F.; Tilly, H.; Chaidos, A.; McKay, P.; Phillips, T.; Assouline, S.; Batlevi, C.L.; Campbell, P.; Ribrag, V.; Damaj, G.L.; et al. Tazemetostat for patients with relapsed or refractory follicular lymphoma: An open-label, single-arm, multicentre, phase 2 trial. Lancet Oncol. 2020, 21, 1433–1442. [Google Scholar] [CrossRef]
- FDA Approves Tazemetostat for Relapsed/Refractory Follicular Lymphoma. Available online: https://www.targetedonc.com/view/fda-approves-tazemetostat-for-relapsed-refractory-follicular-lymphoma (accessed on 29 April 2021).
- Yap, T.A.; Winter, J.N.; Giulino-Roth, L.; Longley, J.; Lopez, J.; Michot, J.M.; Leonard, J.P.; Ribrag, V.; McCabe, M.T.; Creasy, C.L.; et al. Phase I study of the novel enhancer of zeste homolog 2 (EZH2) inhibitor GSK2816126 in patients with advanced hematologic and solid tumors. Clin. Cancer Res. 2019, 25, 7331–7339. [Google Scholar] [CrossRef]
- Huang, S.; Wang, Z.; Zhou, J.; Huang, J.; Zhou, L.; Luo, J.; Wan, Y.Y.; Long, H.; Zhu, B. EZH2 inhibitor GSK126 suppresses antitumor immunity by driving production of myeloid-derived suppressor cells. Cancer Res. 2019, 79, 2009–2020. [Google Scholar] [CrossRef]
- Qi, W.; Chan, H.M.; Teng, L.; Li, L.; Chuai, S.; Zhang, R.; Zeng, J.; Li, M.; Fan, H.; Lin, Y.; et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc. Natl. Acad. Sci. USA 2012, 109, 21360–21365. [Google Scholar] [CrossRef]
- Campbell, J.E.; Kuntz, K.W.; Knutson, S.K.; Warholic, N.M.; Keilhack, H.; Wigle, T.J.; Raimondi, A.; Klaus, C.R.; Rioux, N.; Yokoi, A.; et al. EPZ011989, A potent, orally-available EZH2 inhibitor with robust in vivo activity. ACS Med. Chem. Lett. 2015, 6, 491–495. [Google Scholar] [CrossRef]
- Gehling, V.S.; Vaswani, R.G.; Nasveschuk, C.G.; Duplessis, M.; Iyer, P.; Balasubramanian, S.; Zhao, F.; Good, A.C.; Campbell, R.; Lee, C.; et al. Discovery, design, and synthesis of indole-based EZH2 inhibitors. Bioorganic Med. Chem. Lett. 2015, 25, 3644–3649. [Google Scholar] [CrossRef]
- Chamorro-Jorganes, A.; Ribeiro, M.L.; Profitos-Peleja, N.; Reyes-Garau, D.; Recasens-Zorzo, C.; Valero, J.G.; Armengol, M.; Perez-Galan, P.; Butler, R.; Postigo, A.; et al. Abstract 2925: Safety and efficacy of EZH2 and BRD4 dual targeting in EZH2 Y641mut germinal centre-derived lymphoma. In Proceedings of the Cancer Research, American Association for Cancer Research (AACR). Philadelphia, PA, USA, 22–24 June 2020; Volume 80, p. 2925. [Google Scholar]
- Vaswani, R.G.; Gehling, V.S.; Dakin, L.A.; Cook, A.S.; Nasveschuk, C.G.; Duplessis, M.; Iyer, P.; Balasubramanian, S.; Zhao, F.; Good, A.C.; et al. Identification of (R)-N-((4-Methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1-(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), a Potent and Selective Inhibitor of Histone Methyltransferase EZH2, Suitable for Phase i Clinical Trials for B-Cell Lymphomas. J. Med. Chem. 2016, 59, 9928–9941. [Google Scholar] [PubMed]
- Lakhani, N.J.; Gutierrez, M.; Duska, L.R.; Do, K.T.; Sharma, M.; Gandhi, L.; Papadopoulos, K.P.; Truong, J.; Fan, X.; Lee, J.H.; et al. Phase 1/2 first-in-human (FIH) study of CPI-0209, a novel small molecule inhibitor of enhancer of zeste homolog 2 (EZH2) in patients with advanced tumors. J. Clin. Oncol. 2021, 39, 3104. [Google Scholar] [CrossRef]
- Bisserier, M.; Wajapeyee, N. Mechanisms of resistance to ezh2 inhibitors in diffuse large b-cell lymphomas. Blood 2018, 131, 2125–2137. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Liu, Y.; Hsu, Y.J.; Fujiwara, Y.; Kim, J.; Mao, X.; Yuan, G.C.; Orkin, S.H. EZH1 Mediates Methylation on Histone H3 Lysine 27 and Complements EZH2 in Maintaining Stem Cell Identity and Executing Pluripotency. Mol. Cell 2008, 32, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Konze, K.D.; Ma, A.; Li, F.; Barsyte-Lovejoy, D.; Parton, T.; MacNevin, C.J.; Liu, F.; Gao, C.; Huang, X.P.; Kuznetsova, E.; et al. An orally bioavailable chemical probe of the lysine methyltransferases EZH2 and EZH1. ACS Chem. Biol. 2013, 8, 1324–1334. [Google Scholar] [CrossRef] [PubMed]
- Honma, D.; Kanno, O.; Watanabe, J.; Kinoshita, J.; Hirasawa, M.; Nosaka, E.; Shiroishi, M.; Takizawa, T.; Yasumatsu, I.; Horiuchi, T.; et al. Novel orally bioavailable EZH1/2 dual inhibitors with greater antitumor efficacy than an EZH2 selective inhibitor. Cancer Sci. 2017, 108, 2069–2078. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, D.; Tobinai, K.; Makita, S.; Ishida, T.; Kusumoto, S.; Ishitsuka, K.; Yoshimitsu, M.; Imaizumi, Y.; Sawayama, Y.; Takeuchi, S.; et al. First-in-Human Study of the EZH1/2 Dual Inhibitor DS-3201b in Patients with Relapsed or Refractory Non-Hodgkin Lymphomas—Preliminary Results. Blood 2017, 130, 4070. [Google Scholar]
- Kung, P.P.; Bingham, P.; Brooun, A.; Collins, M.; Deng, Y.L.; Dinh, D.; Fan, C.; Gajiwala, K.S.; Grantner, R.; Gukasyan, H.J.; et al. Optimization of Orally Bioavailable Enhancer of Zeste Homolog 2 (EZH2) Inhibitors Using Ligand and Property-Based Design Strategies: Identification of Development Candidate (R)-5,8-Dichloro-7-(methoxy(oxetan-3-yl)methyl)-2-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-3,4-dihydroisoquinolin-1(2H)-one (PF-06821497). J. Med. Chem. 2018, 61, 650–665. [Google Scholar]
- Luger, K.; Richmond, T.J. The histone tails of the nucleosome. Curr. Opin. Genet. Dev. 1998, 8, 140–146. [Google Scholar] [CrossRef]
- Weake, V.M.; Workman, J.L. Histone Ubiquitination: Triggering Gene Activity. Mol. Cell 2008, 29, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Shiio, Y.; Eisenman, R.N. Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. USA 2003, 100, 13225–13230. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cadahía, B.; Drobic, B.; Davie, J.R. H3 phosphorylation: Dual role in mitosis and interphaseThis paper is one of a selection of papers published in this Special Issue entitled 30th Annual International Asilomar Chromatin and Chromosomes Conference and has undergone the Journal’s usual peer r. Biochem. Cell Biol. 2009, 87, 695–709. [Google Scholar] [CrossRef]
- Hegyi, K.; Méhes, G. Mitotic Failures in Cancer: Aurora B Kinase and its Potential Role in the Development of Aneuploidy. Pathol. Oncol. Res. 2012, 18, 761–769. [Google Scholar] [CrossRef]
- Ramani, P.; Taylor, S.; Miller, E.; Sowa-Avugrah, E.; May, M.T. High Phosphohistone H3 Expression Correlates with Adverse Clinical, Biological, and Pathological Factors in Neuroblastomas. J. Histochem. Cytochem. 2015, 63, 397–407. [Google Scholar] [CrossRef]
- Hale, C.S.; Qian, M.; Ma, M.W.; Scanlon, P.; Berman, R.S.; Shapiro, R.L.; Pavlick, A.C.; Shao, Y.; Polsky, D.; Osman, I.; et al. Mitotic Rate in Melanoma. Am. J. Surg. Pathol. 2013, 37, 882–889. [Google Scholar] [CrossRef]
- Ginter, P.S.; Shin, S.J.; Liu, Y.; Chen, Z.; D’Alfonso, T.M. Phosphohistone H3 expression correlates with manual mitotic counts and aids in identification of “hot spots” in fibroepithelial tumors of the breast. Hum. Pathol. 2016, 49, 90–98. [Google Scholar] [CrossRef]
- Khieu, M.L.; Broadwater, D.R.; Aden, J.K.; Coviello, J.M.; Lynch, D.T.; Hall, J.M. The Utility of Phosphohistone H3 (PHH3) in Follicular Lymphoma Grading: A Comparative Study with Ki-67 and H&E Mitotic Count. Am. J. Clin. Pathol. 2019, 151, 542–550. [Google Scholar]
- Méhes, G.; Hegyi, K.; Jobanputra, R.; Beke, L.; Vereb, G.; Bedekovics, J. Distinct Dynamics of Mitotic Transition in B-Cell Lymphoma and Reactive B-Cell Lymphoproliferations Determined by H3S10 Phosphohistone Immunolabeling. Pathobiology 2017, 84, 243–250. [Google Scholar] [CrossRef]
- Crosio, C.; Fimia, G.M.; Loury, R.; Kimura, M.; Okano, Y.; Zhou, H.; Sen, S.; Allis, C.D.; Sassone-Corsi, P. Mitotic Phosphorylation of Histone H3: Spatio-Temporal Regulation by Mammalian Aurora Kinases. Mol. Cell. Biol. 2002, 22, 874–885. [Google Scholar] [CrossRef]
- Ikezoe, T.; Takeuchi, T.; Yang, J.; Adachi, Y.; Nishioka, C.; Furihata, M.; Koeffler, H.P.; Yokoyama, A. Analysis of Aurora B kinase in non-Hodgkin lymphoma. Lab. Investig. 2009, 89, 1364–1373. [Google Scholar] [CrossRef][Green Version]
- Floc’h, N.; Ashton, S.; Ferguson, D.; Taylor, P.; Carnevalli, L.S.; Hughes, A.M.; Harris, E.; Hattersley, M.; Wen, S.; Curtis, N.J.; et al. Modeling Dose and Schedule Effects of AZD2811 Nanoparticles Targeting Aurora B Kinase for Treatment of Diffuse Large B-cell Lymphoma. Mol. Cancer Ther. 2019, 18, 909–919. [Google Scholar] [CrossRef]
- Kelly, K.R.; Friedberg, J.W.; Park, S.I.; McDonagh, K.; Hayslip, J.; Persky, D.; Ruan, J.; Puvvada, S.; Rosen, P.; Iyer, S.P.; et al. Phase I Study of the Investigational Aurora A Kinase Inhibitor Alisertib plus Rituximab or Rituximab/Vincristine in Relapsed/Refractory Aggressive B-cell Lymphoma. Clin. Cancer Res. 2018, 24, 6150–6159. [Google Scholar] [CrossRef]
- Huertas, D.; Soler, M.; Moreto, J.; Villanueva, A.; Martinez, A.; Vidal, A.; Charlton, M.; Moffat, D.; Patel, S.; McDermott, J.; et al. Antitumor activity of a small-molecule inhibitor of the histone kinase Haspin. Oncogene 2012, 31, 1408–1418. [Google Scholar] [CrossRef]
- Rzymski, T.; Zarebski, A.; Windak, R.; Sibinska, Z.; Klosowska-Wardega, A.; Trebacz, E.; Cholody, M.; Szamborska-Gbur, A.; Milik, M.; Prymula, K.; et al. Abstract 3845: Antitumor activity of SEL120: An orally available dual inhibitors of Haspin/CDK9, for standalone and combination therapy with AuroraB inhibitors in solid tumors and hematopoietic malignancies. In Proceedings of the Experimental and Molecular Therapeutics. Am. Assoc. Cancer Res. 2012, 72, 3845. [Google Scholar]
- Ayyappan, S.; Maddocks, K. Novel and emerging therapies for B cell lymphoma. J. Hematol. Oncol. 2019, 12, 82. [Google Scholar] [CrossRef]
- Chen, E.; Staudt, L.M.; Green, A.R. Janus Kinase Deregulation in Leukemia and Lymphoma. Immunity 2012, 36, 529–541. [Google Scholar] [CrossRef] [PubMed]
- Rui, L.; Drennan, A.C.; Ceribelli, M.; Zhu, F.; Wright, G.W.; Huang, D.W.; Xiao, W.; Li, Y.; Grindle, K.M.; Lu, L.; et al. Epigenetic gene regulation by Janus kinase 1 in diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. USA 2016, 113, E7260–E7267. [Google Scholar] [CrossRef]
- Porpaczy, E.; Tripolt, S.; Hoelbl-Kovacic, A.; Gisslinger, B.; Bago-Horvath, Z.; Casanova-Hevia, E.; Clappier, E.; Decker, T.; Fajmann, S.; Fux, D.A.; et al. Aggressive B-cell lymphomas in patients with myelofibrosis receiving JAK1/2 inhibitor therapy. Blood 2018, 132, 694–706. [Google Scholar] [CrossRef] [PubMed]
- Rumi, E.; Zibellini, S. JAK inhibitors and risk of B-cell lymphomas. Blood 2019, 133, 2251–2253. [Google Scholar] [CrossRef] [PubMed]
- Nocturne, G.; Pascaud, J.; Ly, B.; Tahmasebi, F.; Mariette, X. JAK inhibitors alter NK cell functions and may impair immunosurveillance against lymphomagenesis. Cell. Mol. Immunol. 2020, 17, 552–553. [Google Scholar] [CrossRef] [PubMed]
- Paakinaho, V.; Lempiäinen, J.K.; Sigismondo, G.; Niskanen, E.A.; Malinen, M.; Jääskeläinen, T.; Varjosalo, M.; Krijgsveld, J.; Palvimo, J.J. SUMOylation regulates the protein network and chromatin accessibility at glucocorticoid receptor-binding sites. Nucleic Acids Res. 2021, 49, 1951–1971. [Google Scholar] [CrossRef] [PubMed]
- Qu, C.; Kunkalla, K.; Vaghefi, A.; Frederiksen, J.K.; Liu, Y.; Chapman, J.R.; Blonska, M.; Bernal-Mizrachi, L.; Alderuccio, J.P.; Lossos, I.S.; et al. Smoothened stabilizes and protects TRAF6 from degradation: A novel non-canonical role of smoothened with implications in lymphoma biology. Cancer Lett. 2018, 436, 149–158. [Google Scholar] [CrossRef]
- Pham, L.V.; Zhou, H.-J.; Lin-Lee, Y.-C.; Tamayo, A.T.; Yoshimura, L.C.; Fu, L.; Darnay, B.G.; Ford, R.J. Nuclear Tumor Necrosis Factor Receptor-associated Factor 6 in Lymphoid Cells Negatively Regulates c-Myb-mediated Transactivation through Small Ubiquitin-related Modifier-1 Modification. J. Biol. Chem. 2008, 283, 5081–5089. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.L.; Guppy, B.J.; Sawchuk, L.; Davie, J.R.; McManus, K.J. Regulation of chromatin structure via histone post-translational modification and the link to carcinogenesis. Cancer Metastasis Rev. 2013, 32, 363–376. [Google Scholar] [CrossRef]
- Nishida, Y.; Maeda, A.; Kim, M.J.; Cao, L.; Kubota, Y.; Ishizawa, J.; AlRawi, A.; Kato, Y.; Iwama, A.; Fujisawa, M.; et al. The novel BMI-1 inhibitor PTC596 downregulates MCL-1 and induces p53-independent mitochondrial apoptosis in acute myeloid leukemia progenitor cells. Blood Cancer J. 2017, 7, e527. [Google Scholar] [CrossRef]
- Nagel, S.; Ehrentraut, S.; Tomasch, J.; Quentmeier, H.; Meyer, C.; Kaufmann, M.; Drexler, H.G.; MacLeod, R.A.F. Ectopic Expression of Homeobox Gene NKX2-1 in Diffuse Large B-Cell Lymphoma Is Mediated by Aberrant Chromatin Modifications. PLoS ONE 2013, 8, e61447. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).