R-Spondin2, a Positive Canonical WNT Signaling Regulator, Controls the Expansion and Differentiation of Distal Lung Epithelial Stem/Progenitor Cells in Mice
Abstract
:1. Introduction
2. Results
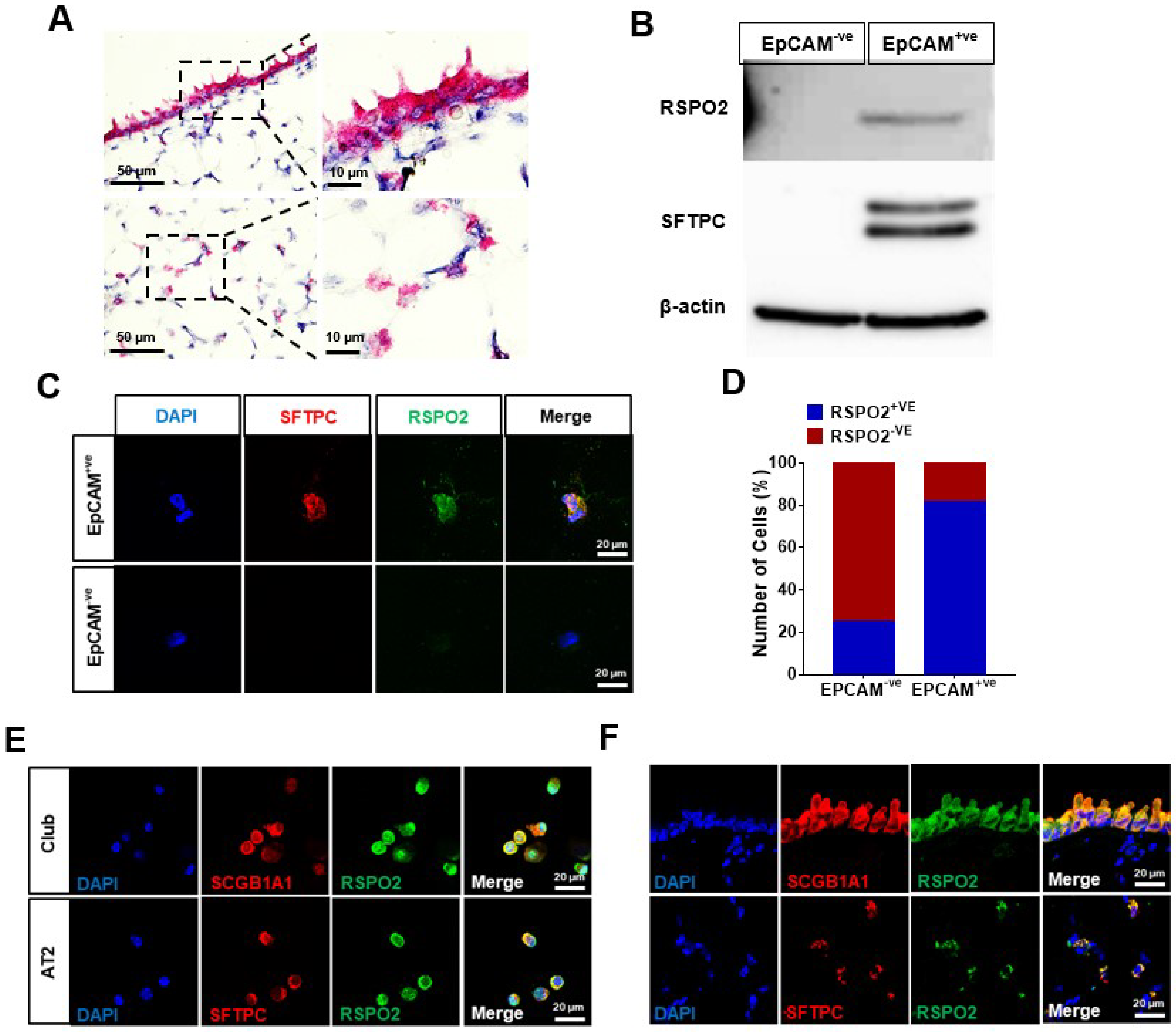
2.1. Rspo2 Is Expressed in Bronchial and Alveolar Epithelium
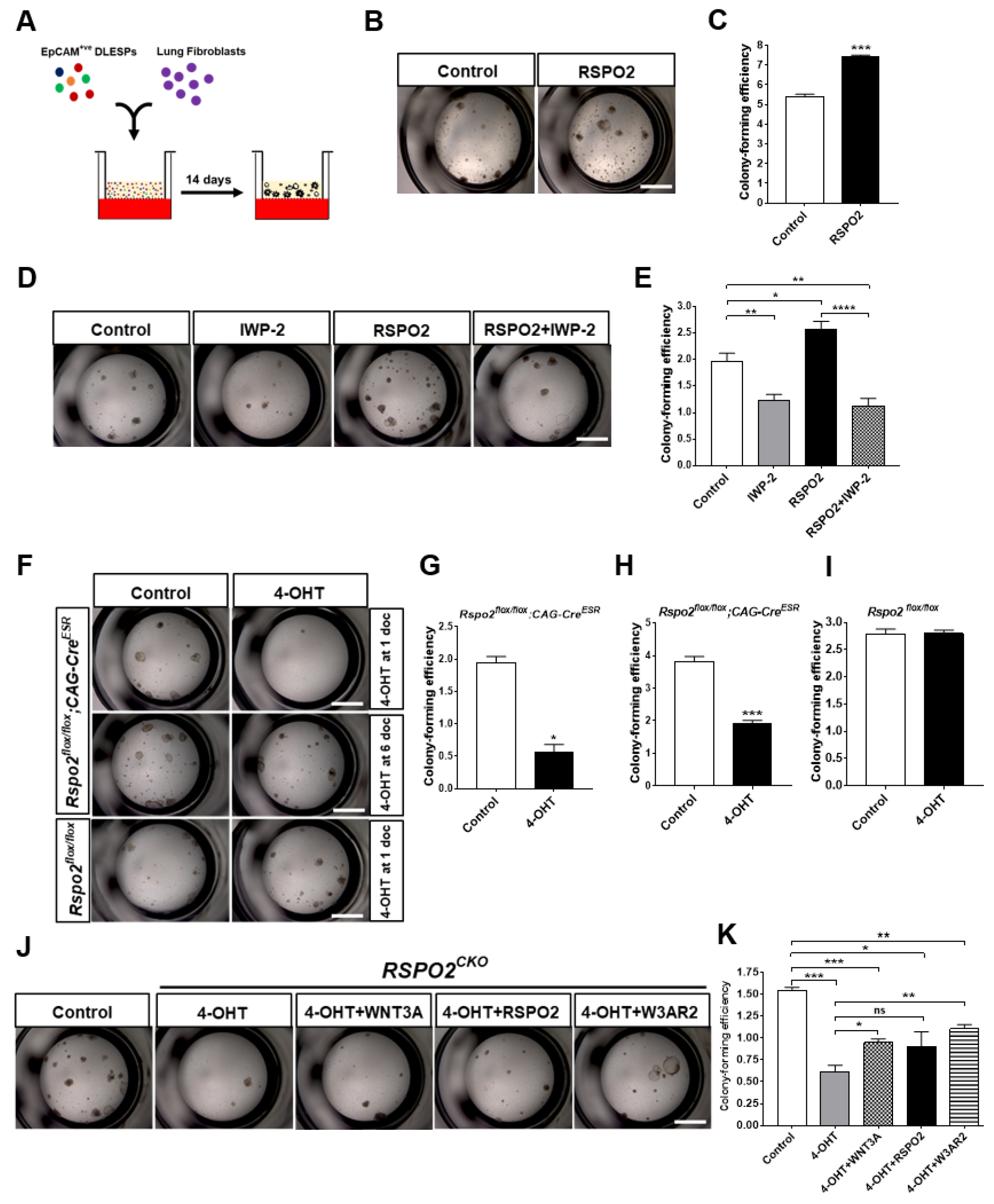
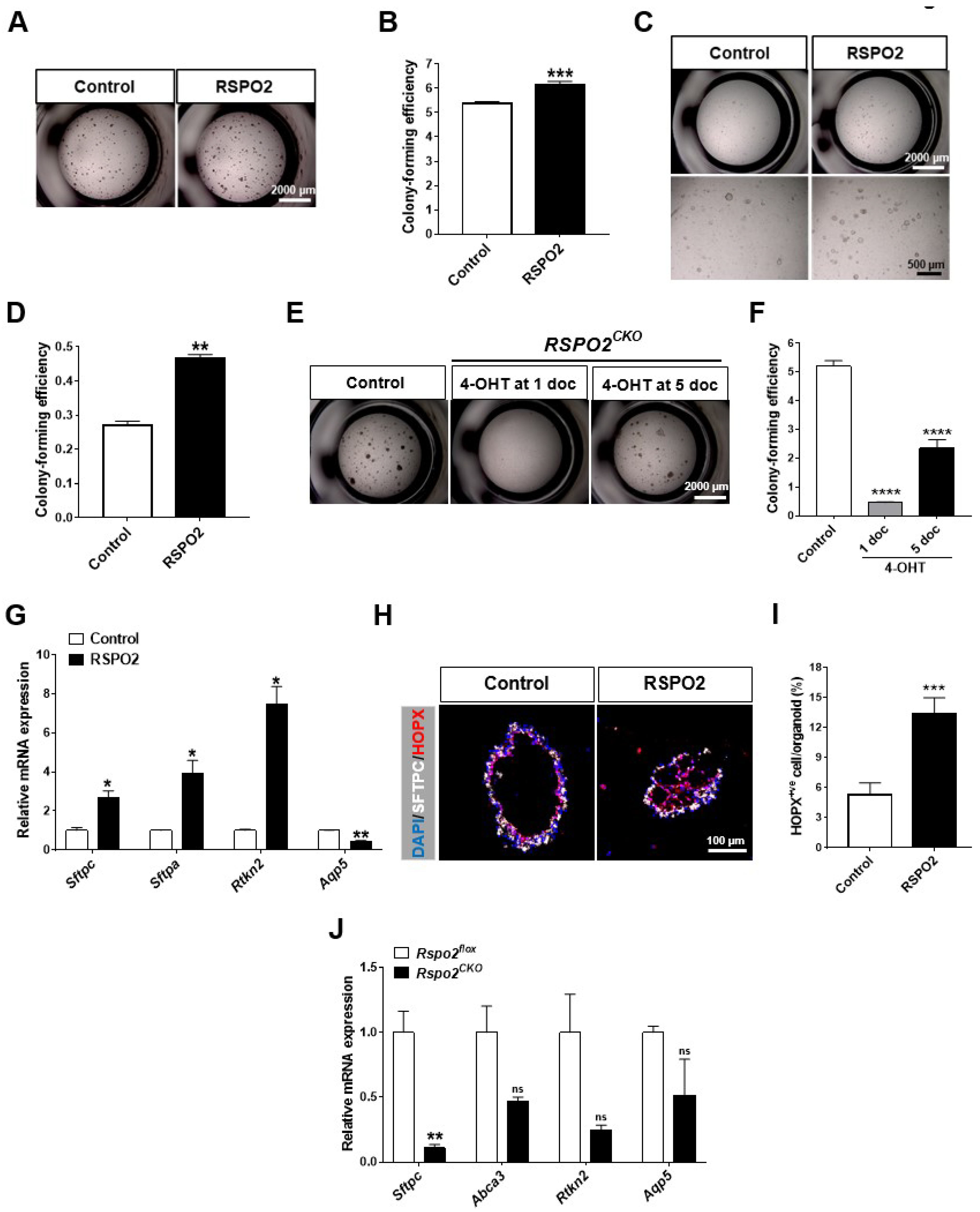
2.2. RSPO2 Is a Key Autocrine Factor Required for the Colony Formation of DLESPs
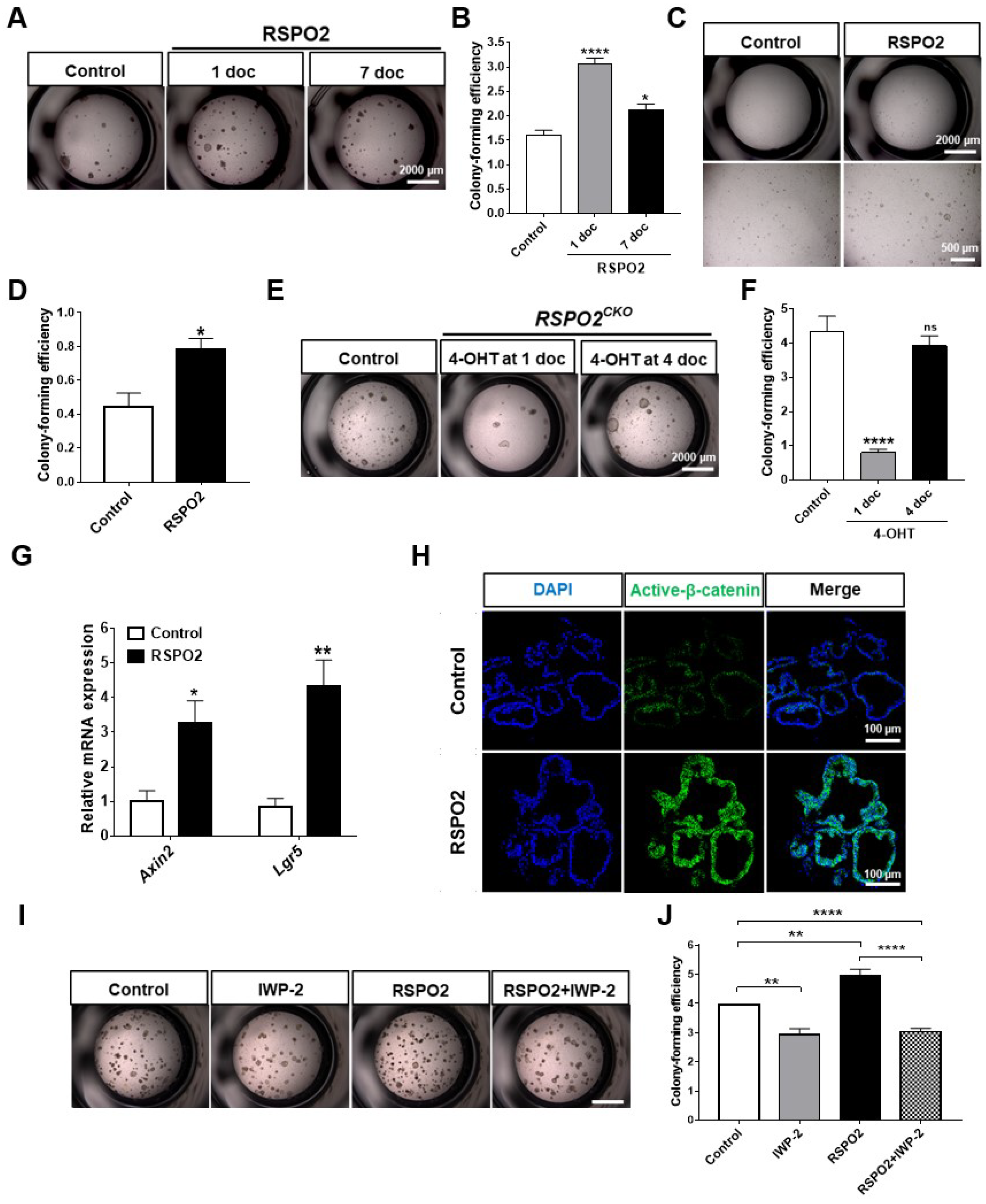
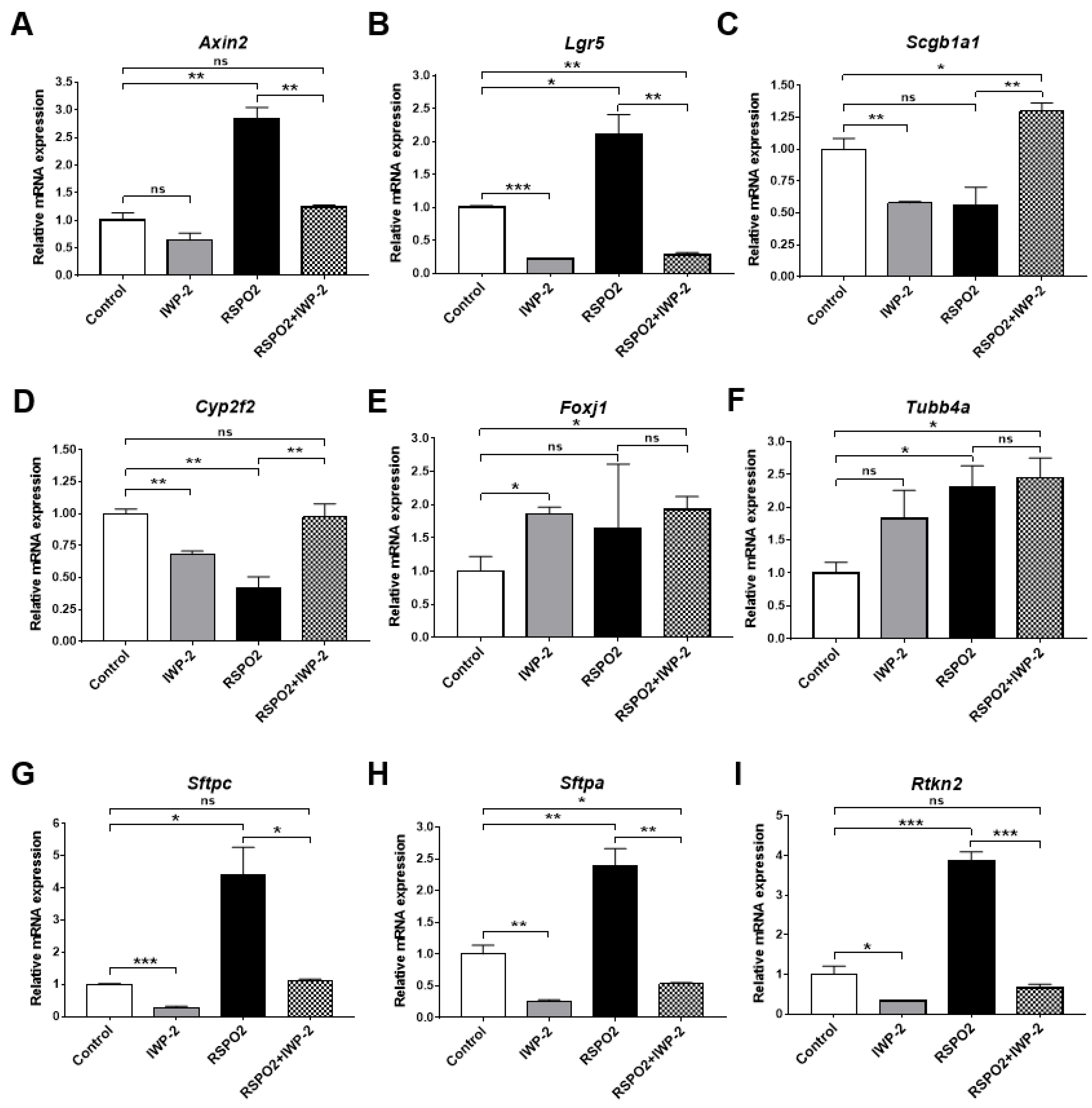
2.3. RSPO2 Supports Colony Formation of Club Cells through the Activation of Endogenous WNT/β-Catenin Signaling
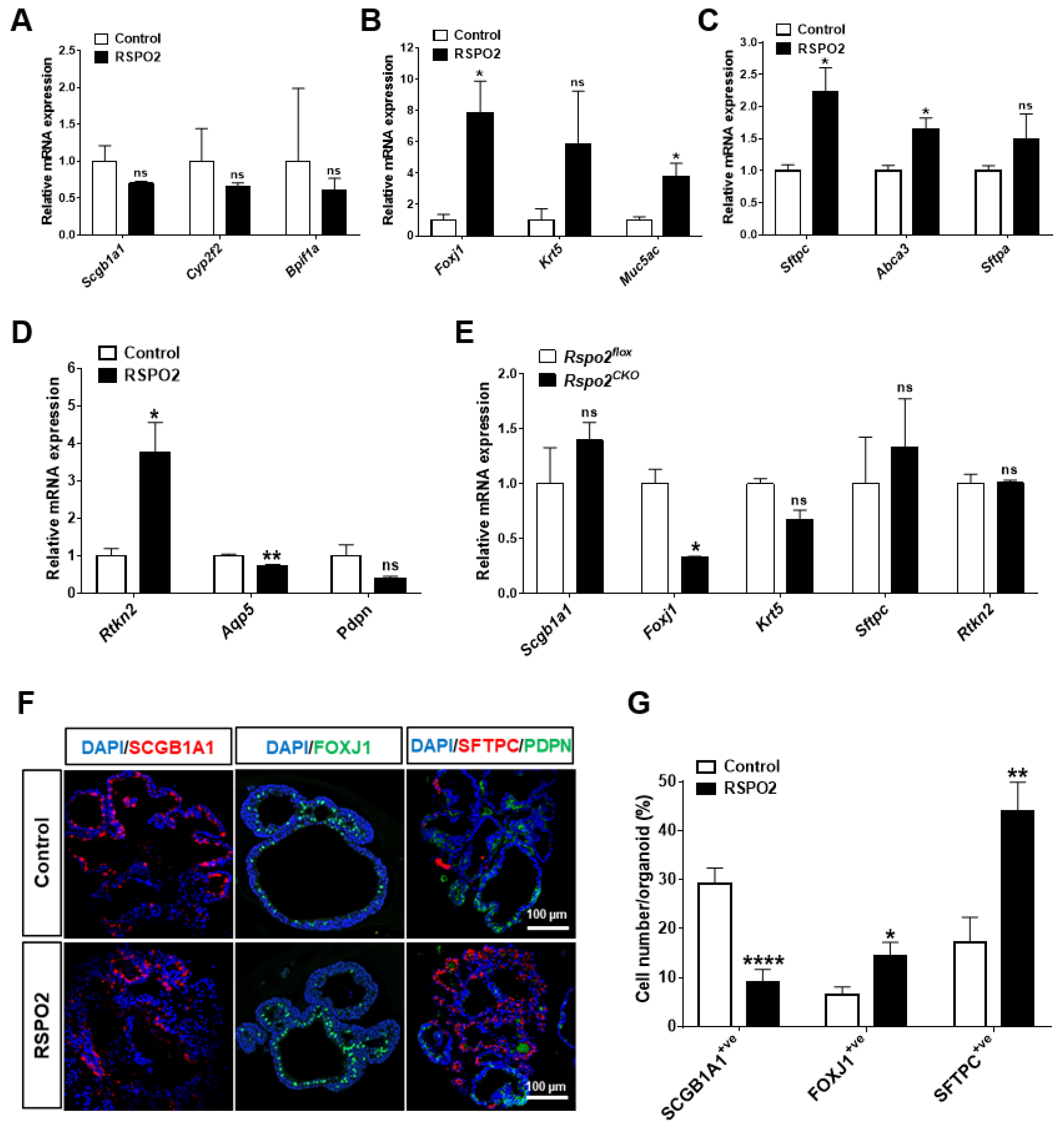
2.4. RSPO2 Promotes the Differentiation of Club Cells into Multiple Epithelial Lineages
2.5. RSPO2 Promotes the Differentiation of Club Cells into Alveolar Lineages in an Endogenous WNT/β-Catenin-Activation Manner
2.6. Rspo2 Is Essential for the Colony Formation and Differentiation of AT2 Cells
2.7. Loss of the Rspo2 gene Impairs Airway Regeneration after Naphthalene-Induced Lung Injury
3. Discussion
3.1. Rspo2 Expression in the Adult Lung Stem/Progenitor Cells
3.2. Autocrine RSPO2 Supports Colony Formation of Lung Stem/Progenitor Cells
3.3. Diverse Regulation of Differentiation by RSPO2 in the Lung Epithelial Progenitor/Stem Cells
3.4. Is RSPO2 a Paracrine Factor for Other Types of Lung Cells?
4. Materials and Methods
4.1. Animals
4.2. Gene Targeting for Conditional Rspo2 Inactivation
4.3. In Vivo Naphthalene-Induced Lung Injury
4.4. Isolation of Primary Lung Stem/Progenitor Cells and Fibroblasts
4.5. Ex Vivo 3D Organoid Culture
4.6. Sample Preparation for Histological and Immunological Staining
4.7. Immunofluorescence Staining, In Situ RNA Hybridization, and Histological Staining
4.8. Western Blot Analysis
4.9. Quantitative PCR (qRT-PCR) Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Raslan, A.A.; Yoon, J.K. WNT Signaling in Lung Repair and Regeneration. Mol. Cells 2020, 43, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Wansleeben, C.; Barkauskas, C.E.; Rock, J.R.; Hogan, B.L. Stem cells of the adult lung: Their development and role in homeostasis, regeneration, and disease. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 131–148. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Rawlins, E.L. Developmental mechanisms and adult stem cells for therapeutic lung regeneration. Dev. Biol. 2018, 433, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Hogan, B.L.; Barkauskas, C.E.; Chapman, H.A.; Epstein, J.A.; Jain, R.; Hsia, C.C.; Niklason, L.; Calle, E.; Le, A.; Randell, S.H.; et al. Repair and regeneration of the respiratory system: Complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 2014, 15, 123–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, T.J.; Anderson, P.J.; Rotti, P.G.; Tyler, S.R.; Crooke, A.K.; Choi, S.H.; Montoro, D.T.; Silverman, C.L.; Shahin, W.; Zhao, R.; et al. Submucosal Gland Myoepithelial Cells Are Reserve Stem Cells That Can Regenerate Mouse Tracheal Epithelium. Cell Stem Cell 2018, 22, 653–667.E5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rawlins, E.L.; Okubo, T.; Xue, Y.; Brass, D.M.; Auten, R.L.; Hasegawa, H.; Wang, F.; Hogan, B.L. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell 2009, 4, 525–534. [Google Scholar] [CrossRef] [Green Version]
- Rock, J.R.; Onaitis, M.W.; Rawlins, E.L.; Lu, Y.; Clark, C.P.; Xue, Y.; Randell, S.H.; Hogan, B.L. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. USA 2009, 106, 12771–12775. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Fine, A. Stem Cells in Lung Injury and Repair. Am. J. Pathol. 2016, 186, 2544–2550. [Google Scholar] [CrossRef] [Green Version]
- Tata, P.R.; Mou, H.; Pardo-Saganta, A.; Zhao, R.; Prabhu, M.; Law, B.M.; Vinarsky, V.; Cho, J.L.; Breton, S.; Sahay, A.; et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature 2013, 503, 218–223. [Google Scholar] [CrossRef] [Green Version]
- Guha, A.; Deshpande, A.; Jain, A.; Sebastiani, P.; Cardoso, W.V. Uroplakin 3a+ Cells Are a Distinctive Population of Epithelial Progenitors that Contribute to Airway Maintenance and Post-injury Repair. Cell Rep. 2017, 19, 246–254. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.T.; Yin, W.; Nakamichi, Y.; Panza, P.; Grohmann, B.; Buettner, C.; Guenther, S.; Ruppert, C.; Kobayashi, Y.; Guenther, A.; et al. WNT/RYK signaling restricts goblet cell differentiation during lung development and repair. Proc. Natl. Acad. Sci. USA 2019, 116, 25697–25706. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Soh, B.S.; Yin, L.; Hu, G.; Chen, Q.; Choi, H.; Han, J.; Chow, V.T.; Chen, J. Differentiation of Club Cells to Alveolar Epithelial Cells In Vitro. Sci. Rep. 2017, 7, 41661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olajuyin, A.M.; Zhang, X.; Ji, H.L. Alveolar type 2 progenitor cells for lung injury repair. Cell Death Discov. 2019, 5, 63. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Liu, K.; Cui, G.; Huang, X.; Yao, S.; Guo, W.; Qin, Z.; Li, Y.; Yang, R.; Pu, W.; et al. Lung regeneration by multipotent stem cells residing at the bronchioalveolar-duct junction. Nat. Genet. 2019, 51, 728–738. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.F.; Jackson, E.L.; Woolfenden, A.E.; Lawrence, S.; Babar, I.; Vogel, S.; Crowley, D.; Bronson, R.T.; Jacks, T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 2005, 121, 823–835. [Google Scholar] [CrossRef] [Green Version]
- Desai, T.J.; Brownfield, D.G.; Krasnow, M.A. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature 2014, 507, 190–194. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Tammela, T.; Hofree, M.; Choi, J.; Marjanovic, N.D.; Han, S.; Canner, D.; Wu, K.; Paschini, M.; Bhang, D.H.; et al. Anatomically and Functionally Distinct Lung Mesenchymal Populations Marked by Lgr5 and Lgr6. Cell 2017, 170, 1149–1163.E12. [Google Scholar] [CrossRef] [Green Version]
- Volckaert, T.; Yuan, T.; Chao, C.M.; Bell, H.; Sitaula, A.; Szimmtenings, L.; El Agha, E.; Chanda, D.; Majka, S.; Bellusci, S.; et al. Fgf10-Hippo Epithelial-Mesenchymal Crosstalk Maintains and Recruits Lung Basal Stem Cells. Dev. Cell 2017, 43, 48–59.E5. [Google Scholar] [CrossRef] [Green Version]
- Zepp, J.A.; Zacharias, W.J.; Frank, D.B.; Cavanaugh, C.A.; Zhou, S.; Morley, M.P.; Morrisey, E.E. Distinct Mesenchymal Lineages and Niches Promote Epithelial Self-Renewal and Myofibrogenesis in the Lung. Cell 2017, 170, 1134–1148.E10. [Google Scholar] [CrossRef] [Green Version]
- Fang, S.; Zhang, S.; Dai, H.; Hu, X.; Li, C.; Xing, Y. The role of pulmonary mesenchymal cells in airway epithelium regeneration during injury repair. Stem Cell Res. Ther. 2019, 10, 366. [Google Scholar] [CrossRef] [Green Version]
- Clevers, H.; Loh, K.M.; Nusse, R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 2014, 346, 1248012. [Google Scholar] [CrossRef] [PubMed]
- Pongracz, J.E.; Stockley, R.A. Wnt signalling in lung development and diseases. Respir. Res. 2006, 7, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussain, M.; Xu, C.; Lu, M.; Wu, X.; Tang, L.; Wu, X. Wnt/β-catenin signaling links embryonic lung development and asthmatic airway remodeling. Biochim. Et Biophys. Acta (BBA)—Mol. Basis Dis. 2017, 1863, 3226–3242. [Google Scholar] [CrossRef] [PubMed]
- Volckaert, T.; Dill, E.; Campbell, A.; Tiozzo, C.; Majka, S.; Bellusci, S.; De Langhe, S.P. Parabronchial smooth muscle constitutes an airway epithelial stem cell niche in the mouse lung after injury. J. Clin. Investig. 2011, 121, 4409–4419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volckaert, T.; Campbell, A.; De Langhe, S. c-Myc regulates proliferation and Fgf10 expression in airway smooth muscle after airway epithelial injury in mouse. PLoS ONE 2013, 8, e71426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zacharias, W.J.; Frank, D.B.; Zepp, J.A.; Morley, M.P.; Alkhaleel, F.A.; Kong, J.; Zhou, S.; Cantu, E.; Morrisey, E.E. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 2018, 555, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Nabhan, A.N.; Brownfield, D.G.; Harbury, P.B.; Krasnow, M.A.; Desai, T.J. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 2018, 359, 1118–1123. [Google Scholar] [CrossRef] [Green Version]
- Kamata, T.; Katsube, K.; Michikawa, M.; Yamada, M.; Takada, S.; Mizusawa, H. R-spondin, a novel gene with thrombospondin type 1 domain, was expressed in the dorsal neural tube and affected in Wnts mutants. Biochim. Biophys. Acta 2004, 1676, 51–62. [Google Scholar] [CrossRef]
- Kim, K.A.; Zhao, J.; Andarmani, S.; Kakitani, M.; Oshima, T.; Binnerts, M.E.; Abo, A.; Tomizuka, K.; Funk, W.D. R-Spondin proteins: A novel link to beta-catenin activation. Cell Cycle 2006, 5, 23–26. [Google Scholar] [CrossRef]
- Jin, Y.R.; Yoon, J.K. The R-spondin family of proteins: Emerging regulators of WNT signaling. Int. J. Biochem. Cell Biol. 2012, 44, 2278–2287. [Google Scholar] [CrossRef] [Green Version]
- Raslan, A.A.; Yoon, J.K. R-spondins: Multi-mode WNT signaling regulators in adult stem cells. Int. J. Biochem. Cell Biol. 2019, 106, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Carmon, K.S.; Gong, X.; Lin, Q.; Thomas, A.; Liu, Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 11452–11457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, H.X.; Jiang, X.; Cong, F. Control of Wnt Receptor Turnover by R-spondin-ZNRF3/RNF43 Signaling Module and Its Dysregulation in Cancer. Cancers 2016, 8, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebensohn, A.M.; Rohatgi, R. R-spondins can potentiate WNT signaling without LGRs. Elife 2018, 7, e33126. [Google Scholar] [CrossRef]
- Szenker-Ravi, E.; Altunoglu, U.; Leushacke, M.; Bosso-Lefevre, C.; Khatoo, M.; Thi Tran, H.; Naert, T.; Noelanders, R.; Hajamohideen, A.; Beneteau, C.; et al. RSPO2 inhibition of RNF43 and ZNRF3 governs limb development independently of LGR4/5/6. Nature 2018, 557, 564–569. [Google Scholar] [CrossRef]
- Glinka, A.; Dolde, C.; Kirsch, N.; Huang, Y.L.; Kazanskaya, O.; Ingelfinger, D.; Boutros, M.; Cruciat, C.M.; Niehrs, C. LGR4 and LGR5 are R-spondin receptors mediating Wnt/beta-catenin and Wnt/PCP signalling. EMBO Rep. 2011, 12, 1055–1061. [Google Scholar] [CrossRef] [Green Version]
- Ohkawara, B.; Glinka, A.; Niehrs, C. Rspo3 binds syndecan 4 and induces Wnt/PCP signaling via clathrin-mediated endocytosis to promote morphogenesis. Dev. Cell 2011, 20, 303–314. [Google Scholar] [CrossRef] [Green Version]
- Bell, S.M.; Schreiner, C.M.; Wert, S.E.; Mucenski, M.L.; Scott, W.J.; Whitsett, J.A. R-spondin 2 is required for normal laryngeal-tracheal, lung and limb morphogenesis. Development 2008, 135, 1049–1058. [Google Scholar] [CrossRef] [Green Version]
- Nam, J.S.; Turcotte, T.J.; Yoon, J.K. Dynamic expression of R-spondin family genes in mouse development. Gene Expr. Patterns 2007, 7, 306–312. [Google Scholar] [CrossRef]
- Munguia-Reyes, A.; Balderas-Martinez, Y.I.; Becerril, C.; Checa, M.; Ramirez, R.; Ortiz, B.; Melendez-Zajgla, J.; Pardo, A.; Selman, M. R-Spondin-2 Is Upregulated in Idiopathic Pulmonary Fibrosis and Affects Fibroblast Behavior. Am. J. Respir. Cell Mol. Biol. 2018, 59, 65–76. [Google Scholar] [CrossRef]
- Wu, L.; Zhang, W.; Qian, J.; Wu, J.; Jiang, L.; Ling, C. R-spondin family members as novel biomarkers and prognostic factors in lung cancer. Oncol. Lett. 2019, 18, 4008–4015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, S.R.; Costa, M.; Pastore, C.F.; Zhao, G.; Weiner, A.I.; Adams, S.; Palashikar, G.; Quansah, K.; Hankenson, K.; Herbert, D.R.; et al. R-spondin 2 mediates neutrophil egress into the alveolar space through increased lung permeability. BMC Res. Notes 2020, 13, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, J.S.; Park, E.; Turcotte, T.J.; Palencia, S.; Zhan, X.; Lee, J.; Yun, K.; Funk, W.D.; Yoon, J.K. Mouse R-spondin2 is required for apical ectodermal ridge maintenance in the hindlimb. Dev. Biol. 2007, 311, 124–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Seidl, C.; Sun, R.; Glinka, A.; Niehrs, C. R-spondins are BMP receptor antagonists in Xenopus early embryonic development. Nat. Commun. 2020, 11, 5570. [Google Scholar] [CrossRef] [PubMed]
- Kotton, D.N.; Morrisey, E.E. Lung regeneration: Mechanisms, applications and emerging stem cell populations. Nat. Med. 2014, 20, 822–832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yildirim, A.Ö.; Veith, M.; Rausch, T.; Müller, B.; Kilb, P.; Van Winkle, L.S.; Fehrenbach, H. Keratinocyte growth factor protects against Clara cell injury induced by naphthalene. Eur. Respir. J. 2008, 32, 694. [Google Scholar] [CrossRef] [Green Version]
- Flozak, A.S.; Lam, A.P.; Russell, S.; Jain, M.; Peled, O.N.; Sheppard, K.A.; Beri, R.; Mutlu, G.M.; Budinger, G.R.; Gottardi, C.J. Beta-catenin/T-cell factor signaling is activated during lung injury and promotes the survival and migration of alveolar epithelial cells. J. Biol. Chem. 2010, 285, 3157–3167. [Google Scholar] [CrossRef] [Green Version]
- Tanjore, H.; Degryse, A.L.; Crossno, P.F.; Xu, X.C.; McConaha, M.E.; Jones, B.R.; Polosukhin, V.V.; Bryant, A.J.; Cheng, D.S.; Newcomb, D.C.; et al. Beta-catenin in the alveolar epithelium protects from lung fibrosis after intratracheal bleomycin. Am. J. Respir. Crit. Care Med. 2013, 187, 630–639. [Google Scholar] [CrossRef] [Green Version]
- Frank, D.B.; Peng, T.; Zepp, J.A.; Snitow, M.; Vincent, T.L.; Penkala, I.J.; Cui, Z.; Herriges, M.J.; Morley, M.P.; Zhou, S.; et al. Emergence of a Wave of Wnt Signaling that Regulates Lung Alveologenesis by Controlling Epithelial Self-Renewal and Differentiation. Cell Rep. 2016, 17, 2312–2325. [Google Scholar] [CrossRef] [Green Version]
- Andersson-Sjoland, A.; Karlsson, J.C.; Rydell-Tormanen, K. ROS-induced endothelial stress contributes to pulmonary fibrosis through pericytes and Wnt signaling. Lab. Investig. 2016, 96, 206–217. [Google Scholar] [CrossRef]
- Majidinia, M.; Aghazadeh, J.; Jahanban-Esfahlani, R.; Yousefi, B. The roles of Wnt/beta-catenin pathway in tissue development and regenerative medicine. J. Cell Physiol. 2018, 233, 5598–5612. [Google Scholar] [CrossRef] [PubMed]
- Zemke, A.C.; Teisanu, R.M.; Giangreco, A.; Drake, J.A.; Brockway, B.L.; Reynolds, S.D.; Stripp, B.R. Beta-Catenin is not necessary for maintenance or repair of the bronchiolar epithelium. Am. J. Respir. Cell Mol. Biol. 2009, 41, 535–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Ng-Blichfeldt, J.P.; Ota, C.; Ciminieri, C.; Ren, W.; Hiemstra, P.S.; Stolk, J.; Gosens, R.; Konigshoff, M. Wnt/beta-catenin signaling is critical for regenerative potential of distal lung epithelial progenitor cells in homeostasis and emphysema. Stem Cells 2020, 38, 1467–1478. [Google Scholar] [CrossRef] [PubMed]
- Skronska-Wasek, W.; Mutze, K.; Königshoff, M. R-spondins induce canonical Wnt signaling in lung epithelial cells. Eur. Respir. J. 2014, 44, P1838. [Google Scholar]
- Zhao, R.; Fallon, T.R.; Saladi, S.V.; Pardo-Saganta, A.; Villoria, J.; Mou, H.; Vinarsky, V.; Gonzalez-Celeiro, M.; Nunna, N.; Hariri, L.P.; et al. Yap tunes airway epithelial size and architecture by regulating the identity, maintenance, and self-renewal of stem cells. Dev. Cell 2014, 30, 151–165. [Google Scholar] [CrossRef] [Green Version]
- Sinha, M.; Lowell, C.A. Isolation of Highly Pure Primary Mouse Alveolar Epithelial Type II Cells by Flow Cytometric Cell Sorting. Bio. Protoc. 2016, 6, e2013. [Google Scholar] [CrossRef] [Green Version]
- Bertoncello, I.; McQualter, J. Isolation and clonal assay of adult lung epithelial stem/progenitor cells. Curr. Protoc. Stem Cell Biol. 2011, 16, 2G.1. [Google Scholar] [CrossRef]
- Hegab, A.E.; Arai, D.; Gao, J.; Kuroda, A.; Yasuda, H.; Ishii, M.; Naoki, K.; Soejima, K.; Betsuyaku, T. Mimicking the niche of lung epithelial stem cells and characterization of several effectors of their in vitro behavior. Stem Cell Res. 2015, 15, 109–121. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Flanagan, J.; Su, N.; Wang, L.C.; Bui, S.; Nielson, A.; Wu, X.; Vo, H.T.; Ma, X.J.; Luo, Y. RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 2012, 14, 22–29. [Google Scholar] [CrossRef] [Green Version]
- Arlt, M.J.; Born, W.; Fuchs, B. Improved visualization of lung metastases at single cell resolution in mice by combined in-situ perfusion of lung tissue and X-Gal staining of lacZ-tagged tumor cells. J. Vis. Exp. 2012, 66, e4162. [Google Scholar] [CrossRef] [Green Version]
- Sah, J.P.; Hao, N.T.T.; Han, X.; Tran, T.T.T.; McCarthy, S.; Oh, Y.; Yoon, J.K. Ectonucleotide pyrophosphatase 2 (ENPP2) plays a crucial role in myogenic differentiation through the regulation by WNT/beta-Catenin signaling. Int. J. Biochem. Cell Biol. 2020, 118, 105661. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raslan, A.A.; Oh, Y.J.; Jin, Y.R.; Yoon, J.K. R-Spondin2, a Positive Canonical WNT Signaling Regulator, Controls the Expansion and Differentiation of Distal Lung Epithelial Stem/Progenitor Cells in Mice. Int. J. Mol. Sci. 2022, 23, 3089. https://doi.org/10.3390/ijms23063089
Raslan AA, Oh YJ, Jin YR, Yoon JK. R-Spondin2, a Positive Canonical WNT Signaling Regulator, Controls the Expansion and Differentiation of Distal Lung Epithelial Stem/Progenitor Cells in Mice. International Journal of Molecular Sciences. 2022; 23(6):3089. https://doi.org/10.3390/ijms23063089
Chicago/Turabian StyleRaslan, Ahmed A., Youn Jeong Oh, Yong Ri Jin, and Jeong Kyo Yoon. 2022. "R-Spondin2, a Positive Canonical WNT Signaling Regulator, Controls the Expansion and Differentiation of Distal Lung Epithelial Stem/Progenitor Cells in Mice" International Journal of Molecular Sciences 23, no. 6: 3089. https://doi.org/10.3390/ijms23063089
APA StyleRaslan, A. A., Oh, Y. J., Jin, Y. R., & Yoon, J. K. (2022). R-Spondin2, a Positive Canonical WNT Signaling Regulator, Controls the Expansion and Differentiation of Distal Lung Epithelial Stem/Progenitor Cells in Mice. International Journal of Molecular Sciences, 23(6), 3089. https://doi.org/10.3390/ijms23063089





