Glycosylphosphatidylinositol Anchor Biosynthesis Pathway-Related Protein GPI7 Is Required for the Vegetative Growth and Pathogenicity of Colletotrichum graminicola
Abstract
:1. Introduction
2. Results
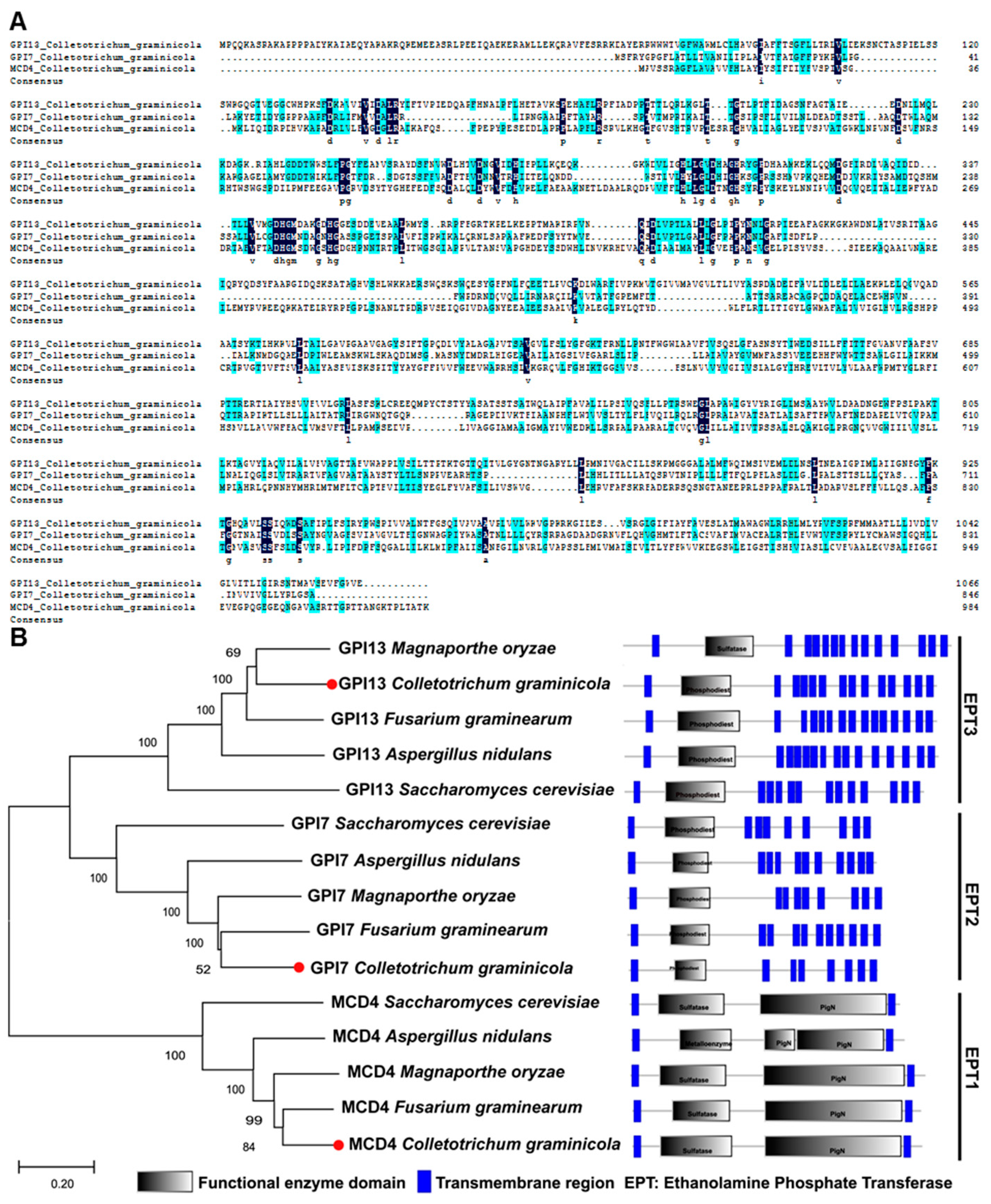
2.1. Identification of GPI Anchor Synthesis Pathway Proteins in C. graminicola
2.2. Phylogenetic Analysis of CgEPTs in C. graminicola
2.3. CgGPI7 Is Essential for Vegetative Growth and Conidia Development of C. graminicola
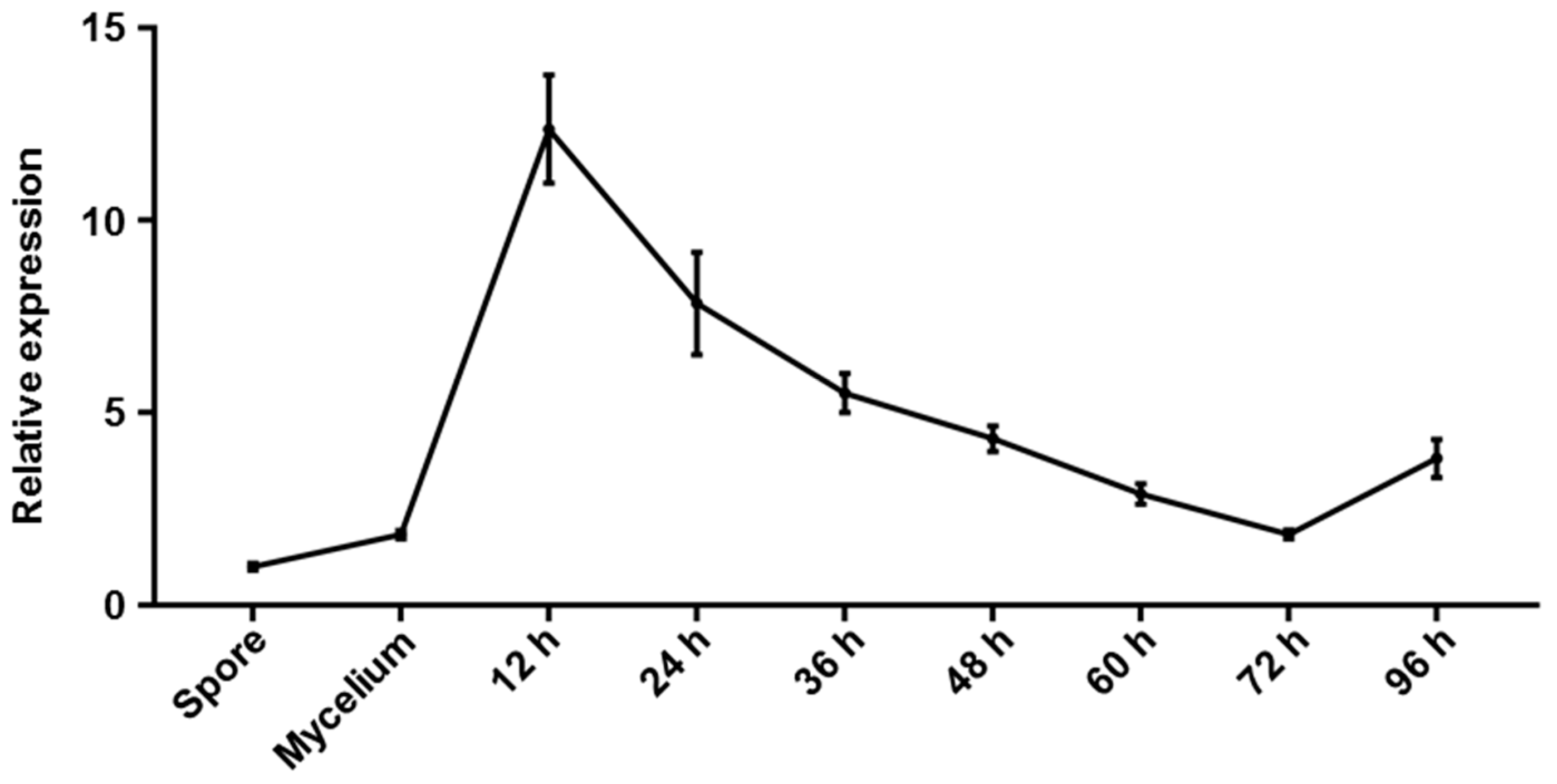
2.4. Gene Expression Pattern of CgGPI7 in C. graminicola
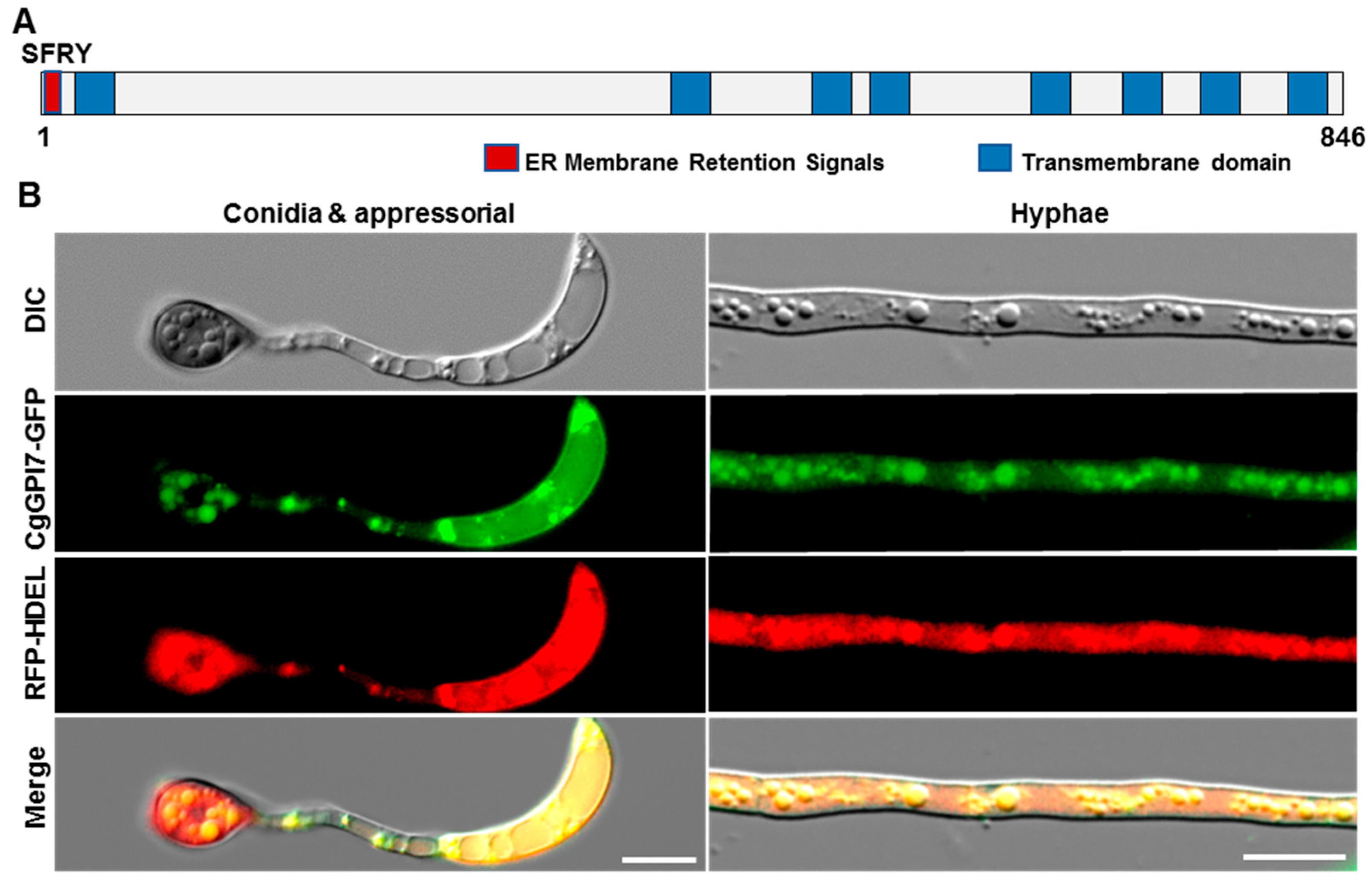
2.5. CgGPI7 Localized in the Endoplasmic Reticulum (ER)
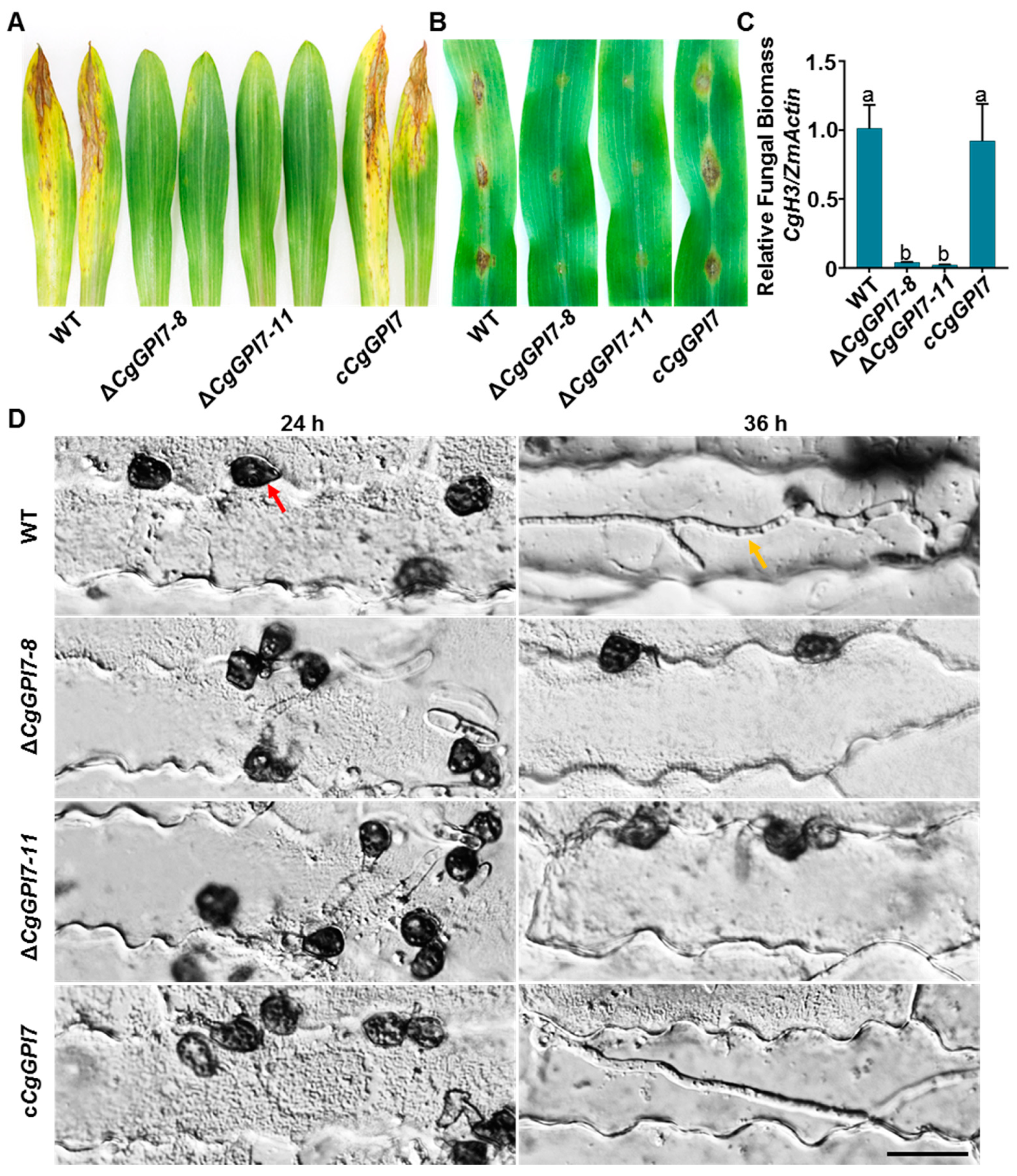
2.6. CgGPI7 Is Essential for Pathogenicity of C. graminicola
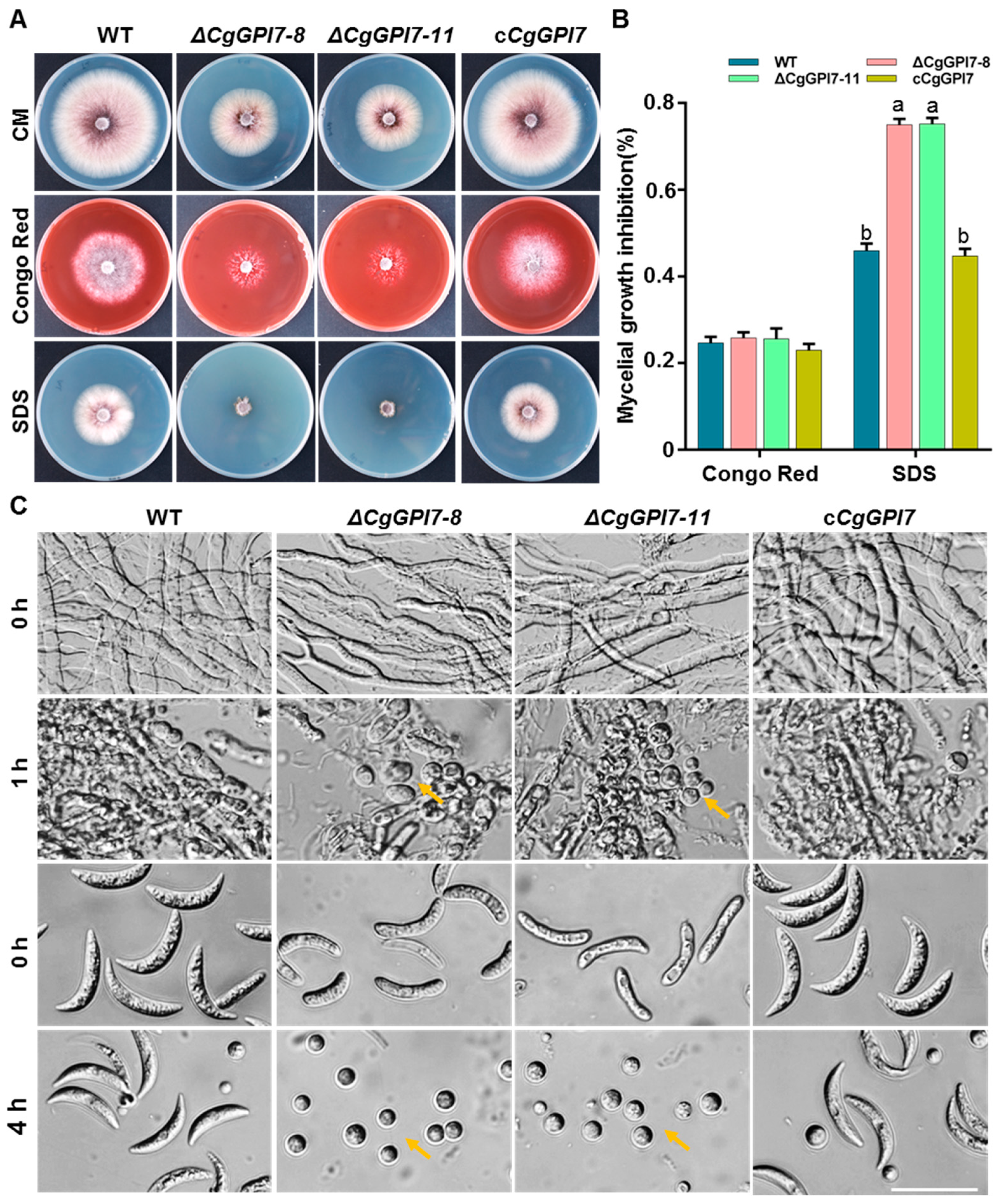
2.7. CgGPI7 Affects the Cell Wall Integrity of C. graminicola
2.8. Deletion of CgGPI7 Resulted in the Up-Regulation of Cell Wall Synthesis-Related Genes in C. graminicola
3. Discussion
4. Materials and Methods
4.1. Fungal Strains and Plant Culture Conditions
4.2. Bioinformatic and Phylogenetic Analysis
4.3. Gene Disruption and Complementation
4.4. Biological Phenotypic Analysis
4.5. Cell Wall Integrity Analysis
4.6. Subcellular Localization Analysis of CgGPI7
4.7. Pathogenicity Analysis
4.8. qRT-PCR Assays
4.9. Statistical Analysis Method
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferguson, M.A.; Kinoshita, T.; Hart, G.W. Glycosylphosphatidylinositol Anchors. In Essentials of Glycobiology; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2009; pp. 227–234. ISBN 9780879697709. [Google Scholar]
- Mayor, S.; Riezman, H. Sorting GPI-anchored proteins. Nat. Rev. Mol. Cell Biol. 2004, 5, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Pittet, M.; Conzelmann, A. Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 2007, 1771, 405–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebreton, S.; Zurzolo, C.; Paladino, S. Organization of GPI-anchored proteins at the cell surface and its physiopathological relevance. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 403–419. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Lee, C.; Sabharwal, P.; Zhang, M.; Meyers, C.L.F.; Sockanathan, S. GDE2 promotes neurogenesis by glycosylphosphatidylinositol-anchor cleavage of RECK. Science 2013, 339, 324–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murata, D.; Nomura, K.H.; Dejima, K.; Mizuguchi, S.; Kawasaki, N.; Matsuishi-Nakajima, Y.; Ito, S.; Gengyo-Ando, K.; Kage-Nakadai, E.; Mitani, S.; et al. GPI-anchor synthesis is indispensable for the germline development of the nematode Caenorhabditis elegans. Mol. Biol. Cell. 2012, 23, 982–995. [Google Scholar] [CrossRef] [PubMed]
- Inoue, N.; Murakami, Y.; Kinoshita, T. Molecular genetics of paroxysmal nocturnal hemoglobinuria. Int. J. Hematol. 2003, 77, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Harris, D.A.; Gorodinsky, A.; Lehmann, S.; Moulder, K.; Shyng, S.L. Cell biology of the prion protein. Curr. Top. Microbiol. Immunol. 1996, 207, 77–93. [Google Scholar] [CrossRef] [PubMed]
- Kelemen, L.E. The role of folate receptor alpha in cancer development, progression and treatment: Cause, consequence or innocent bystander? Int. J. Cancer 2006, 119, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Duggan, C.; Maguire, T.; McDermott, E.; O’Higgins, N.; Fennelly, J.J.; Duffy, M.J. Urokinase plasminogen activator and urokinase plasminogen activator receptor in breast cancer. Int. J. Cancer 1995, 61, 597–600. [Google Scholar] [CrossRef] [PubMed]
- Klis, F.M.; Sosinska, G.J.; de Groot, P.W.J.; Brul, S. Covalently linked cell wall proteins of Candida albicans and their role in fitness and virulence. FEMS Yeast Res. 2009, 9, 1013–1028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Xing, J.; Cai, X.; Hendy, A.; He, W.; Yang, J.; Huang, J.; Peng, Y.-L.; Ryder, L.; Chen, X.-L. GPI7-mediated glycosylphosphatidylinositol anchoring regulates appressorial penetration and immune evasion during infection of Magnaporthe oryzae. Environ. Microbiol. 2020, 22, 2581–2595. [Google Scholar] [CrossRef] [PubMed]
- Mann, P.A.; McLellan, C.A.; Koseoglu, S.; Si, Q.; Kuzmin, E.; Flattery, A.; Harris, G.; Sher, X.; Murgolo, N.; Wang, H.; et al. Chemical Genomics-Based Antifungal Drug Discovery: Targeting Glycosylphosphatidylinositol (GPI) Precursor Biosynthesis. ACS Infect. Dis. 2015, 1, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Newman, H.A.; Romeo, M.J.; Lewis, S.E.; Yan, B.C.; Orlean, P.; Levin, D.E. Gpi19, the Saccharomyces cerevisiae homologue of mammalian PIG-P, is a subunit of the initial enzyme for glycosylphosphatidylinositol anchor biosynthesis. Eukaryot. Cell 2005, 4, 1801–1807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leidich, S.D.; Kostova, Z.; Latek, R.R.; Costello, L.C.; Drapp, D.A.; Gray, W.; Fassler, J.S.; Orlean, P. Temperature-sensitive yeast GPI anchoring mutants gpi2 and gpi3 are defective in the synthesis of N-acetylglucosaminyl phosphatidylinositol. Cloning of the GPI2 gene. J. Biol. Chem. 1995, 270, 13029–13035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sobering, A.K.; Watanabe, R.; Romeo, M.J.; Yan, B.C.; Specht, C.A.; Orlean, P.; Riezman, H.; Levin, D.E. Yeast Ras regulates the complex that catalyzes the first step in GPI-anchor biosynthesis at the ER. Cell 2004, 117, 637–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, B.C.; Westfall, B.A.; Orlean, P. Ynl038wp (Gpi15p) is the Saccharomyces cerevisiae homologue of human Pig-Hp and participates in the first step in glycosylphosphatidylinositol assembly. Yeast 2001, 18, 1383–1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umemura, M.; Okamoto, M.; Nakayama, K.; Sagane, K.; Tsukahara, K.; Hata, K.; Jigami, Y. GWT1 gene is required for inositol acylation of glycosylphosphatidylinositol anchors in yeast. J. Biol. Chem. 2003, 278, 23639–23647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, R.; Ohishi, K.; Maeda, Y.; Nakamura, N.; Kinoshita, T. Mammalian PIG-L and its yeast homologue Gpi12p are N-acetylglucosaminylphosphatidylinositol de-N-acetylases essential in glycosylphosphatidylinositol biosynthesis. Biochem. J. 1999, 339, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Benachour, A.; Sipos, G.; Flury, I.; Reggiori, F.; Canivenc-Gansel, E.; Vionnet, C.; Conzelmann, A.; Benghezal, M. Deletion of GPI7, a yeast gene required for addition of a side chain to the glycosylphosphatidylinositol (GPI) core structure, affects GPI protein transport, remodeling, and cell wall integrity. J. Biol. Chem. 1999, 274, 15251–15261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grimme, S.J.; Westfall, B.A.; Wiedman, J.M.; Taron, C.H.; Orlean, P. The essential Smp3 protein is required for addition of the side-branching fourth mannose during assembly of yeast glycosylphosphatidylinositols. J. Biol. Chem. 2001, 276, 27731–27739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taron, C.H.; Wiedman, J.M.; Grimme, S.J.; Orlean, P. Glycosylphosphatidylinositol biosynthesis defects in Gpi11p- and Gpi13p-deficient yeast suggest a branched pathway and implicate gpi13p in phosphoethanolamine transfer to the third mannose. Mol. Biol. Cell 2000, 11, 1611–1630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaynor, E.C.; Mondésert, G.; Grimme, S.J.; Reed, S.I.; Orlean, P.; Emr, S.D. MCD4 encodes a conserved endoplasmic reticulum membrane protein essential for glycosylphosphatidylinositol anchor synthesis in yeast. Mol. Biol. Cell 1999, 10, 627–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeda, Y.; Watanabe, R.; Harris, C.L.; Hong, Y.; Ohishi, K.; Kinoshita, K.; Kinoshita, T. PIG-M transfers the first mannose to glycosylphosphatidylinositol on the lumenal side of the ER. EMBO J. 2001, 20, 250–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, J.Y.; Hong, Y.; Ashida, H.; Shishioh, N.; Murakami, Y.; Morita, Y.S.; Maeda, Y.; Kinoshita, T. PIG-V involved in transferring the second mannose in glycosylphosphatidylinositol. J. Biol. Chem. 2005, 280, 9489–9497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sütterlin, C.; Escribano, M.V.; Gerold, P.; Maeda, Y.; Mazon, M.J.; Kinoshita, T.; Schwarz, R.T.; Riezman, H. Saccharomyces cerevisiae GPI10, the functional homologue of human PIG-B, is required for glycosylphosphatidylinositol-anchor synthesis. Biochem. J. 1998, 332 Pt 1, 153–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nuoffer, C.; Horvath, A.; Riezman, H. Analysis of the sequence requirements for glycosylphosphatidylinositol anchoring of Saccharomyces cerevisiae Gas1 protein. J. Biol. Chem. 1993, 268, 10558–10563. [Google Scholar] [CrossRef]
- Moran, P.; Caras, I.W. Fusion of sequence elements from non-anchored proteins to generate a fully functional signal for glycophosphatidylinositol membrane anchor attachment. J. Cell Biol. 1991, 115, 1595–1600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kinoshita, T.; Fujita, M. Biosynthesis of GPI-anchored proteins: Special emphasis on GPI lipid remodeling. J. Lipid Res. 2016, 57, 6–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richard, M.L.; Plaine, A. Comprehensive analysis of glycosylphosphatidylinositol-anchored proteins in Candida albicans. Eukaryot. Cell 2007, 6, 119–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisenhaber, B.; Maurer-Stroh, S.; Novatchkova, M.; Schneider, G.; Eisenhaber, F. Enzymes and auxiliary factors for GPI lipid anchor biosynthesis and post-translational transfer to proteins. Bioessays 2003, 25, 367–385. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, H.; Du, T.; Zhou, H.; Wilson, I.B.H.; Yang, J.; Latgé, J.-P.; Jin, C. Aspergillus fumigatus phosphoethanolamine transferase gene gpi7 is required for proper transportation of the cell wall GPI-anchored proteins and polarized growth. Sci. Rep. 2019, 9, 5857. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhou, H.; Luo, Y.; Ouyang, H.; Hu, H.; Jin, C. Glycosylphosphatidylinositol (GPI) anchor is required in Aspergillus fumigatus for morphogenesis and virulence. Mol. Microbiol. 2007, 64, 1014–1027. [Google Scholar] [CrossRef] [PubMed]
- de Jesus Miranda, V.; Porto, W.F.; da Rocha Fernandes, G.; Pogue, R.; Nolasco, D.O.; Araujo, A.C.G.; Cota, L.V.; de Freitas, C.G.; Dias, S.C.; Franco, O.L. Comparative transcriptomic analysis indicates genes associated with local and systemic resistance to Colletotrichum graminicola in maize. Sci. Rep. 2017, 7, 2483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balmer, D.; de Papajewski, D.V.; Planchamp, C.; Glauser, G.; Mauch-Mani, B. Induced resistance in maize is based on organ-specific defence responses. Plant J. 2013, 74, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Oliveira-Garcia, E.; Deising, H.B. The Glycosylphosphatidylinositol Anchor Biosynthesis Genes GPI12, GAA1, and GPI8 Are Essential for Cell-Wall Integrity and Pathogenicity of the Maize Anthracnose Fungus Colletotrichum graminicola. Mol. Plant. Microbe. Interact. 2016, 29, 889–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richard, M.; de Groot, P.; Courtin, O.; Poulain, D.; Klis, F.; Gaillardin, C. GPI7 affects cell-wall protein anchorage in Saccharomyces cerevisiae and Candida albicans. Microbiology 2002, 148, 2125–2133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tada, R.; Aimanianda, V. The Biosynthetic Pathway of 1,6-Branched β-(1,3)-Glucan, the Biopolymer That Constitutes the Core Structure of Fungal Cell Walls. Trends Glycosci. Glycotechnol. 2020, 32, E119–E125. [Google Scholar] [CrossRef]
- Farkaš, V. Fungal cell walls: Their structure, biosynthesis and biotechnological aspects. Acta Biotechnol. 1990, 10, 225–238. [Google Scholar] [CrossRef]
- Hong, Y.; Maeda, Y.; Watanabe, R.; Ohishi, K.; Mishkind, M.; Riezman, H.; Kinoshita, T. Pig-n, a mammalian homologue of yeast Mcd4p, is involved in transferring phosphoethanolamine to the first mannose of the glycosylphosphatidylinositol. J. Biol. Chem. 1999, 274, 35099–35106. [Google Scholar] [CrossRef] [Green Version]
- Imhof, I.; Flury, I.; Vionnet, C.; Roubaty, C.; Egger, D.; Conzelmann, A. Glycosylphosphatidylinositol (GPI) proteins of Saccharomyces cerevisiae contain ethanolamine phosphate groups on the alpha1,4-linked mannose of the GPI anchor. J. Biol. Chem. 2004, 279, 19614–19627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, Y.; Maeda, Y.; Watanabe, R.; Inoue, N.; Ohishi, K.; Kinoshita, T. Requirement of PIG-F and PIG-O for transferring phosphoethanolamine to the third mannose in glycosylphosphatidylinositol. J. Biol. Chem. 2000, 275, 20911–20919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benghezal, M.; Lipke, P.N.; Conzelmann, A. Identification of six complementation classes involved in the biosynthesis of glycosylphosphatidylinositol anchors in Saccharomyces cerevisiae. J. Cell Biol. 1995, 130, 1333–1344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horiuchi, H.; Katayama, T. Protein Kinase C of Filamentous Fungi and Its Roles in the Stresses Affecting Hyphal Morphogenesis and Conidiation. In Stress Biology of Yeasts and Fungi; Springer: Berlin/Heidelberg, Germany, 2015; pp. 185–198. [Google Scholar] [CrossRef]
- Munro, C.A.; Selvaggini, S.; de Bruijn, I.; Walker, L.; Lenardon, M.D.; Gerssen, B.; Milne, S.; Brown, A.J.P.; Gow, N.A.R. The PKC, HOG and Ca2+ signalling pathways co-ordinately regulate chitin synthesis in Candida albicans. Mol. Microbiol. 2007, 63, 1399–1413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popolo, L.; Gualtieri, T.; Ragni, E. The yeast cell-wall salvage pathway. Med. Mycol. 2001, 39 (Suppl. S1), 111–121. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mei, J.; Ning, N.; Wu, H.; Chen, X.; Li, Z.; Liu, W. Glycosylphosphatidylinositol Anchor Biosynthesis Pathway-Related Protein GPI7 Is Required for the Vegetative Growth and Pathogenicity of Colletotrichum graminicola. Int. J. Mol. Sci. 2022, 23, 2985. https://doi.org/10.3390/ijms23062985
Mei J, Ning N, Wu H, Chen X, Li Z, Liu W. Glycosylphosphatidylinositol Anchor Biosynthesis Pathway-Related Protein GPI7 Is Required for the Vegetative Growth and Pathogenicity of Colletotrichum graminicola. International Journal of Molecular Sciences. 2022; 23(6):2985. https://doi.org/10.3390/ijms23062985
Chicago/Turabian StyleMei, Jie, Na Ning, Hanxiang Wu, Xiaolin Chen, Zhiqiang Li, and Wende Liu. 2022. "Glycosylphosphatidylinositol Anchor Biosynthesis Pathway-Related Protein GPI7 Is Required for the Vegetative Growth and Pathogenicity of Colletotrichum graminicola" International Journal of Molecular Sciences 23, no. 6: 2985. https://doi.org/10.3390/ijms23062985
APA StyleMei, J., Ning, N., Wu, H., Chen, X., Li, Z., & Liu, W. (2022). Glycosylphosphatidylinositol Anchor Biosynthesis Pathway-Related Protein GPI7 Is Required for the Vegetative Growth and Pathogenicity of Colletotrichum graminicola. International Journal of Molecular Sciences, 23(6), 2985. https://doi.org/10.3390/ijms23062985






