Arabidopsis Plasma Membrane ATPase AHA5 Is Negatively Involved in PAMP-Triggered Immunity
Abstract
1. Introduction
2. Results
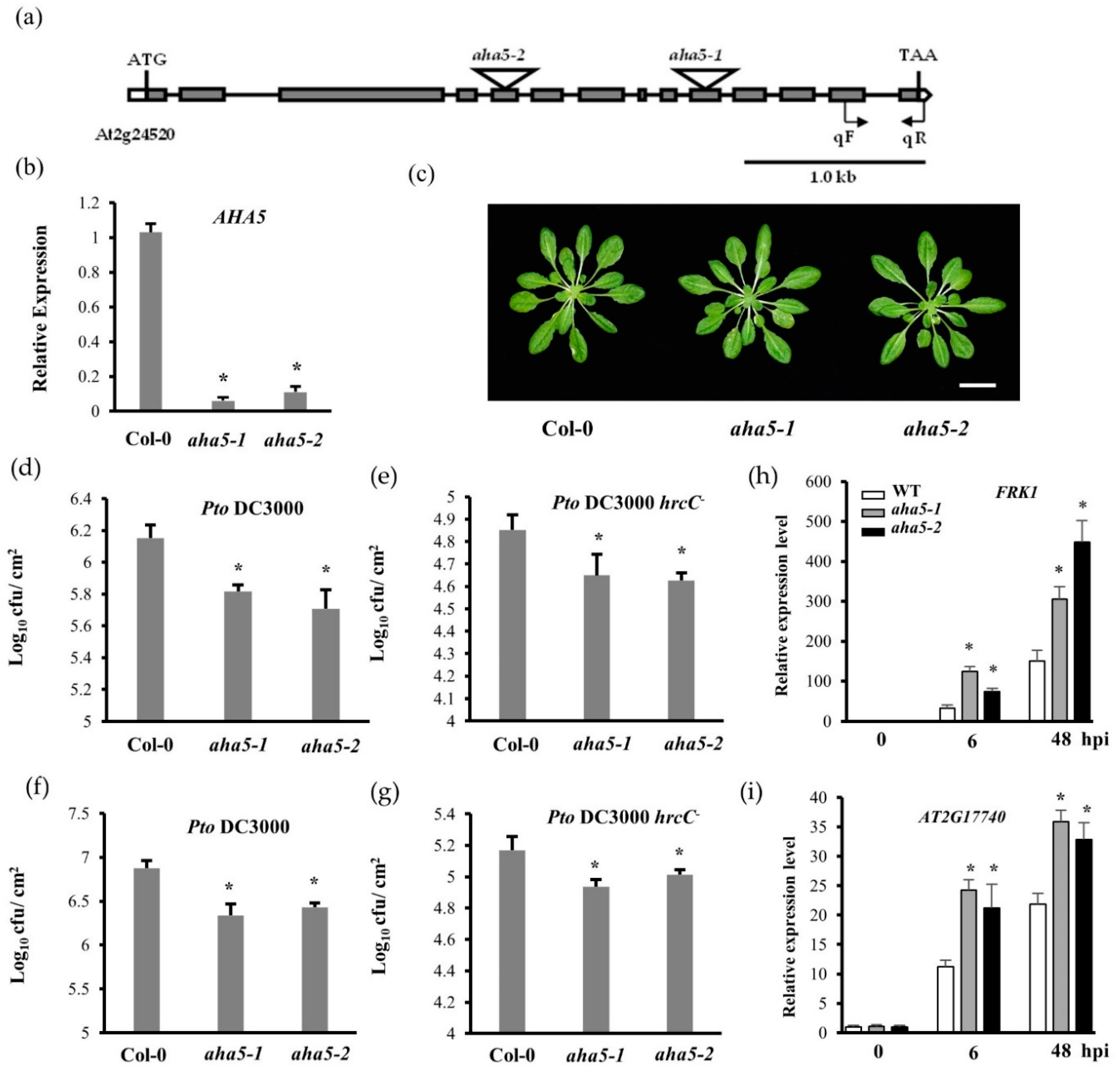
2.1. The aha5 Mutants Displayed an Enhanced Resistance against the Pto Pathogens
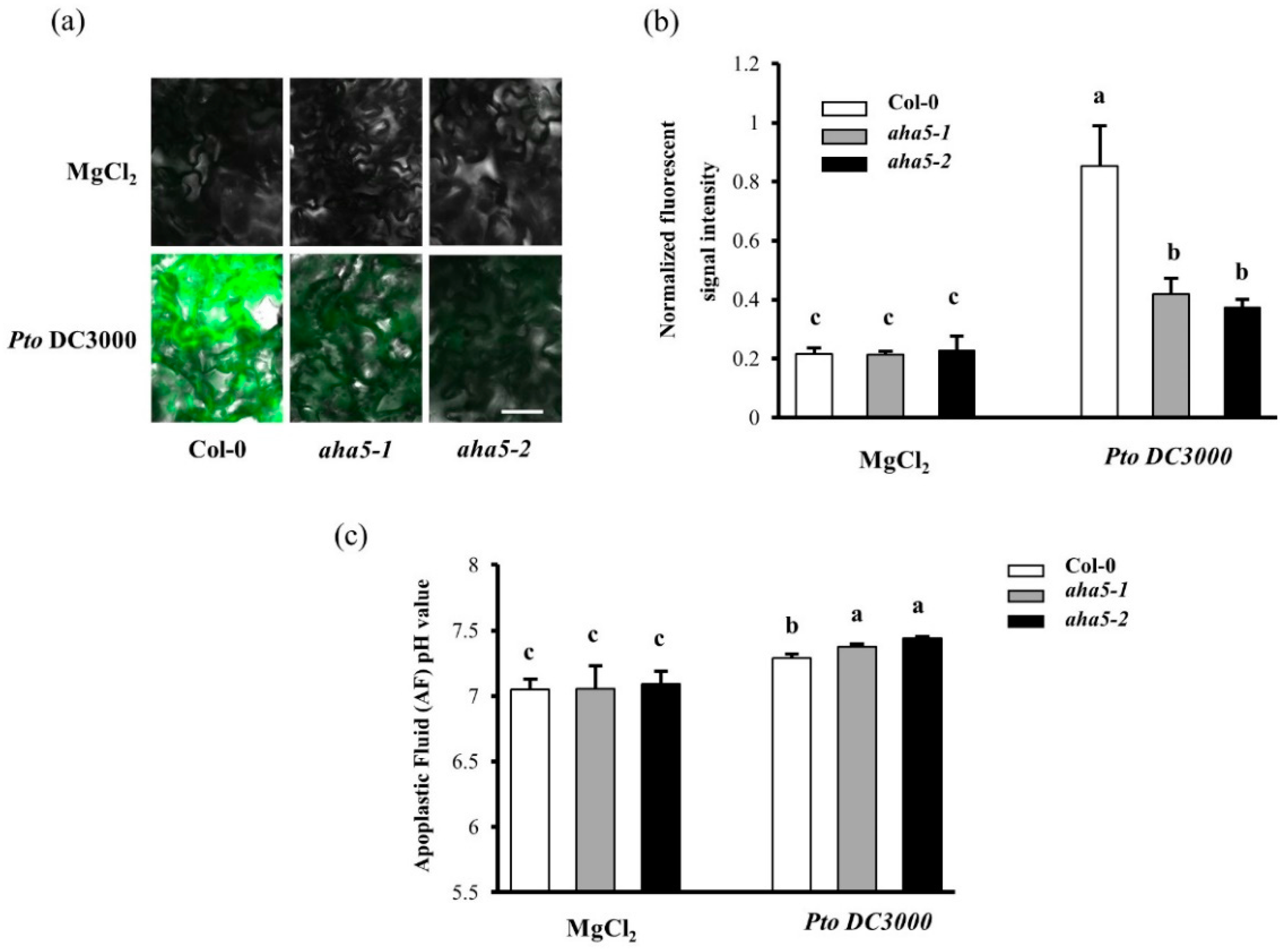
2.2. AHA5 Is Involved in the pH Homeostasis of the Cytoplasm and Apoplast during the PTI
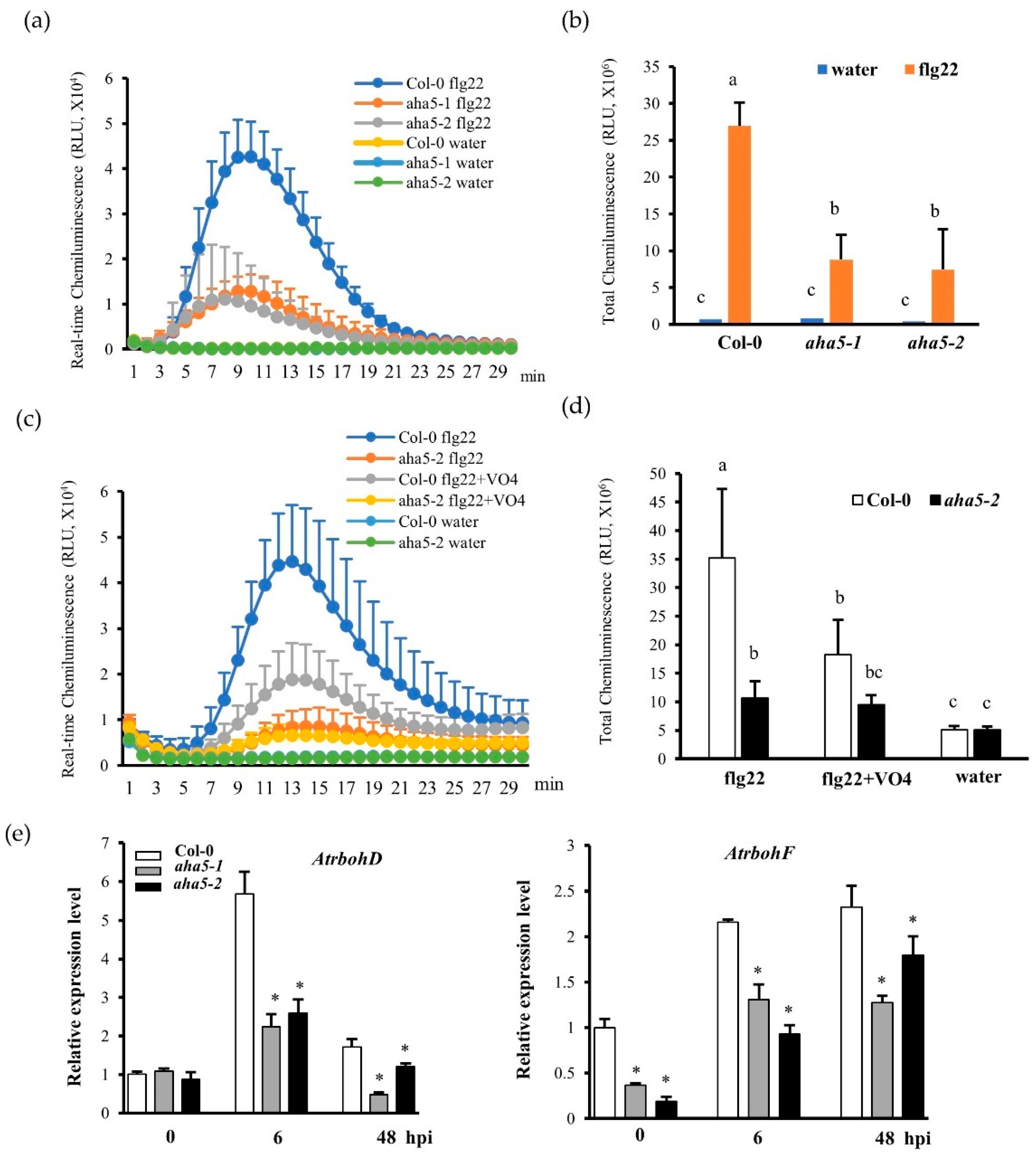
2.3. AHA5 Is Required for the PTI-Induced Apoplastic H2O2 Accumulation
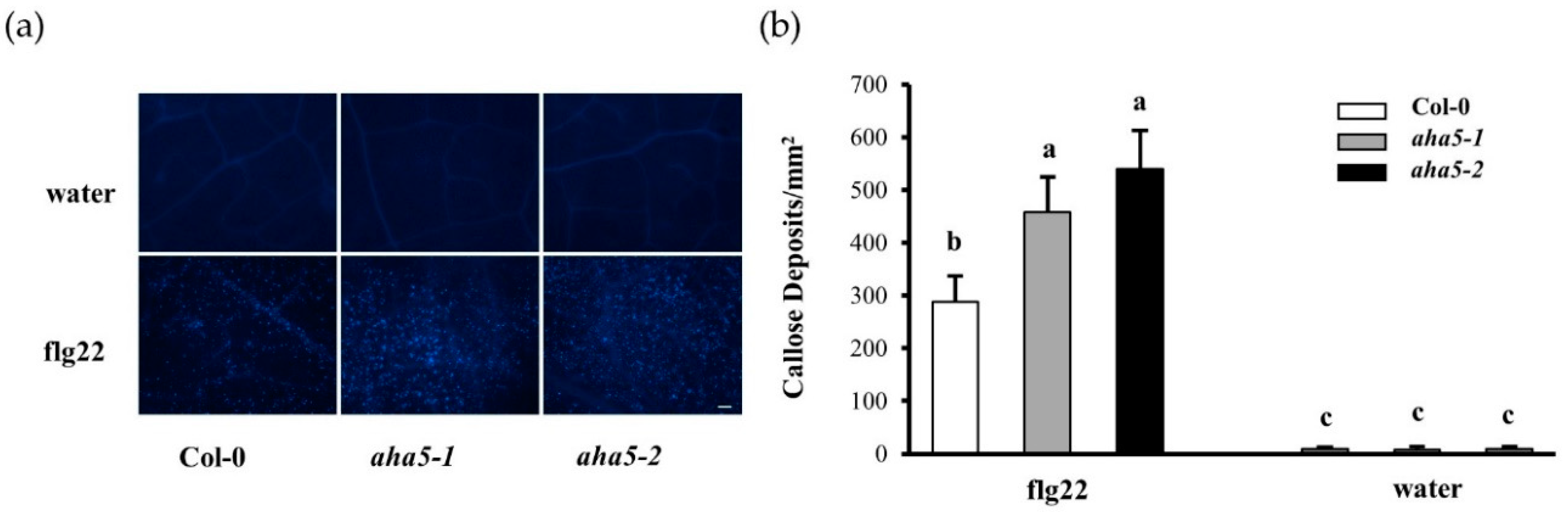
2.4. The aha5 Mutant Plants Showed Enhanced Callose Deposition upon the PAMP Treatment
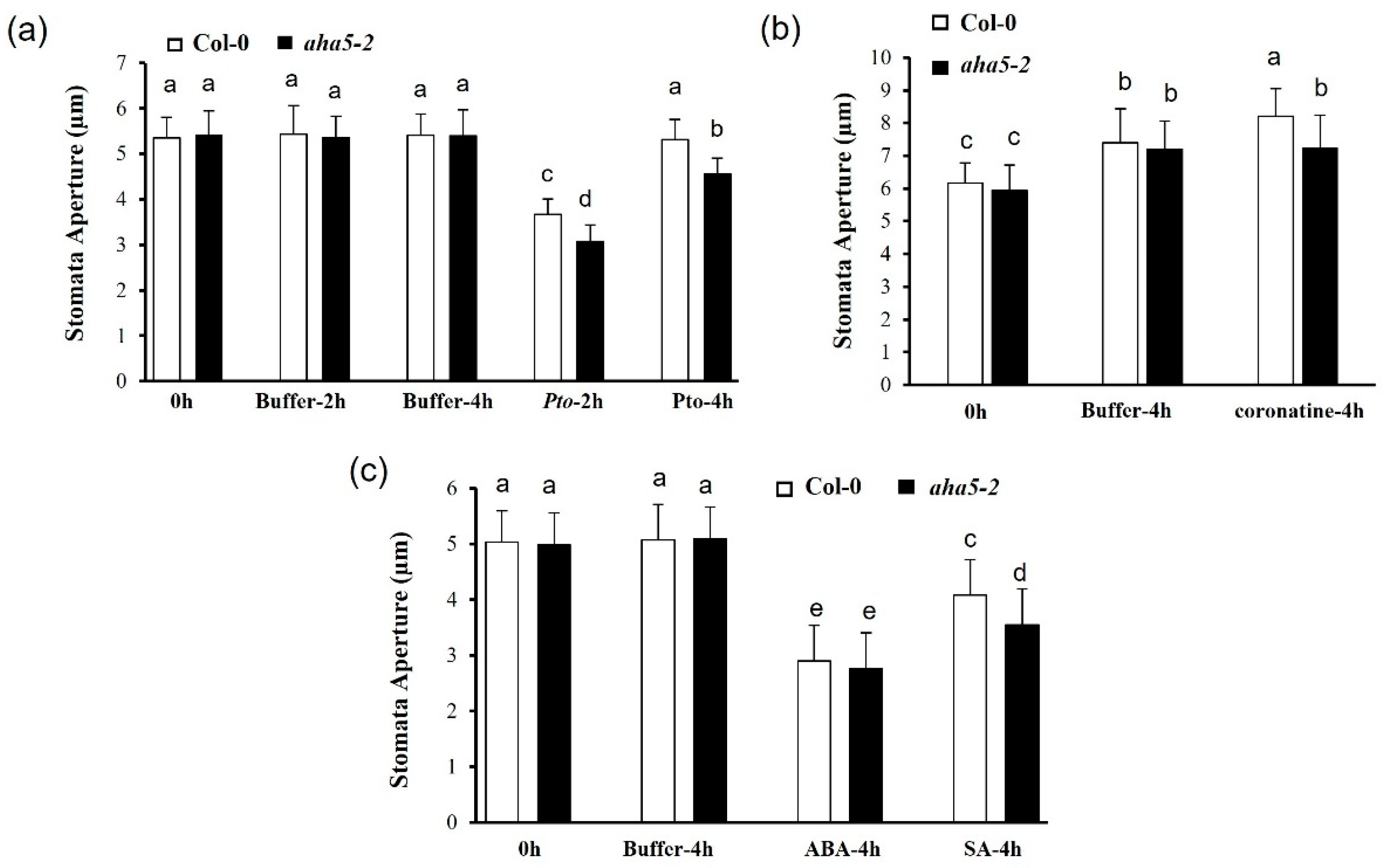
2.5. The aha5 Mutant Plants Affected the Stomata Apertures in the Pathogen and Hormone Treatments
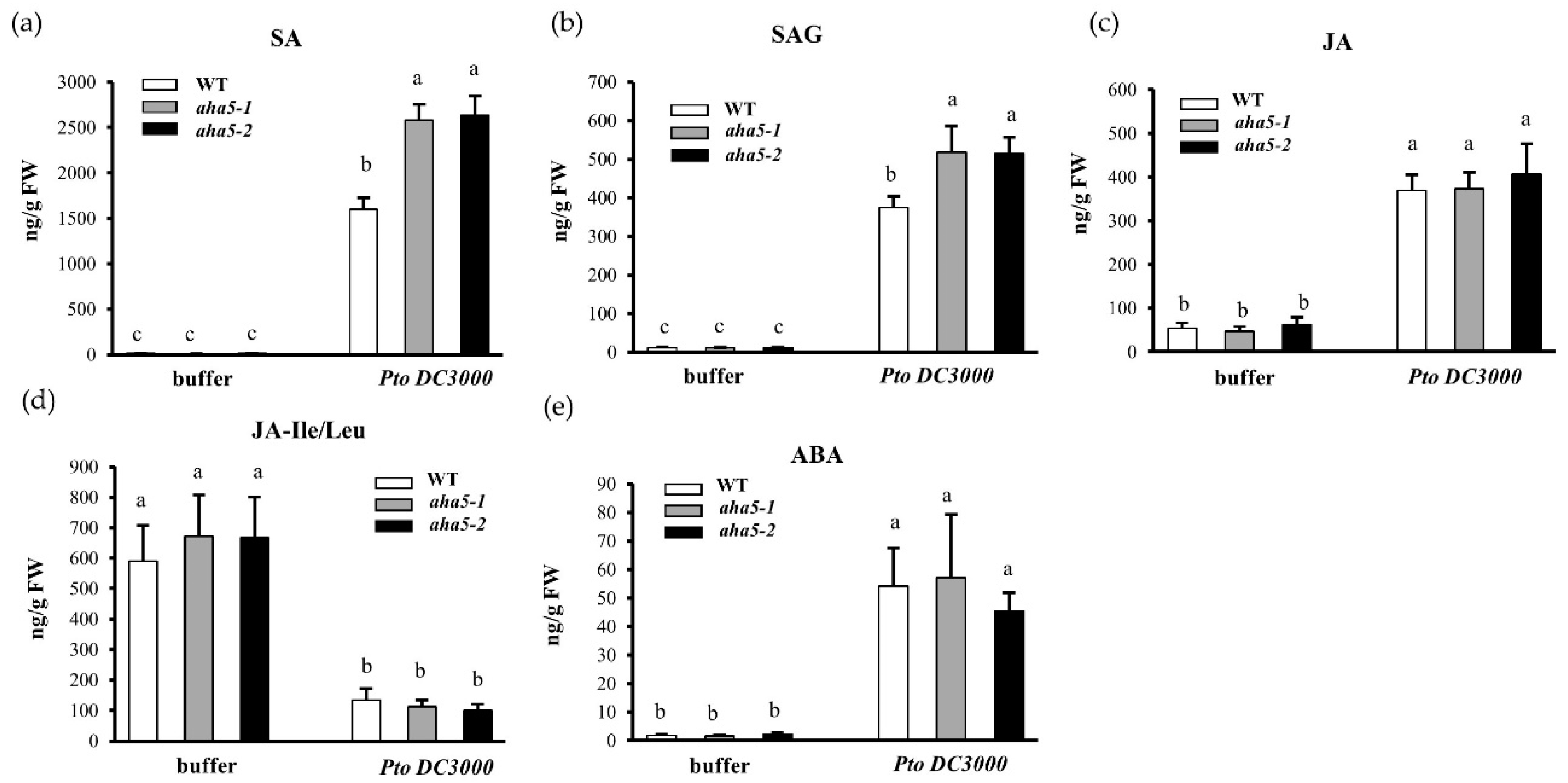
2.6. The aha5 Mutants Accumulate Higher Levels of SA and Induce SA-Responsive Defense Genes in Response to the Pto DC3000 Infection
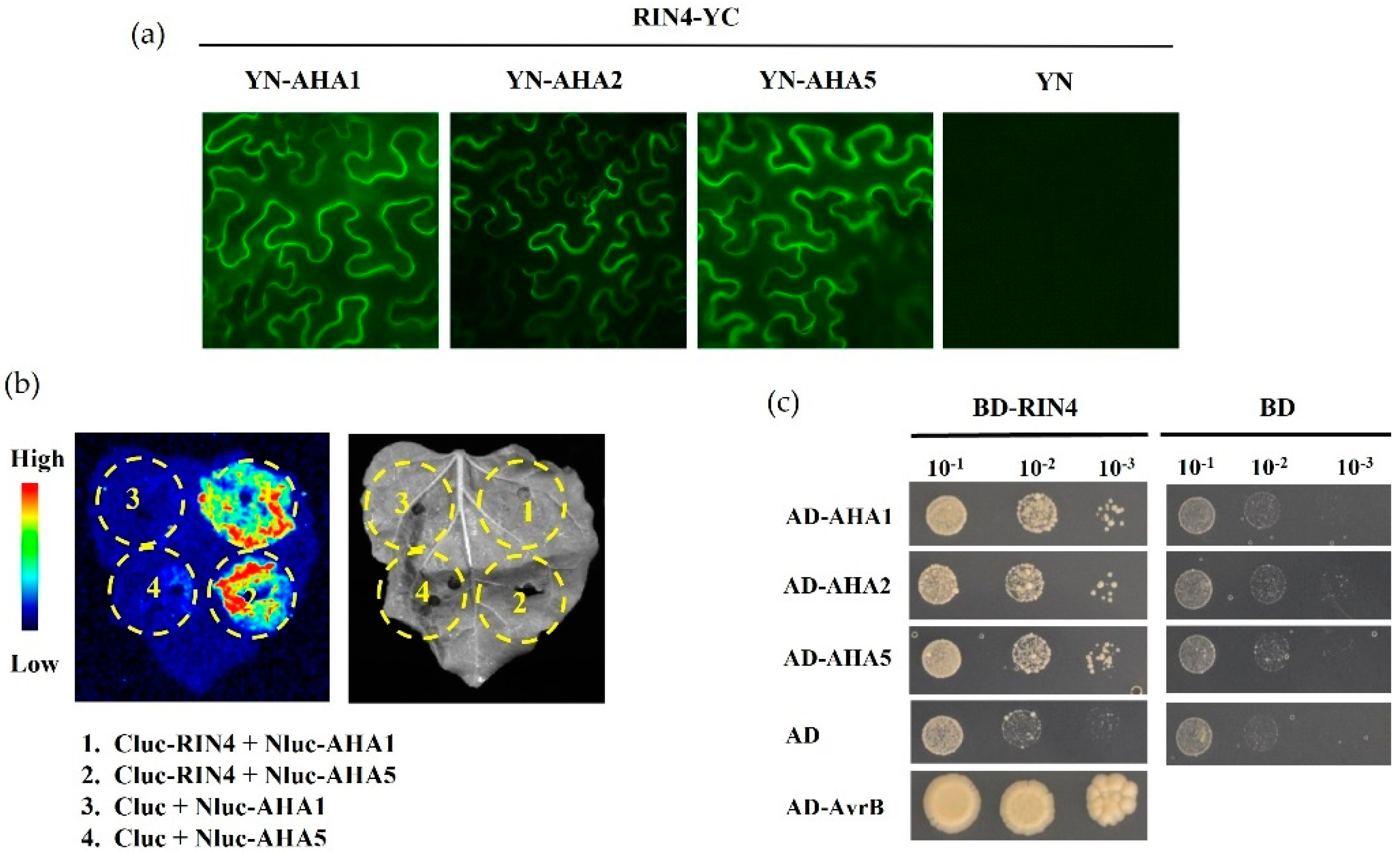
2.7. AHA5 Interacts with RIN4 In Vitro and In Vivo
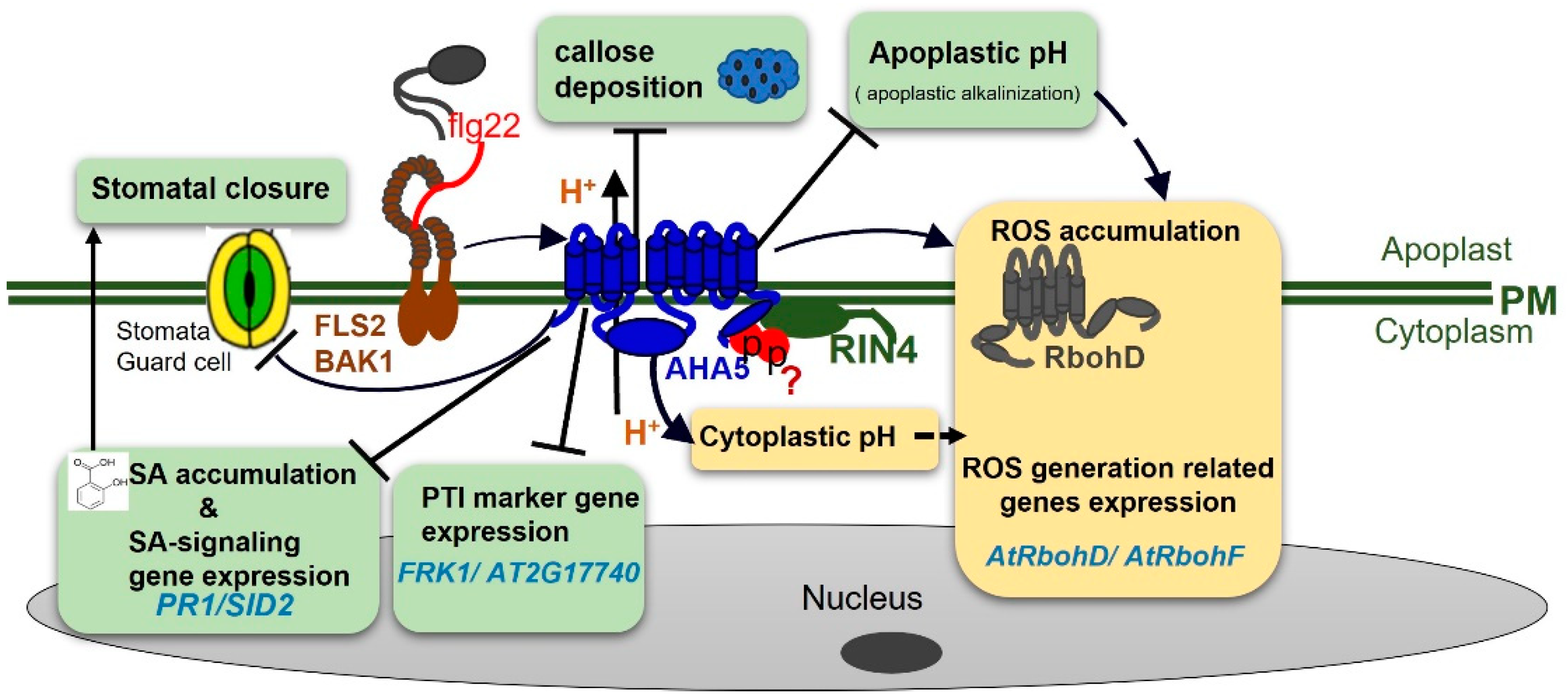
3. Discussion
4. Materials and Methods
4.1. Plant Growth and Pathogen Inoculations
4.2. Phylogenetic and Alignment Analysis
4.3. qPCR Analysis of Gene Expression
4.4. The Cytoplasmic pH Gradient Staining
4.5. Apoplastic Fluid Extraction and Apoplastic pH Evaluation
4.6. H2O2 Assay
4.7. Callose Deposition Assay
4.8. Stomata Closure Assays
4.9. Phytohormone Quantification
4.10. Bimolecular Fluorescence Complementation (BiFC) Assay
4.11. Luciferase Complementation Imaging (LCI) Assay
4.12. Yeast Two-Hybrid (Y2H) Assay
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BCECF-AM | [2′, 7′-Bis (2-carboxyethyl)-5 (6)-carboxyfluorescein tetrakis (acetoxymethyl) ester. |
| PM H+-ATPases | plasma membrane H+ pumps |
References
- Yu, X.; Feng, B.; He, P.; Shan, L. From Chaos to Harmony: Responses and Signaling upon Microbial Pattern Recognition. Annu. Rev. Phytopathol. 2017, 55, 109–137. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Vance, R.E.; Dangl, J.L. Intracellular innate immune surveillance devices in plants and animals. Science 2016, 354, aaf6395. [Google Scholar] [CrossRef] [PubMed]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.M.; He, S.Y.; Xin, X.F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2021, 592, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Ngou, B.P.M.; Ahn, H.-K.; Ding, P.; Jones, J.D.G. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 2021, 592, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Toruño, T.Y.; Shen, M.; Coaker, G.; Mackey, D. Regulated disorder: Posttranslational modifications control the RIN4 plant immune signaling hub. Mol. Plant-Microbe Interact. 2019, 32, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.K.; Macoy, D.M.; Kim, W.Y.; Lee, S.Y.; Kim, M.G. Role of RIN4 in Regulating PAMP-Triggered Immunity and Effector-Triggered Immunity: Current Status and Future Perspectives. Mol. Cells 2019, 42, 503–511. [Google Scholar] [CrossRef]
- Liu, J.; Elmore, J.M.; Fuglsang, A.T.; Palmgren, M.G.; Staskawicz, B.J.; Coaker, G. RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol. 2009, 7, e1000139. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Bourdais, G.; Yu, G.; Robatzek, S.; Coaker, G. Phosphorylation of the Plant Immune Regulator RPM1-INTERACTING PROTEIN4 Enhances Plant Plasma Membrane H(+)-ATPase Activity and Inhibits Flagellin-Triggered Immune Responses in Arabidopsis. Plant Cell 2015, 27, 2042–2056. [Google Scholar] [CrossRef]
- Zhou, Z.; Wu, Y.; Yang, Y.; Du, M.; Zhang, X.; Guo, Y.; Li, C.; Zhou, J.-M. An Arabidopsis Plasma Membrane Proton ATPase Modulates JA Signaling and Is Exploited by the Pseudomonas syringae Effector Protein AvrB for Stomatal Invasion. Plant Cell 2015, 27, 2032–2041. [Google Scholar] [CrossRef] [PubMed]
- Day, B.; Dahlbeck, D.; Huang, J.; Chisholm, S.T.; Li, D.; Staskawicz, B.J. Molecular basis for the RIN4 negative regulation of RPS2 disease resistance. Plant Cell 2005, 17, 1292–1305. [Google Scholar] [CrossRef]
- Kim, M.G.; da Cunha, L.; McFall, A.J.; Belkhadir, Y.; DebRoy, S.; Dangl, J.L.; Mackey, D. Two Pseudomonas syringae Type III Effectors Inhibit RIN4-Regulated Basal Defense in Arabidopsis. Cell 2005, 121, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, R. Structure, function and regulation of the plant vacuolar H(+)-translocating ATPase. Biochim. Biophys. Acta 2000, 1465, 17–36. [Google Scholar] [CrossRef]
- Morsomme, P.; Boutry, M. The plant plasma membrane H(+)-ATPase: Structure, function and regulation. Biochim. Biophys. Acta 2000, 1465, 1–16. [Google Scholar] [CrossRef]
- Portillo, F. Regulation of plasma membrane H(+)-ATPase in fungi and plants. Biochim. Biophys. Acta 2000, 1469, 31–42. [Google Scholar] [CrossRef]
- Falhof, J.; Pedersen, J.T.; Fuglsang, A.T.; Palmgren, M. Plasma Membrane H(+)-ATPase Regulation in the Center of Plant Physiology. Mol. Plant 2016, 9, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Kaundal, A.; Ramu, V.S. GENERAL CONTROL NONREPRESSIBLE4 Degrades 14-3-3 and the RIN4 Complex to Regulate Stomatal Aperture with Implications on Nonhost Disease Resistance and Drought Tolerance. Plant Cell 2017, 29, 2233–2248. [Google Scholar] [CrossRef]
- Ueno, K.; Kinoshita, T.; Inoue, S.; Emi, T.; Shimazaki, K. Biochemical characterization of plasma membrane H+-ATPase activation in guard cell protoplasts of Arabidopsis thaliana in response to blue light. Plant Cell Physiol. 2005, 46, 955–963. [Google Scholar] [CrossRef] [PubMed]
- Robertson, W.R.; Clark, K.; Young, J.C.; Sussman, M.R. An Arabidopsis thaliana plasma membrane proton pump is essential for pollen development. Genetics 2004, 168, 1677–1687. [Google Scholar] [CrossRef][Green Version]
- Caesar, K.; Elgass, K.; Chen, Z.; Huppenberger, P.; Witthöft, J.; Schleifenbaum, F.; Blatt, M.R.; Oecking, C.; Harter, K. A fast brassinolide-regulated response pathway in the plasma membrane of Arabidopsis thaliana. Plant J. 2011, 66, 528–540. [Google Scholar] [CrossRef]
- Santi, S.; Schmidt, W. Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 2009, 183, 1072–1084. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Zhang, D.; Song, T.; Xu, F.; Lin, S.; Xu, W.; Li, Q.; Zhu, Y.; Liang, J.; Zhang, J. Arabidopsis plasma membrane H+-ATPase genes AHA2 and AHA7 have distinct and overlapping roles in the modulation of root tip H+ efflux in response to low-phosphorus stress. J. Exp. Bot. 2017, 68, 1731–1741. [Google Scholar] [CrossRef]
- Gévaudant, F.; Duby, G.; von Stedingk, E.; Zhao, R.; Morsomme, P.; Boutry, M. Expression of a constitutively activated plasma membrane H+-ATPase alters plant development and increases salt tolerance. Plant Physiol. 2007, 144, 1763–1776. [Google Scholar] [CrossRef] [PubMed]
- Baxter, I.R.; Young, J.C.; Armstrong, G.; Foster, N.; Bogenschutz, N.; Cordova, T.; Peer, W.A.; Hazen, S.P.; Murphy, A.S.; Harper, J.F. A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2005, 102, 2649–2654. [Google Scholar] [CrossRef] [PubMed]
- Ando, E. Red Light-Induced Phosphorylation of Plasma Membrane H(+)-ATPase in Stomatal Guard Cells. PLoS Biol. 2018, 178, 838–849. [Google Scholar] [CrossRef] [PubMed]
- Inoue, S.I.; Kinoshita, T. Blue Light Regulation of Stomatal Opening and the Plasma Membrane H(+)-ATPase. Plant Cell 2017, 174, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Zhang, L. Stomatal Defense a Decade Later. Plant Physiol. 2017, 174, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Underwood, W.; Koczan, J.; Nomura, K.; He, S.Y. Plant stomata function in innate immunity against bacterial invasion. Cell 2006, 126, 969–980. [Google Scholar] [CrossRef]
- Laboratory, S.I.G.A. T-DNA Primer Design. Available online: http://signal.salk.edu/tdnaprimers.2.html (accessed on 28 July 2005).
- Smith, J.M.; Leslie, M.E.; Robinson, S.J.; Korasick, D.A.; Zhang, T.; Backues, S.K.; Cornish, P.V.; Koo, A.J.; Bednarek, S.Y.; Heese, A. Loss of Arabidopsis thaliana Dynamin-Related Protein 2B reveals separation of innate immune signaling pathways. PLoS Pathog. 2014, 10, e1004578. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Y.; Wang, Y. Apoplastic Proteases: Powerful Weapons against Pathogen Infection in Plants. Plant Commun. 2020, 1, 100085. [Google Scholar] [CrossRef] [PubMed]
- Geilfus, C.M. The pH of the Apoplast: Dynamic Factor with Functional Impact Under Stress. Mol. Plant 2017, 10, 1371–1386. [Google Scholar] [CrossRef] [PubMed]
- Song, C.P.; Guo, Y.; Qiu, Q.; Lambert, G.; Galbraith, D.W.; Jagendorf, A.; Zhu, J.K. A probable Na+(K+)/H+ exchanger on the chloroplast envelope functions in pH homeostasis and chloroplast development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2004, 101, 10211–10216. [Google Scholar] [CrossRef] [PubMed]
- Andres, Z.; Perez-Hormaeche, J.; Leidi, E.O.; Schlucking, K.; Steinhorst, L.; McLachlan, D.H.; Schumacher, K.; Hetherington, A.M.; Kudla, J.; Cubero, B.; et al. Control of vacuolar dynamics and regulation of stomatal aperture by tonoplast potassium uptake. Proc. Natl. Acad. Sci. USA 2014, 111, E1806–E1814. [Google Scholar] [CrossRef] [PubMed]
- Gentzel, I.; Giese, L.; Zhao, W.; Alonso, A.P.; Mackey, D. A Simple Method for Measuring Apoplast Hydration and Collecting Apoplast Contents. Plant Physiol. 2019, 179, 1265–1272. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Wang, J.; Gong, Z.; Zhou, J.M. Apoplastic ROS signaling in plant immunity. Curr. Opin. Plant Biol. 2017, 38, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Lamb, C.; Dixon, R.A. THE OXIDATIVE BURST IN PLANT DISEASE RESISTANCE. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 251–275. [Google Scholar] [CrossRef]
- Baker, C.J.; Orlandi, E.W. Active oxygen in plant pathogenesis. Annu. Rev. Phytopathol. 1995, 33, 299–321. [Google Scholar] [CrossRef]
- Yan, F.; Zhu, Y.; Müller, C.; Zörb, C.; Schubert, S. Adaptation of H+-Pumping and Plasma Membrane H+ ATPase Activity in Proteoid Roots of White Lupin under Phosphate Deficiency. Plant Physiol. 2002, 129, 50–63. [Google Scholar] [CrossRef]
- Majumdar, A.; Kar, R.K. Congruence between PM H(+)-ATPase and NADPH oxidase during root growth: A necessary probability. Protoplasma 2018, 255, 1129–1137. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Yan, F.; Zorb, C.; Schubert, S. A link between citrate and proton release by proteoid roots of white lupin (Lupinus albus L.) grown under phosphorus-deficient conditions? Plant Cell Physiol. 2005, 46, 892–901. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yang, G. Signal function studies of ROS, especially RBOH-dependent ROS, in plant growth, development and environmental stress. J. Plant Growth Regul. 2020, 39, 157–171. [Google Scholar] [CrossRef]
- Torres, M.A.; Dangl, J.L.; Jones, J.D. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 2002, 99, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Cheng, J.; Gangadharan, A.; Mackey, D. The coronatine toxin of Pseudomonas syringae is a multifunctional suppressor of Arabidopsis defense. Plant Cell 2012, 24, 4763–4774. [Google Scholar] [CrossRef]
- Wei, H.; Jing, Y.; Zhang, L.; Kong, D. Phytohormones and their crosstalk in regulating stomatal development and patterning. J. Exp. Bot. 2021, 72, 2356–2370. [Google Scholar] [CrossRef] [PubMed]
- Acharya, B.R.; Assmann, S.M. Hormone interactions in stomatal function. Plant Mol. Biol. 2009, 69, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Yang, Y.; Yang, Z.; Wang, X.; Guo, Y. VAMP711 Is Required for Abscisic Acid-Mediated Inhibition of Plasma Membrane H(+)-ATPase Activity. Plant Physiol. 2018, 178, 1332–1343. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, S.; Takemiya, A.; Sakamoto, T.; Kurata, T.; Tsutsumi, T.; Kinoshita, T.; Shimazaki, K. The Plasma Membrane H+-ATPase AHA1 Plays a Major Role in Stomatal Opening in Response to Blue Light. Plant Physiol. 2016, 171, 2731–2743. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Noguchi, K.; Ono, N.; Inoue, S.; Terashima, I.; Kinoshita, T. Overexpression of plasma membrane H+-ATPase in guard cells promotes light-induced stomatal opening and enhances plant growth. Proc. Natl. Acad. Sci. USA 2014, 111, 533–538. [Google Scholar] [CrossRef]
- Keinath, N.F.; Kierszniowska, S.; Lorek, J.; Bourdais, G.; Kessler, S.A.; Shimosato-Asano, H.; Grossniklaus, U.; Schulze, W.X.; Robatzek, S.; Panstruga, R. PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J. Biol. Chem. 2010, 285, 39140–39149. [Google Scholar] [CrossRef]
- Merlot, S.; Leonhardt, N.; Fenzi, F.; Valon, C.; Costa, M.; Piette, L.; Vavasseur, A.; Genty, B.; Boivin, K.; Muller, A.; et al. Constitutive activation of a plasma membrane H(+)-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J. 2007, 26, 3216–3226. [Google Scholar] [CrossRef]
- Haruta, M.; Burch, H.L.; Nelson, R.B.; Barrett-Wilt, G.; Kline, K.G.; Mohsin, S.B.; Young, J.C.; Otegui, M.S.; Sussman, M.R. Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J. Biol. Chem. 2010, 285, 17918–17929. [Google Scholar] [CrossRef] [PubMed]
- Haruta, M.; Sussman, M.R. The effect of a genetically reduced plasma membrane protonmotive force on vegetative growth of Arabidopsis. Plant Physiol. 2012, 158, 1158–1171. [Google Scholar] [CrossRef]
- Yan, S.; McLamore, E.S.; Dong, S.; Gao, H.; Taguchi, M.; Wang, N.; Zhang, T.; Su, X.; Shen, Y. The role of plasma membrane H(+)-ATPase in jasmonate-induced ion fluxes and stomatal closure in Arabidopsis thaliana. Plant J. 2015, 83, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y.; Macho, A.P. Analysis of PAMP-Triggered ROS Burst in Plant Immunity. In Plant Pattern Recognition Receptors: Methods and Protocols; Shan, L., He, P., Eds.; Springer: New York, NY, USA, 2017; pp. 143–153. [Google Scholar] [CrossRef]
- Yi, S.Y.; Shirasu, K.; Moon, J.S.; Lee, S.-G.; Kwon, S.-Y. The activated SA and JA signaling pathways have an influence on flg22-triggered oxidative burst and callose deposition. PLoS ONE 2014, 9, e88951. [Google Scholar]
- Chater, C.C.; Oliver, J.; Casson, S.; Gray, J.E. Putting the brakes on: Abscisic acid as a central environmental regulator of stomatal development. New Phytol. 2014, 202, 376–391. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.S.; Lakshmanan, V.; Caplan, J.L.; Powell, D.; Czymmek, K.J.; Levia, D.F.; Bais, H.P. Rhizobacteria Bacillus subtilis restricts foliar pathogen entry through stomata. Plant J. 2012, 72, 694–706. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Gao, Q.M.; Yu, K.; Lapchyk, L.; Navarre, D.; Hildebrand, D.; Kachroo, A.; Kachroo, P. An intact cuticle in distal tissues is essential for the induction of systemic acquired resistance in plants. Cell Host Microbe 2009, 5, 151–165. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Yang, X.; Lü, S.; Fan, J.; Opiyo, S.; Yang, P.; Mangold, J.; Mackey, D.; Xia, Y. Deciphering the novel role of AtMIN7 in cuticle formation and defense against the bacterial pathogen infection. Int. J. Mol. Sci. 2020, 21, 5547. [Google Scholar] [CrossRef] [PubMed]
- Knepper, C.; Savory, E.A.; Day, B. Arabidopsis NDR1 is an integrin-like protein with a role in fluid loss and plasma membrane-cell wall adhesion. Plant Physiol. 2011, 156, 286–300. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Bellizzi Mdel, R.; Ning, Y.; Meyers, B.C.; Wang, G.L. HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice. Plant Cell 2012, 24, 3783–3794. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.G.; Mackey, D. Measuring cell-wall-based defenses and their effect on bacterial growth in Arabidopsis. In Innate Immunity; Springer: Totowa, NJ, USA, 2008; pp. 443–452. [Google Scholar]
- Jin, L.; Mackey, D.M. Measuring Callose Deposition, an Indicator of Cell Wall Reinforcement, During Bacterial Infection in Arabidopsis. Methods Mol. Biol. 2017, 1578, 195–205. [Google Scholar] [CrossRef]
- Zheng, X.Y.; Spivey, N.W.; Zeng, W.; Liu, P.P.; Fu, Z.Q.; Klessig, D.F.; He, S.Y.; Dong, X. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 2012, 11, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-Q.; Sun, L.-P.; Wang, L.-X.; Fang, X.-W.; Li, Z.-Q.; Zhang, F.-F.; Hu, X.; Qi, C.; He, J.-M. Ethylene mediates salicylic-acid-induced stomatal closure by controlling reactive oxygen species and nitric oxide production in Arabidopsis. Plant Sci. 2020, 294, 110464. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Ishiga, Y.; Clermont, K.; Mysore, K.S. Coronatine inhibits stomatal closure and delays hypersensitive response cell death induced by nonhost bacterial pathogens. PeerJ 2013, 1, e34. [Google Scholar] [CrossRef] [PubMed]
- Forcat, S.; Bennett, M.H.; Mansfield, J.W.; Grant, M.R. A rapid and robust method for simultaneously measuring changes in the phytohormones ABA, JA and SA in plants following biotic and abiotic stress. Plant Methods 2008, 4, 16. [Google Scholar] [CrossRef] [PubMed]
- Gehl, C.; Waadt, R.; Kudla, J.; Mendel, R.-R.; Hänsch, R. New GATEWAY vectors for High Throughput Analyses of Protein–Protein Interactions by Bimolecular Fluorescence Complementation. Mol. Plant 2009, 2, 1051–1058. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Bi, G.; Zhou, J.M. Luciferase Complementation Assay for Protein-Protein Interactions in Plants. Curr. Protoc. Plant Biol. 2018, 3, 42–50. [Google Scholar] [CrossRef]
- Park, C.-H.; Chen, S.; Shirsekar, G.; Zhou, B.; Khang, C.H.; Songkumarn, P.; Afzal, A.J.; Ning, Y.; Wang, R.; Bellizzi, M. The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern–triggered immunity in rice. Plant Cell 2012, 24, 4748–4762. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Z.; Fan, J.; Gao, Y.G.; Wang, Z.; Yang, P.; Liang, Y.; Opiyo, S.; Xia, Y. Arabidopsis Plasma Membrane ATPase AHA5 Is Negatively Involved in PAMP-Triggered Immunity. Int. J. Mol. Sci. 2022, 23, 3857. https://doi.org/10.3390/ijms23073857
Zhao Z, Fan J, Gao YG, Wang Z, Yang P, Liang Y, Opiyo S, Xia Y. Arabidopsis Plasma Membrane ATPase AHA5 Is Negatively Involved in PAMP-Triggered Immunity. International Journal of Molecular Sciences. 2022; 23(7):3857. https://doi.org/10.3390/ijms23073857
Chicago/Turabian StyleZhao, Zhenzhen, Jiangbo Fan, Yu G. Gao, Zonghua Wang, Piao Yang, Yinping Liang, Stephen Opiyo, and Ye Xia. 2022. "Arabidopsis Plasma Membrane ATPase AHA5 Is Negatively Involved in PAMP-Triggered Immunity" International Journal of Molecular Sciences 23, no. 7: 3857. https://doi.org/10.3390/ijms23073857
APA StyleZhao, Z., Fan, J., Gao, Y. G., Wang, Z., Yang, P., Liang, Y., Opiyo, S., & Xia, Y. (2022). Arabidopsis Plasma Membrane ATPase AHA5 Is Negatively Involved in PAMP-Triggered Immunity. International Journal of Molecular Sciences, 23(7), 3857. https://doi.org/10.3390/ijms23073857










