A Zebrafish Model for a Rare Genetic Disease Reveals a Conserved Role for FBXL3 in the Circadian Clock System
Abstract
:1. Introduction
2. Results
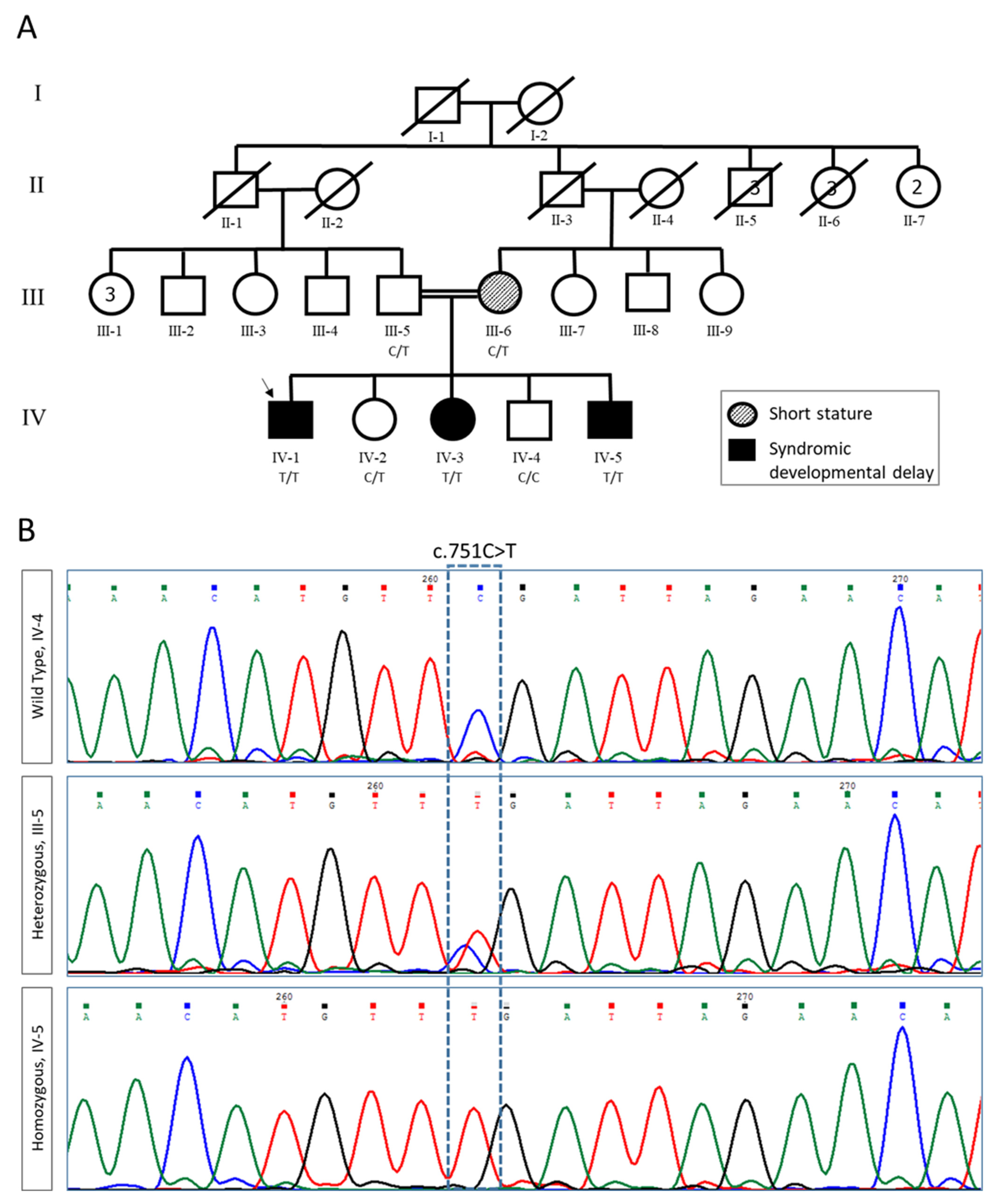
2.1. Clinical Evaluation and Exome Analysis
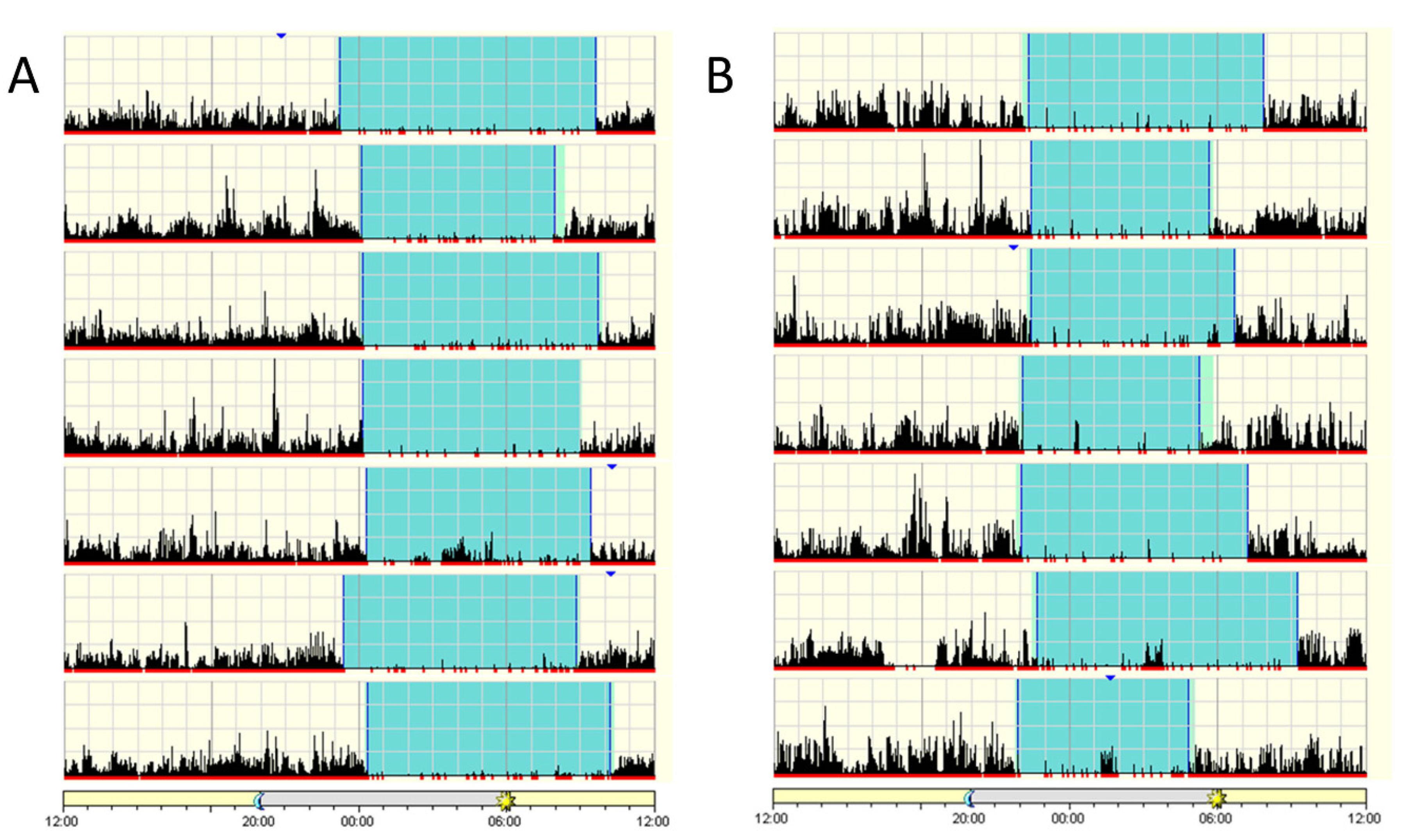
2.2. Sleep Assessment of Patients
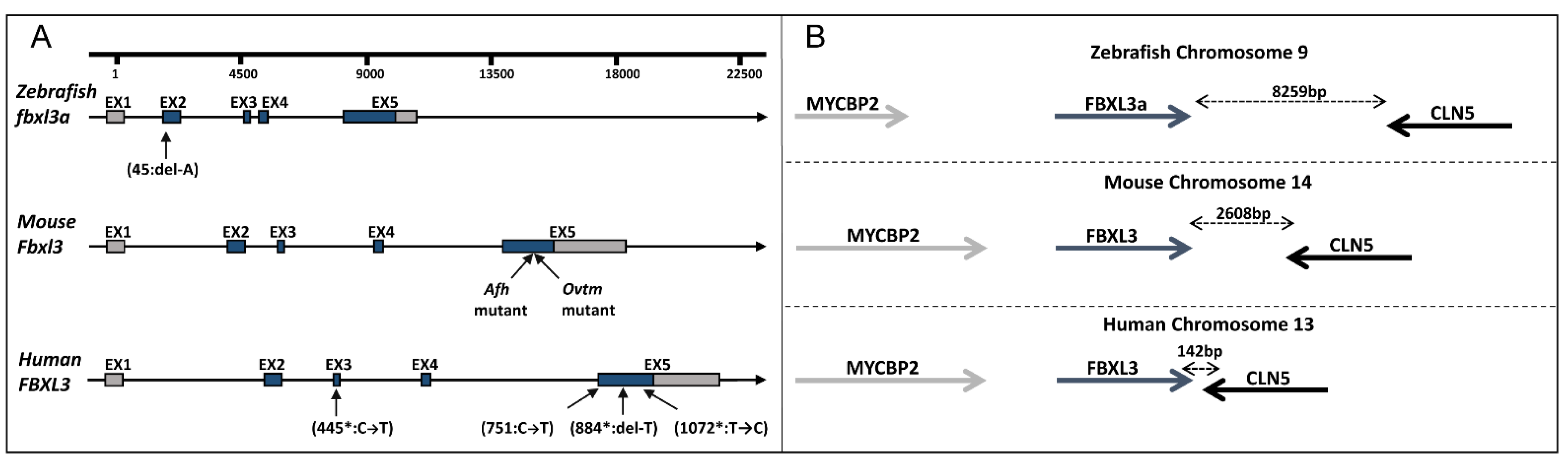
2.3. Establishing a FBXL3-Deficient Zebrafish Line
2.4. FBXL3a Deficiency Does Not Affect Zebrafish Morphology
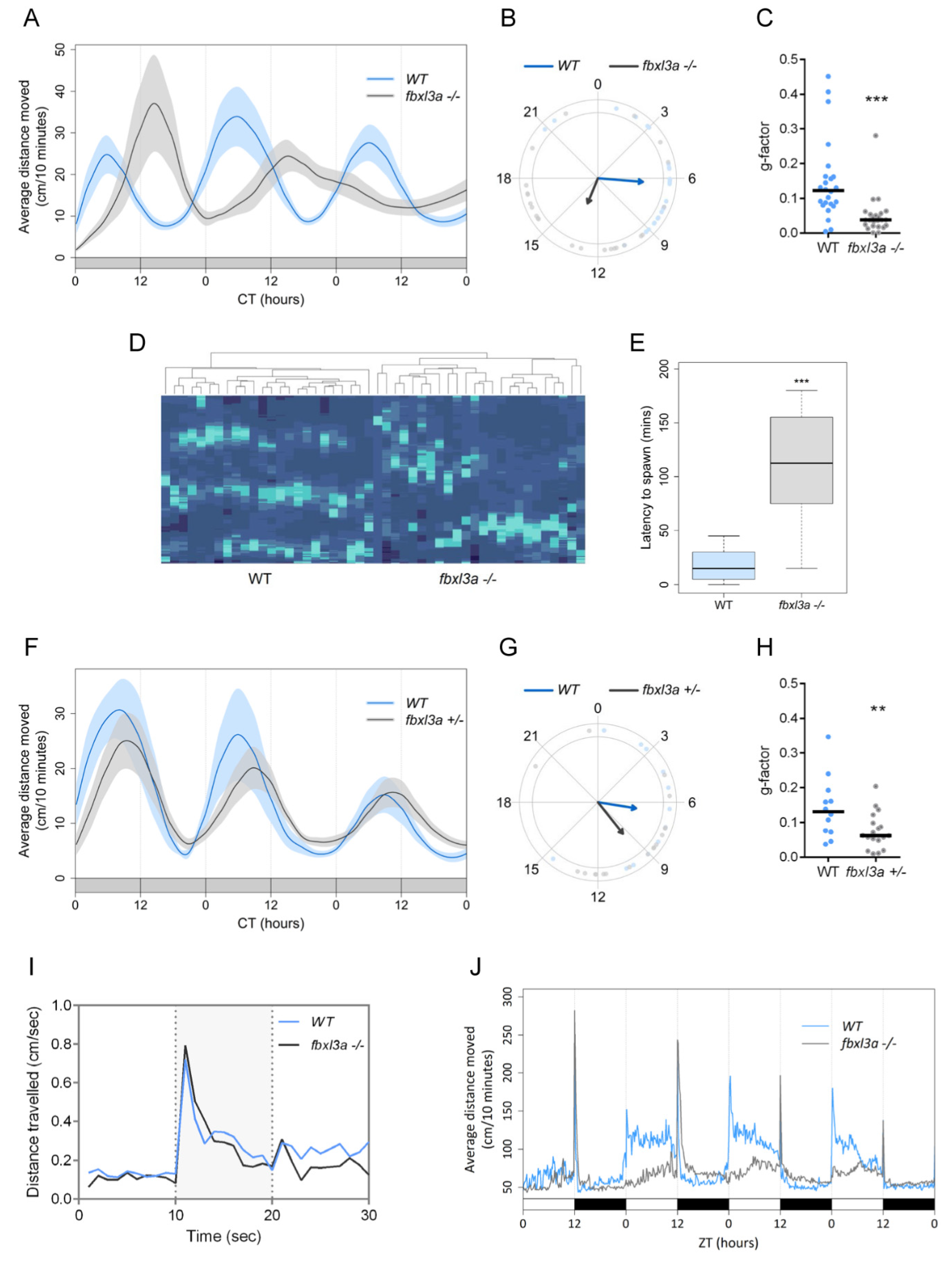
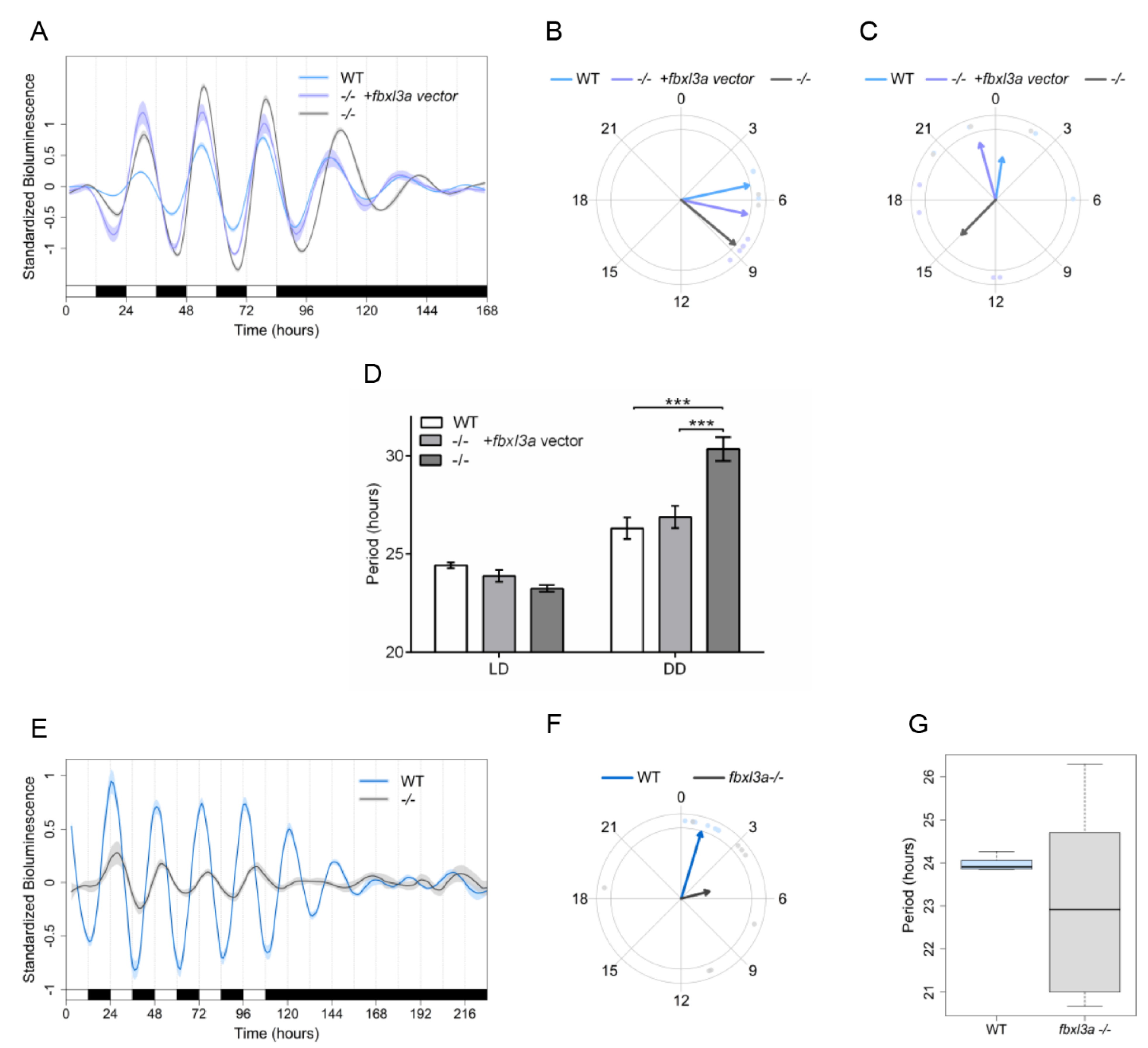
2.5. FBXL3a Deficiency Affects Circadian Rhythms of Locomotor Activity in Zebrafish
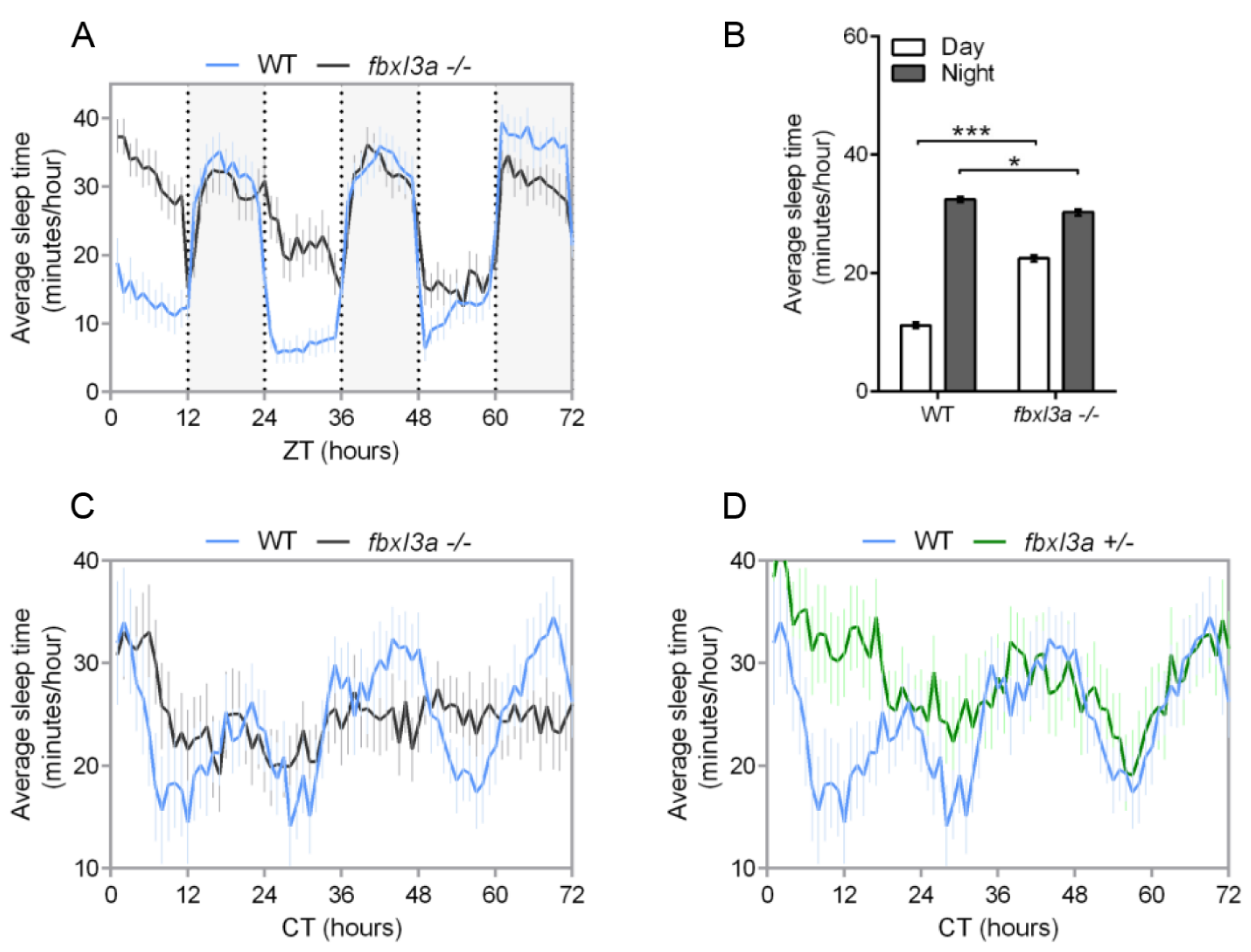
2.6. FBXL3a Deficiency Affects Sleep–Wake Cycles in Zebrafish
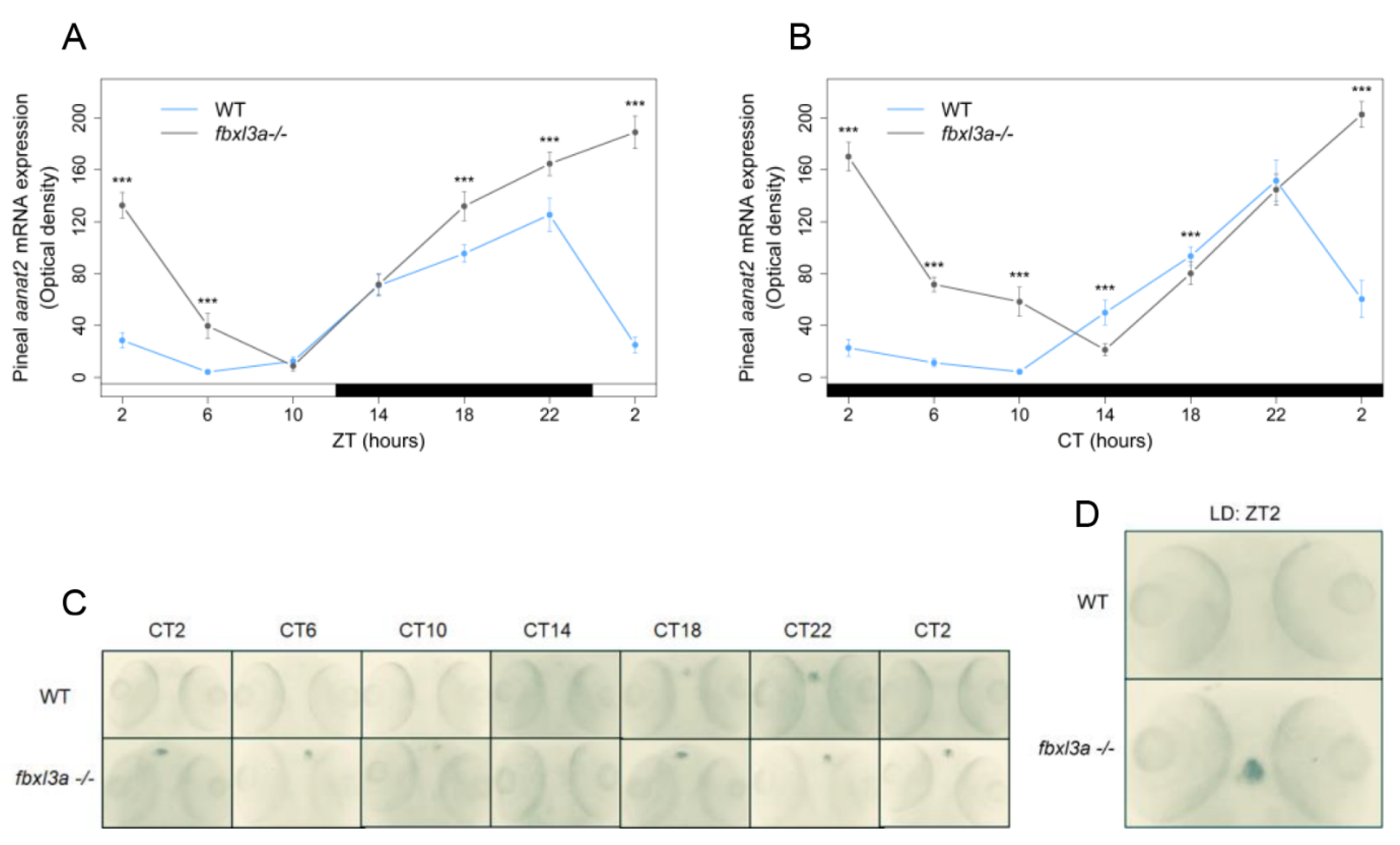
2.7. FBXL3a Deficiency Affects the Rhythmic Expression of Pineal aanat2
2.8. FBXL3a Deficiency Disrupts Molecular Circadian Oscillations
3. Discussion
4. Materials and Methods
4.1. Exome Analysis
4.2. Segregation Analysis
4.3. Actigraphy and Patients Sleep Pattern Detection
4.4. Establishment of the fbxl3a Mutant Zebrafish Line
4.5. Establishment of Cell Lines and Bioluminescence Assay
4.6. Analysis of Bioluminescence
4.7. Cloning and Transfection of Zebrafish fbxl3a Coding Sequence
4.8. Rhythmic Locomotor Activity Measurements in Zebrafish
4.9. Sleep Measurements in Zebrafish
4.10. Zebrafish Morphological Examination
4.11. Whole Mount in Situ Hybridization
4.12. Fish and Embryos
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Takahashi, J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017, 18, 164–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Partch, C.L.; Green, C.B.; Takahashi, J.S. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014, 24, 90–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ko, C.H.; Takahashi, J.S. Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 2006, 15, R271–R277. [Google Scholar] [CrossRef]
- Lowrey, P.L.; Takahashi, J.S. Genetics of circadian rhythms in Mammalian model organisms. Adv. Genet. 2011, 74, 175–230. [Google Scholar]
- Dunlap, J.C. Molecular bases for circadian clocks. Cell 1999, 96, 271–290. [Google Scholar] [CrossRef] [Green Version]
- Godinho, S.I.; Maywood, E.S.; Shaw, L.; Tucci, V.; Barnard, A.R.; Busino, L.; Pagano, M.; Kendall, R.; Quwailid, M.M.; Romero, M.R.; et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 2007, 316, 897–900. [Google Scholar] [CrossRef] [Green Version]
- Xing, W.; Busino, L.; Hinds, T.R.; Marionni, S.T.; Saifee, N.H.; Bush, M.F.; Pagano, M.; Zheng, N. SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature 2013, 496, 64–68. [Google Scholar] [CrossRef]
- Siepka, S.M.; Yoo, S.H.; Park, J.; Song, W.; Kumar, V.; Hu, Y.; Lee, C.; Takahashi, J.S. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 2007, 129, 1011–1023. [Google Scholar] [CrossRef] [Green Version]
- Wegner, S.; Belle, M.D.C.; Hughes, A.T.L.; Diekman, C.O.; Piggins, H.D. Delayed Cryptochrome Degradation Asymmetrically Alters the Daily Rhythm in Suprachiasmatic Clock Neuron Excitability. J. Neurosci. 2017, 37, 7824–7836. [Google Scholar] [CrossRef] [Green Version]
- Ansar, M.; Paracha, S.A.; Serretti, A.; Sarwar, M.T.; Khan, J.; Ranza, E.; Falconnet, E.; Iwaszkiewicz, J.; Shah, S.F.; Qaisar, A.A.; et al. Biallelic variants in FBXL3 cause intellectual disability, delayed motor development and short stature. Hum. Mol. Genet. 2019, 28, 972–979. [Google Scholar] [CrossRef]
- Hirano, A.; Shi, G.; Jones, C.R.; Lipzen, A.; Pennacchio, L.A.; Xu, Y.; Hallows, W.C.; McMahon, T.; Yamazaki, M.; Ptacek, L.J.; et al. A Cryptochrome 2 mutation yields advanced sleep phase in humans. Elife 2016, 5, e16695. [Google Scholar] [CrossRef]
- Machado, R.G.; Eames, B.F. Using Zebrafish to Test the Genetic Basis of Human Craniofacial Diseases. J. Dent. Res. 2017, 96, 1192–1199. [Google Scholar] [CrossRef] [PubMed]
- Carnovali, M.; Banfi, G.; Mariotti, M. Zebrafish Models of Human Skeletal Disorders: Embryo and Adult Swimming Together. Biomed Res. Int. 2019, 2019, 1253710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornille, M.; Dambroise, E.; Komla-Ebri, D.; Kaci, N.; Biosse-Duplan, M.; Di Rocco, F.; Legeai-Mallet, L. Animal models of craniosynostosis. Neurochirurgie 2019, 65, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Vatine, G.; Vallone, D.; Gothilf, Y.; Foulkes, N.S. It’s time to swim! Zebrafish and the circadian clock. FEBS Lett. 2011, 585, 1485–1494. [Google Scholar] [CrossRef]
- Froland Steindal, I.A.; Whitmore, D. Circadian Clocks in Fish-What Have We Learned so far? Biology 2019, 8, 17. [Google Scholar] [CrossRef] [Green Version]
- Foulkes, N.S.; Whitmore, D.; Vallone, D.; Bertolucci, C. Studying the Evolution of the Vertebrate Circadian Clock: The Power of Fish as Comparative Models. Adv. Genet. 2016, 95, 1–30. [Google Scholar]
- Tovin, A.; Alon, S.; Ben-Moshe, Z.; Mracek, P.; Vatine, G.; Foulkes, N.S.; Jacob-Hirsch, J.; Rechavi, G.; Toyama, R.; Coon, S.L.; et al. Systematic identification of rhythmic genes reveals camk1gb as a new element in the circadian clockwork. PLoS Genet. 2012, 8, e1003116. [Google Scholar] [CrossRef]
- Smadja Storz, S.; Tovin, A.; Mracek, P.; Alon, S.; Foulkes, N.S.; Gothilf, Y. Casein kinase 1delta activity: A key element in the zebrafish circadian timing system. PLoS ONE 2013, 8, e54189. [Google Scholar] [CrossRef]
- Ben-Moshe Livne, Z.; Alon, S.; Vallone, D.; Bayleyen, Y.; Tovin, A.; Shainer, I.; Nisembaum, L.G.; Aviram, I.; Smadja-Storz, S.; Fuentes, M.; et al. Genetically Blocking the Zebrafish Pineal Clock Affects Circadian Behavior. PLoS Genet. 2016, 12, e1006445. [Google Scholar] [CrossRef]
- Burgess, H.A.; Granato, M. Modulation of locomotor activity in larval zebrafish during light adaptation. J. Exp. Biol. 2007, 210, 2526–2539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prober, D.A.; Rihel, J.; Onah, A.A.; Sung, R.J.; Schier, A.F. Hypocretin/orexin overexpression induces an insomnia-like phenotype in zebrafish. J. Neurosci. 2006, 26, 13400–13410. [Google Scholar] [CrossRef]
- Falcon, J.; Besseau, L.; Fuentes, M.; Sauzet, S.; Magnanou, E.; Boeuf, G. Structural and functional evolution of the pineal melatonin system in vertebrates. Ann. N. Y. Acad. Sci. 2009, 1163, 101–111. [Google Scholar] [CrossRef]
- Appelbaum, L.; Wang, G.X.; Maro, G.S.; Mori, R.; Tovin, A.; Marin, W.; Yokogawa, T.; Kawakami, K.; Smith, S.J.; Gothilf, Y.; et al. Sleep-wake regulation and hypocretin-melatonin interaction in zebrafish. Proc. Natl. Acad. Sci. USA 2009, 106, 21942–21947. [Google Scholar] [CrossRef] [Green Version]
- Gandhi, A.V.; Mosser, E.A.; Oikonomou, G.; Prober, D.A. Melatonin is required for the circadian regulation of sleep. Neuron 2015, 85, 1193–1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhdanova, I.V. Sleep and its regulation in zebrafish. Rev. Neurosci. 2011, 22, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Zhdanova, I.V.; Wang, S.Y.; Leclair, O.U.; Danilova, N.P. Melatonin promotes sleep-like state in zebrafish. Brain Res. 2001, 903, 263–268. [Google Scholar] [CrossRef]
- Ziv, L.; Levkovitz, S.; Toyama, R.; Falcon, J.; Gothilf, Y. Functional development of the zebrafish pineal gland: Light-induced expression of period2 is required for onset of the circadian clock. J. Neuroendocrinol. 2005, 17, 314–320. [Google Scholar] [CrossRef]
- Gothilf, Y.; Coon, S.L.; Toyama, R.; Chitnis, A.; Namboodiri, M.A.; Klein, D.C. Zebrafish serotonin N-acetyltransferase-2: Marker for development of pineal photoreceptors and circadian clock function. Endocrinology 1999, 140, 4895–4903. [Google Scholar] [CrossRef]
- Vuilleumier, R.; Besseau, L.; Boeuf, G.; Piparelli, A.; Gothilf, Y.; Gehring, W.G.; Klein, D.C.; Falcon, J. Starting the zebrafish pineal circadian clock with a single photic transition. Endocrinology 2006, 147, 2273–2279. [Google Scholar] [CrossRef] [Green Version]
- Vallone, D.; Gondi, S.B.; Whitmore, D.; Foulkes, N.S. E-box function in a period gene repressed by light. Proc. Natl. Acad. Sci. USA 2004, 101, 4106–4111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patke, A.; Murphy, P.J.; Onat, O.E.; Krieger, A.C.; Ozcelik, T.; Campbell, S.S.; Young, M.W. Mutation of the Human Circadian Clock Gene CRY1 in Familial Delayed Sleep Phase Disorder. Cell 2017, 169, 203–215.e13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirota, T.; Lee, J.W.; St John, P.C.; Sawa, M.; Iwaisako, K.; Noguchi, T.; Pongsawakul, P.Y.; Sonntag, T.; Welsh, D.K.; Brenner, D.A.; et al. Identification of small molecule activators of cryptochrome. Science 2012, 337, 1094–1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, H.; Li, Y.; Malek, S.N.; Kim, Y.C.; Xu, J.; Chen, P.; Xiao, F.; Huang, X.; Zhou, X.; Xuan, Z.; et al. New fusion transcripts identified in normal karyotype acute myeloid leukemia. PLoS ONE 2012, 7, e51203. [Google Scholar] [CrossRef]
- Correia, S.P.; Chan, A.B.; Vaughan, M.; Zolboot, N.; Perea, V.; Huber, A.L.; Kriebs, A.; Moresco, J.J.; Yates, J.R., 3rd; Lamia, K.A. The circadian E3 ligase complex SCF(FBXL3+CRY) targets TLK2. Sci. Rep. 2019, 9, 198. [Google Scholar] [CrossRef] [Green Version]
- Huber, A.L.; Papp, S.J.; Chan, A.B.; Henriksson, E.; Jordan, S.D.; Kriebs, A.; Nguyen, M.; Wallace, M.; Li, Z.; Metallo, C.M.; et al. CRY2 and FBXL3 Cooperatively Degrade c-MYC. Mol. Cell 2016, 64, 774–789. [Google Scholar] [CrossRef] [Green Version]
- Abu-Libdeh, B.; Mor-Shaked, H.; Atawna, A.A.; Gillis, D.; Halstuk, O.; Shaul-Lotan, N.; Slae, M.; Sultan, M.; Meiner, V.; Elpeleg, O.; et al. Homozygous variant in MADD, encoding a Rab guanine nucleotide exchange factor, results in pleiotropic effects and a multisystemic disorder. Eur. J. Hum. Genet. 2021, 29, 977–987. [Google Scholar] [CrossRef]
- Daum, H.; Ganapathi, M.; Hirsch, Y.; Griffin, E.L.; LeDuc, C.A.; Hagen, J.; Yagel, S.; Meiner, V.; Chung, W.K.; Mor-Shaked, H. Bi-allelic PAGR1 variants are associated with microcephaly and a severe neurodevelopmental disorder: Genetic evidence from two families. Am. J. Med. Genet. A 2022, 188, 336–342. [Google Scholar] [CrossRef]
- Mor-Shaked, H.; Salah, S.; Yanovsky-Dagan, S.; Meiner, V.; Atawneh, O.M.; Abu-Libdeh, B.; Elpeleg, O.; Harel, T. Biallelic deletion in a minimal CAPN15 intron in siblings with a recognizable syndrome of congenital malformations and developmental delay. Clin. Genet. 2021, 99, 577–582. [Google Scholar] [CrossRef]
- Vallone, D.; Santoriello, C.; Gondi, S.B.; Foulkes, N.S. Basic protocols for zebrafish cell lines: Maintenance and transfection. Methods Mol. Biol. 2007, 362, 429–441. [Google Scholar]
- Jammalamadaka, S.R.; Sengupta, A. Topics in Circular Statistics; World Scientific: Singapore, 2001. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Cavallari, N.; Frigato, E.; Vallone, D.; Frohlich, N.; Lopez-Olmeda, J.F.; Foa, A.; Berti, R.; Sanchez-Vazquez, F.J.; Bertolucci, C.; Foulkes, N.S. A blind circadian clock in cavefish reveals that opsins mediate peripheral clock photoreception. PLoS Biol. 2011, 9, e1001142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Fonio, E.; Galili, T.; Havkin, G.Z.; Golani, I. Quantifying the buildup in extent and complexity of free exploration in mice. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S3), 15580–15587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elbaz, I.; Yelin-Bekerman, L.; Nicenboim, J.; Vatine, G.; Appelbaum, L. Genetic ablation of hypocretin neurons alters behavioral state transitions in zebrafish. J. Neurosci. 2012, 32, 12961–12972. [Google Scholar] [CrossRef] [Green Version]
- Gavaia, P.J.; Sarasquete, C.; Cancela, M.L. Detection of mineralized structures in early stages of development of marine Teleostei using a modified alcian blue-alizarin red double staining technique for bone and cartilage. Biotech. Histochem. 2000, 75, 79–84. [Google Scholar] [CrossRef]
- Walker, M.B.; Kimmel, C.B. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem. 2007, 82, 23–28. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Confino, S.; Dor, T.; Tovin, A.; Wexler, Y.; Ben-Moshe Livne, Z.; Kolker, M.; Pisanty, O.; Park, S.K.; Geyer, N.; Reiter, J.; et al. A Zebrafish Model for a Rare Genetic Disease Reveals a Conserved Role for FBXL3 in the Circadian Clock System. Int. J. Mol. Sci. 2022, 23, 2373. https://doi.org/10.3390/ijms23042373
Confino S, Dor T, Tovin A, Wexler Y, Ben-Moshe Livne Z, Kolker M, Pisanty O, Park SK, Geyer N, Reiter J, et al. A Zebrafish Model for a Rare Genetic Disease Reveals a Conserved Role for FBXL3 in the Circadian Clock System. International Journal of Molecular Sciences. 2022; 23(4):2373. https://doi.org/10.3390/ijms23042373
Chicago/Turabian StyleConfino, Shir, Talya Dor, Adi Tovin, Yair Wexler, Zohar Ben-Moshe Livne, Michaela Kolker, Odelia Pisanty, Sohyun Kathy Park, Nathalie Geyer, Joel Reiter, and et al. 2022. "A Zebrafish Model for a Rare Genetic Disease Reveals a Conserved Role for FBXL3 in the Circadian Clock System" International Journal of Molecular Sciences 23, no. 4: 2373. https://doi.org/10.3390/ijms23042373
APA StyleConfino, S., Dor, T., Tovin, A., Wexler, Y., Ben-Moshe Livne, Z., Kolker, M., Pisanty, O., Park, S. K., Geyer, N., Reiter, J., Edvardson, S., Mor-Shaked, H., Elpeleg, O., Vallone, D., Appelbaum, L., Foulkes, N. S., & Gothilf, Y. (2022). A Zebrafish Model for a Rare Genetic Disease Reveals a Conserved Role for FBXL3 in the Circadian Clock System. International Journal of Molecular Sciences, 23(4), 2373. https://doi.org/10.3390/ijms23042373





