Calcium Signaling in the Cerebellar Radial Glia and Its Association with Morphological Changes during Zebrafish Development
Abstract
:1. Introduction
2. Results
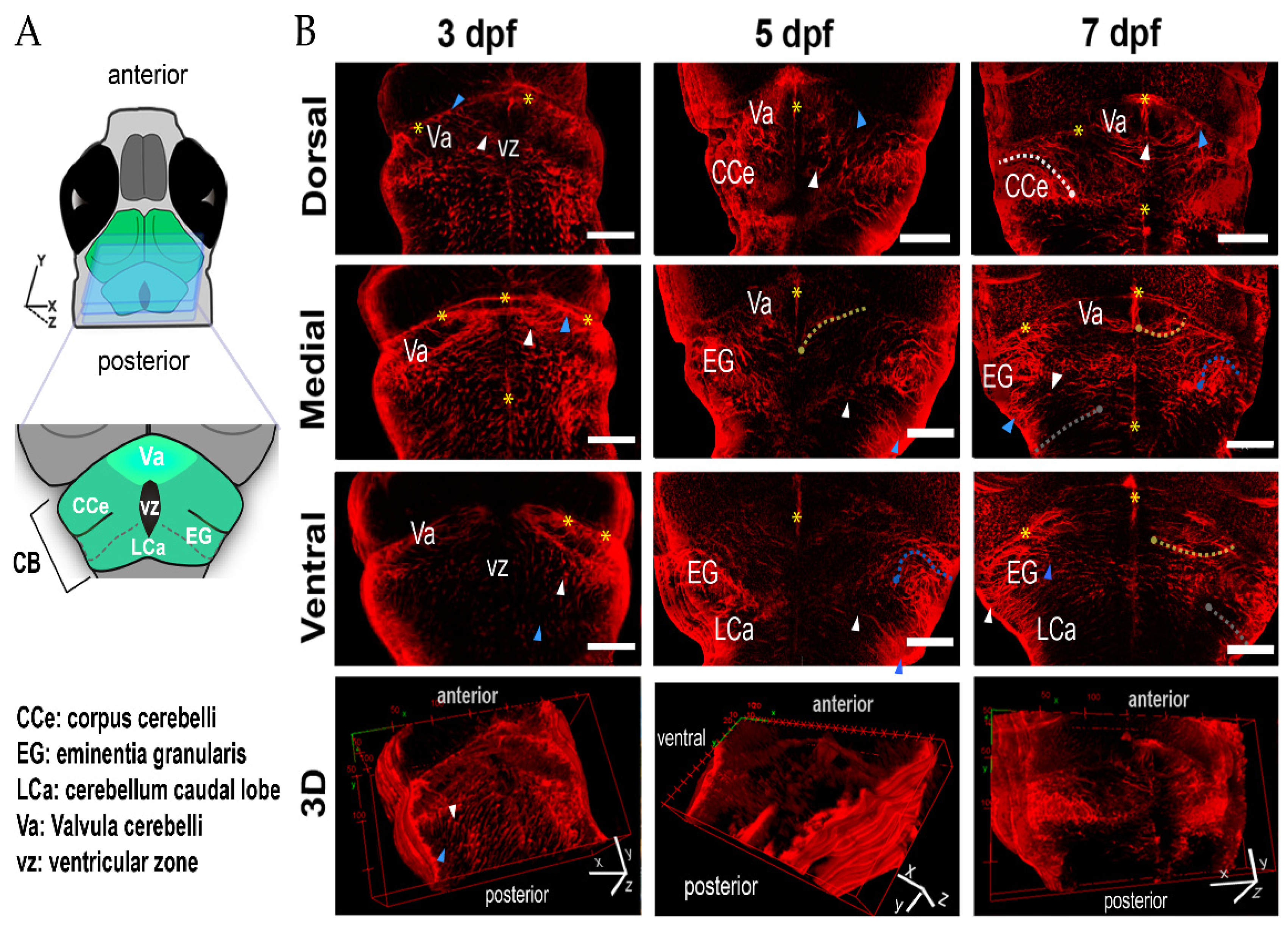
2.1. Distribution of Radial Glia in the Larval Zebrafish Cerebellum
2.2. Morphometric Analysis of Radial Glia during Development
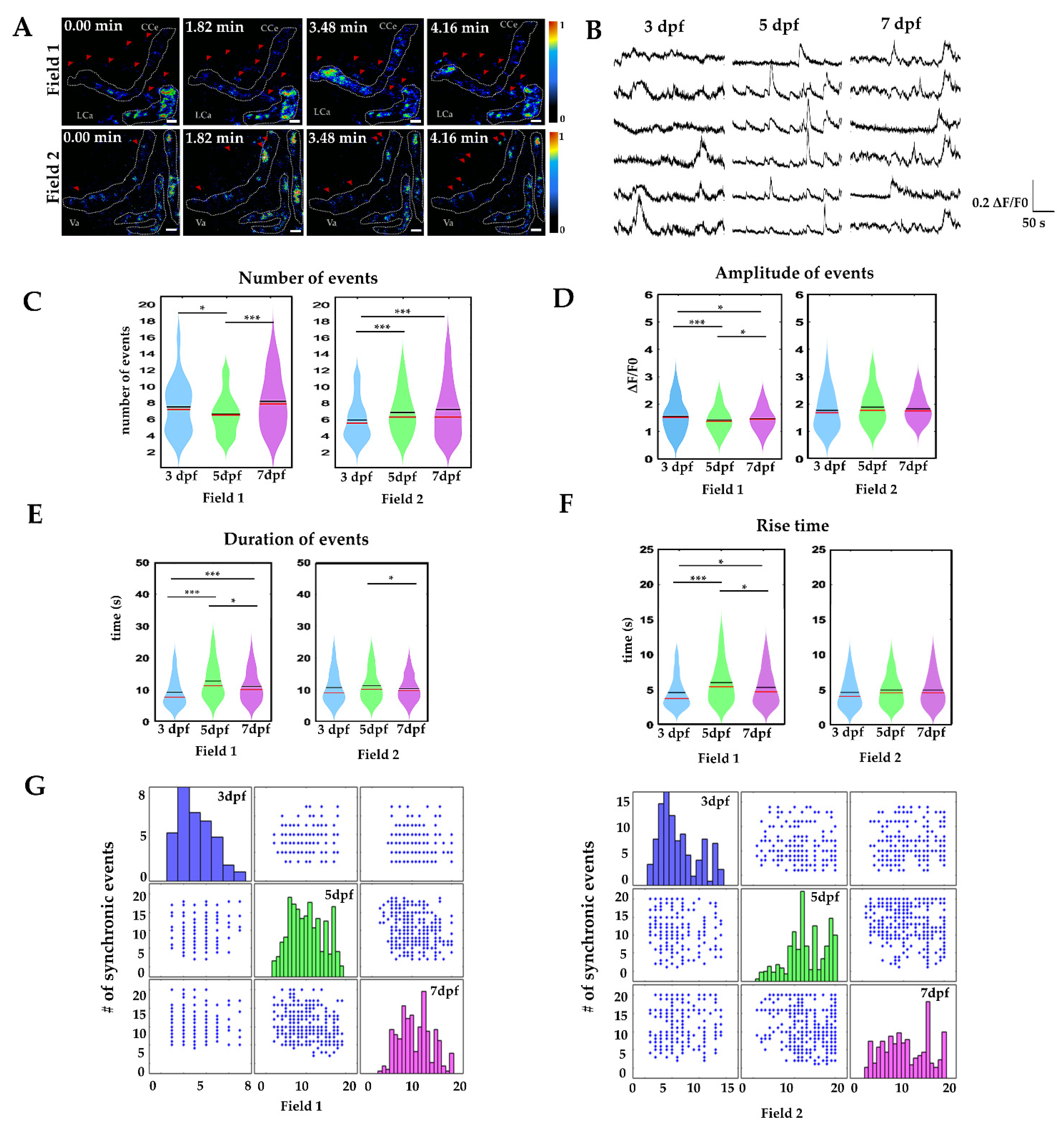
2.3. Calcium Signaling in Radial Glia during Development
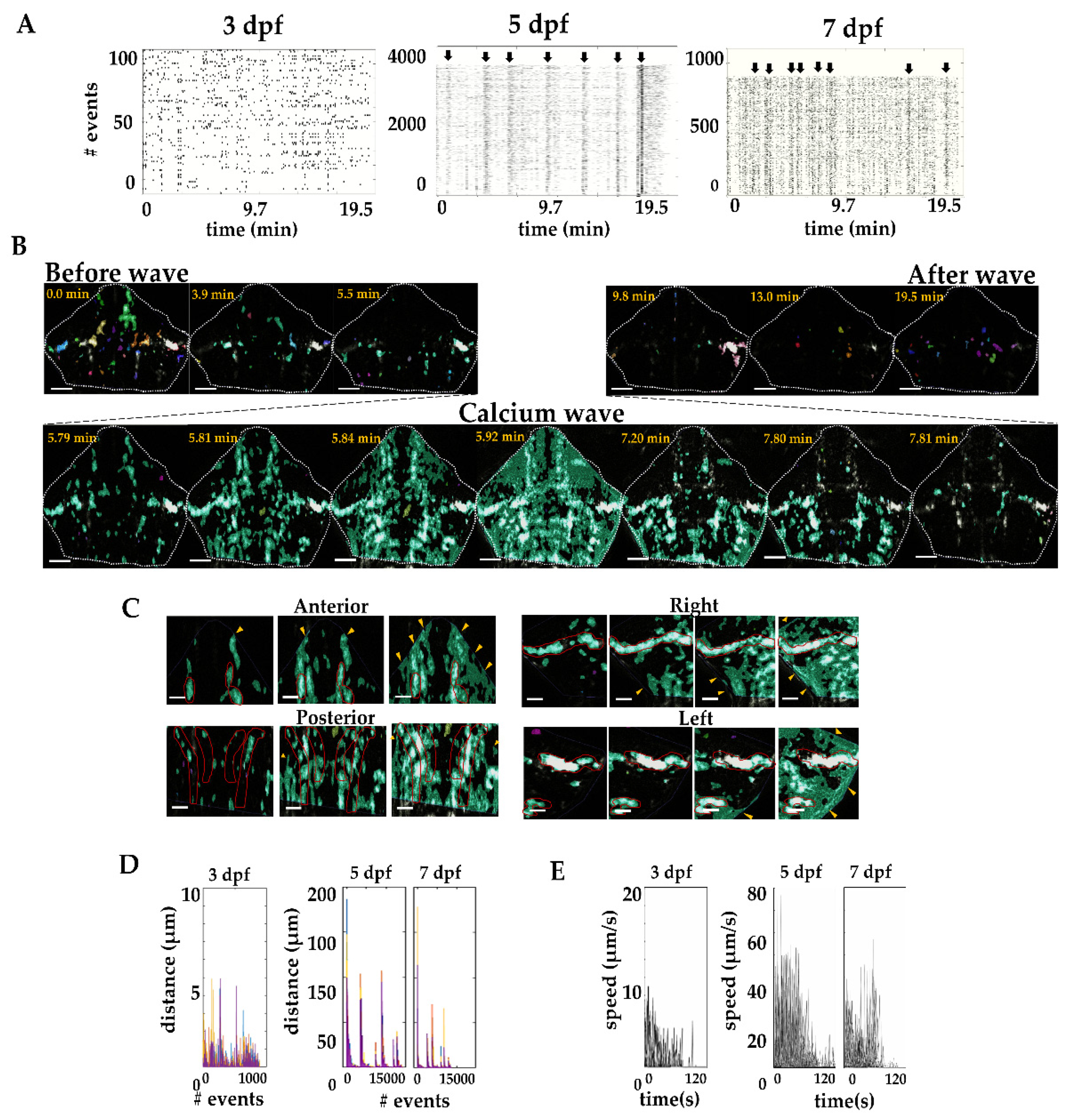
2.4. Calcium Waves in the Developing Cerebellum
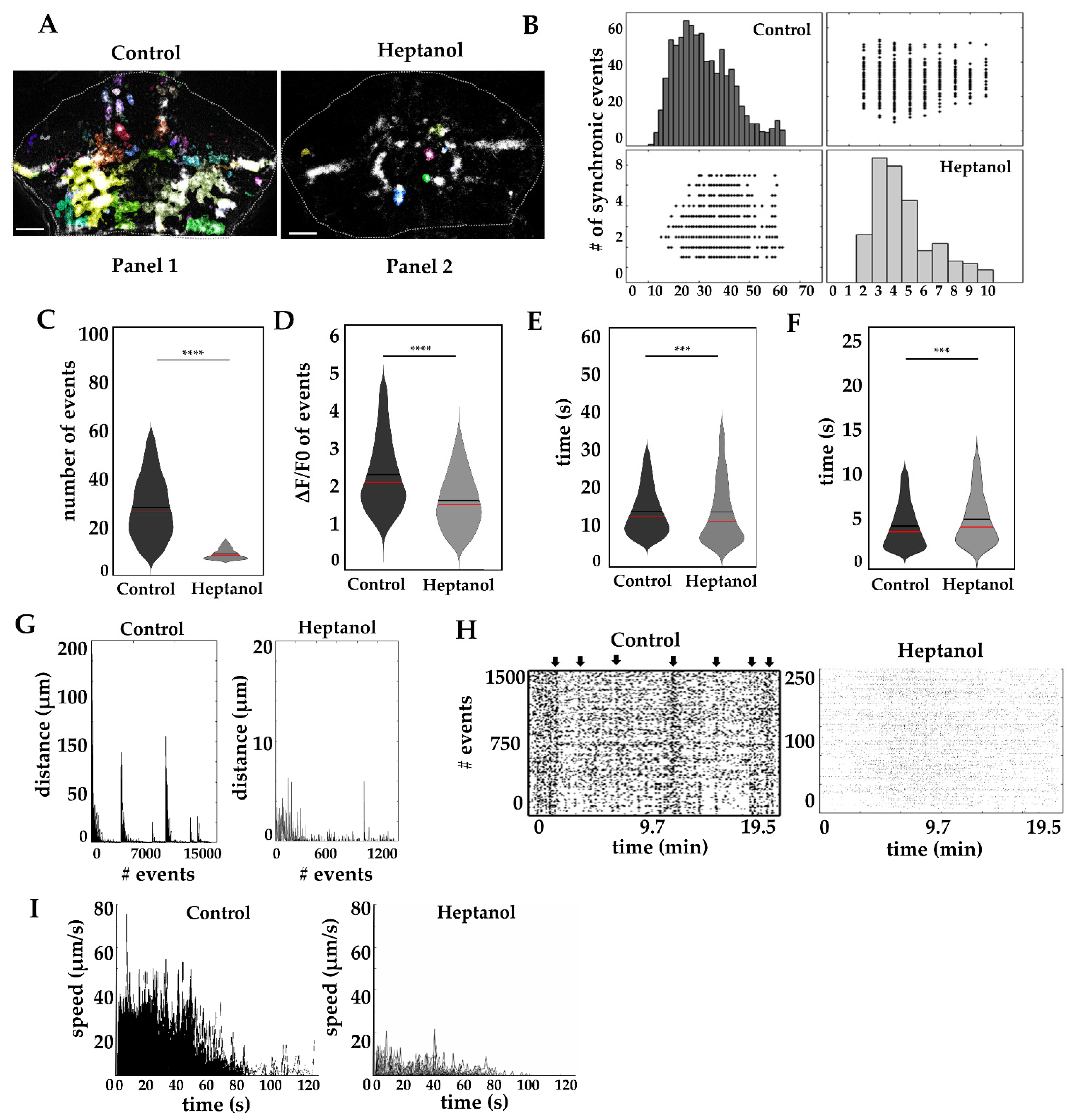
2.5. Effect of Gap Junction Blockage on Calcium Waves
3. Discussion
4. Materials and Methods
4.1. Zebrafish Housing Care
4.2. Molecular Biology
4.3. Generation of the Tg (pTol2-gfap-GCaMP6s)
4.4. Whole Mount Immunohistochemistry
4.5. Confocal Imaging
4.6. Morphological Analysis
4.7. Statistical Analysis of Radial Glial Cell Morphology
4.8. In Vivo Calcium Imaging
4.9. In Vivo Pharmacological Blockage of Gap Junctions
4.10. Analysis and Statistics of Calcium Imaging
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koziol, L.F.; Budding, D.; Andreasen, N.; D’Arrigo, S.; Bulgheroni, S.; Imamizu, H.; Ito, M.; Manto, M.; Marvel, C.; Parker, K.; et al. Consensus Paper: The Cerebellum’s Role in Movement and Cognition. Cerebellum 2014, 13, 151–177. [Google Scholar] [CrossRef]
- Schmahmann, J.D. The Cerebellum and Cognition. Neurosci. Lett. 2019, 688, 62–75. [Google Scholar] [CrossRef]
- Manto, M.; Gandini, J.; Feil, K.; Strupp, M. Cerebellar Ataxias: An Update. Curr. Opin. Neurol. 2020, 33, 150–160. [Google Scholar] [CrossRef] [PubMed]
- van der Heijden, M.E.; Gill, J.S.; Sillitoe, R.V. Abnormal Cerebellar Development in Autism Spectrum Disorders. Dev. Neurosci. 2021, 43, 181–190. [Google Scholar] [CrossRef]
- Wang, S.S.-H.; Kloth, A.D.; Badura, A. The Cerebellum, Sensitive Periods, and Autism. Neuron 2014, 83, 518–532. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, M.; Hibi, M. Development and Evolution of Cerebellar Neural Circuits. Dev. Growth Differ. 2012, 54, 373–389. [Google Scholar] [CrossRef] [PubMed]
- Hibi, M.; Matsuda, K.; Takeuchi, M.; Shimizu, T.; Murakami, Y. Evolutionary Mechanisms That Generate Morphology and Neural-Circuit Diversity of the Cerebellum. Dev. Growth Differ. 2017, 59, 228–243. [Google Scholar] [CrossRef]
- Matsui, H.; Namikawa, K.; Babaryka, A.; Koster, R.W. Functional Regionalization of the Teleost Cerebellum Analyzed in Vivo. Proc. Natl. Acad. Sci. USA 2014, 111, 11846–11851. [Google Scholar] [CrossRef]
- Jurisch-Yaksi, N.; Yaksi, E.; Kizil, C. Radial Glia in the Zebrafish Brain: Functional, Structural, and Physiological Comparison with the Mammalian Glia. Glia 2020, 68, 2451–2470. [Google Scholar] [CrossRef]
- Takeuchi, M.; Matsuda, K.; Yamaguchi, S.; Asakawa, K.; Miyasaka, N.; Lal, P.; Yoshihara, Y.; Koga, A.; Kawakami, K.; Shimizu, T.; et al. Establishment of Gal4 Transgenic Zebrafish Lines for Analysis of Development of Cerebellar Neural Circuitry. Dev. Biol. 2015, 397, 1–17. [Google Scholar] [CrossRef]
- Hamling, K.R.; Tobias, Z.J.C.; Weissman, T.A. Mapping the Development of Cerebellar Purkinje Cells in Zebrafish: Purkinje Cell Development in Zebrafish. Dev. Neurobiol. 2015, 75, 1174–1188. [Google Scholar] [CrossRef]
- Kaslin, J.; Kroehne, V.; Ganz, J.; Hans, S.; Brand, M. Distinct Roles of Neuroepithelial-like and Radial Glia-like Progenitor Cells in Cerebellar Regeneration. Development 2017, 144, 1462–1471. [Google Scholar] [CrossRef]
- Than-Trong, E.; Bally-Cuif, L. Radial Glia and Neural Progenitors in the Adult Zebrafish Central Nervous System: Adult Radial Glia in Zebrafish. Glia 2015, 63, 1406–1428. [Google Scholar] [CrossRef]
- Bae, Y.K.; Kani, S.; Shimizu, T.; Tanabe, K.; Nojima, H.; Kimura, Y.; Higashijima, S.; Hibi, M. Anatomy of zebrafish cerebellum and screen for mutations affecting its development. Dev. Biol. 2009, 330, 406–426. [Google Scholar] [CrossRef]
- Hsieh, J.-Y.; Ulrich, B.; Issa, F.A.; Wan, J.; Papazian, D.M. Rapid Development of Purkinje Cell Excitability, Functional Cerebellar Circuit, and Afferent Sensory Input to Cerebellum in Zebrafish. Front. Neural Circuits 2014, 8, 147. [Google Scholar] [CrossRef]
- Aizenberg, M.; Schuman, E.M. Cerebellar-Dependent Learning in Larval Zebrafish. J. Neurosci. 2011, 31, 8708–8712. [Google Scholar] [CrossRef]
- Hibi, M.; Shimizu, T. Development of the Cerebellum and Cerebellar Neural Circuits. Dev. Neurobiol. 2012, 72, 282–301. [Google Scholar] [CrossRef] [PubMed]
- Ekström, P.; Johnsson, C.-M.; Ohlin, L.-M. Ventricular Proliferation Zones in the Brain of an Adult Teleost Fish and Their Relation to Neuromeres and Migration (Secondary Matrix) Zones: Proliferation Zones in Adult Teleost Brain. J. Comp. Neurol. 2001, 436, 92–110. [Google Scholar] [CrossRef] [PubMed]
- Lange, C.; Rost, F.; Machate, A.; Reinhardt, S.; Lesche, M.; Weber, A.; Kuscha, V.; Dahl, A.; Rulands, S.; Brand, M. Single Cell Sequencing of Radial Glia Progeny Reveals Diversity of Newborn Neurons in the Adult Zebrafish Brain. Development 2020, 147, dev185595. [Google Scholar] [CrossRef] [PubMed]
- Zupanc, G.K.H.; Hinsch, K.; Gage, F.H. Proliferation, Migration, Neuronal Differentiation, and Long-Term Survival of New Cells in the Adult Zebrafish Brain. J. Comp. Neurol. 2005, 488, 290–319. [Google Scholar] [CrossRef] [PubMed]
- Semyanov, A.; Henneberger, C.; Agarwal, A. Making Sense of Astrocytic Calcium Signals—From Acquisition to Interpretation. Nat. Rev. Neurosci. 2020, 21, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Shigetomi, E.; Patel, S.; Khakh, B.S. Probing the Complexities of Astrocyte Calcium Signaling. Trends Cell Biol. 2016, 26, 300–312. [Google Scholar] [CrossRef]
- Weissman, T.A.; Riquelme, P.A.; Ivic, L.; Flint, A.C.; Kriegstein, A.R. Calcium Waves Propagate through Radial Glial Cells and Modulate Proliferation in the Developing Neocortex. Neuron 2004, 43, 647–661. [Google Scholar] [CrossRef]
- Denizot, A.; Arizono, M.; Nägerl, U.V.; Soula, H.; Berry, H. Simulation of Calcium Signaling in Fine Astrocytic Processes: Effect of Spatial Properties on Spontaneous Activity. PLoS Comput. Biol. 2019, 15, e1006795. [Google Scholar] [CrossRef] [PubMed]
- Nett, W.J.; Oloff, S.H.; McCarthy, K.D. Hippocampal Astrocytes In Situ Exhibit Calcium Oscillations That Occur Independent of Neuronal Activity. J. Neurophysiol. 2002, 87, 528–537. [Google Scholar] [CrossRef]
- Nimmerjahn, A.; Mukamel, E.A.; Schnitzer, M.J. Motor Behavior Activates Bergmann Glial Networks. Neuron 2009, 62, 400–412. [Google Scholar] [CrossRef]
- Morita, M.; Higuchi, C.; Moto, T.; Kozuka, N.; Susuki, J.; Itofusa, R.; Yamashita, J.; Kudo, Y. Dual Regulation of Calcium Oscillation in Astrocytes by Growth Factors and Pro-Inflammatory Cytokines via the Mitogen-Activated Protein Kinase Cascade. J. Neurosci. 2003, 23, 10944–10952. [Google Scholar] [CrossRef]
- Lines, J.; Martin, E.D.; Kofuji, P.; Aguilar, J.; Araque, A. Astrocytes Modulate Sensory-Evoked Neuronal Network Activity. Nat. Commun. 2020, 11, 3689. [Google Scholar] [CrossRef]
- Perea, G. Properties of Synaptically Evoked Astrocyte Calcium Signal Reveal Synaptic Information Processing by Astrocytes. J. Neurosci. 2005, 25, 2192–2203. [Google Scholar] [CrossRef] [PubMed]
- Hoogland, T.M.; Kuhn, B.; Gobel, W.; Huang, W.; Nakai, J.; Helmchen, F.; Flint, J.; Wang, S.S.-H. Radially Expanding Transglial Calcium Waves in the Intact Cerebellum. Proc. Natl. Acad. Sci. USA 2009, 106, 3496–3501. [Google Scholar] [CrossRef]
- Mathiesen, C.; Brazhe, A.; Thomsen, K.; Lauritzen, M. Spontaneous Calcium Waves in Bergman Glia Increase with Age and Hypoxia and May Reduce Tissue Oxygen. J. Cereb. Blood Flow Metab. 2013, 33, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Plattner, H.; Verkhratsky, A. Evolution of Calcium Signalling. Cell Calcium 2015, 57, 121–122. [Google Scholar] [CrossRef]
- Liu, J.; Salvati, K.A.; Baraban, S.C. In Vivo Calcium Imaging Reveals Disordered Interictal Network Dynamics in Epileptic Stxbp1b Zebrafish. iScience 2021, 24, 102558. [Google Scholar] [CrossRef] [PubMed]
- Arellano, J.I.; Morozov, Y.M.; Micali, N.; Rakic, P. Radial Glial Cells: New Views on Old Questions. Neurochem. Res. 2021, 46, 2512–2524. [Google Scholar] [CrossRef]
- Betizeau, M.; Cortay, V.; Patti, D.; Pfister, S.; Gautier, E.; Bellemin-Ménard, A.; Afanassieff, M.; Huissoud, C.; Douglas, R.J.; Kennedy, H.; et al. Precursor Diversity and Complexity of Lineage Relationships in the Outer Subventricular Zone of the Primate. Neuron 2013, 80, 442–457. [Google Scholar] [CrossRef]
- Nowakowski, T.J.; Pollen, A.A.; Sandoval-Espinosa, C.; Kriegstein, A.R. Transformation of the Radial Glia Scaffold Demarcates Two Stages of Human Cerebral Cortex Development. Neuron 2016, 91, 1219–1227. [Google Scholar] [CrossRef]
- Rakic, P. Mode of Cell Migration to the Superficial Layers of Fetal Monkey Neocortex. J. Comp. Neurol. 1972, 145, 61–83. [Google Scholar] [CrossRef]
- Nowakowski, T.J.; Bhaduri, A.; Pollen, A.A.; Alvarado, B.; Mostajo-Radji, M.A.; Di Lullo, E.; Haeussler, M.; Sandoval-Espinosa, C.; Liu, S.J.; Velmeshev, D.; et al. Spatiotemporal Gene Expression Trajectories Reveal Developmental Hierarchies of the Human Cortex. Science 2017, 358, 1318–1323. [Google Scholar] [CrossRef]
- Raj, B.; Wagner, D.E.; McKenna, A.; Pandey, S.; Klein, A.M.; Shendure, J.; Gagnon, J.A.; Schier, A.F. Simultaneous Single-Cell Profiling of Lineages and Cell Types in the Vertebrate Brain. Nat. Biotechnol. 2018, 36, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Ashworth, R.; Devogelaere, B.; Fabes, J.; Tunwell, R.E.; Koh, K.R.; De Smedt, H.; Patel, S. Molecular and Functional Characterization of Inositol Trisphosphate Receptors during Early Zebrafish Development. J. Biol. Chem. 2007, 282, 13984–13993. [Google Scholar] [CrossRef]
- Chatterjee, B.; Chin, A.J.; Valdimarsson, G.; Finis, C.; Sonntag, J.M.; Choi, B.Y.; Tao, L.; Balasubramanian, K.; Bell, C.; Krufka, A.; et al. Developmental Regulation and Expression of the Zebrafish Connexin43 Gene. Dev. Dyn. 2005, 233, 890–906. [Google Scholar] [CrossRef] [PubMed]
- Lo, J. 15,000 Unique Zebrafish EST Clusters and Their Future Use in Microarray for Profiling Gene Expression Patterns During Embryogenesis. Genome Res. 2003, 13, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Poskanzer, K.E.; Freeman, M.R.; Monk, K.R. Live-Imaging of Astrocyte Morphogenesis and Function in Zebrafish Neural Circuits. Nat. Neurosci. 2020, 23, 1297–1306. [Google Scholar] [CrossRef]
- Gu, X.; Spitzer, N.C. Distinct Aspects of Neuronal Differentiation Encoded by Frequency of Spontaneous Ca2+ Transients. Nature 1995, 375, 784–787. [Google Scholar] [CrossRef]
- Scemes, E.; Duval, N.; Meda, P. Reduced Expression of P2Y1 Receptors in Connexin43-Null Mice Alters Calcium Signaling and Migration of Neural Progenitor Cells. J. Neurosci. 2003, 23, 11444–11452. [Google Scholar] [CrossRef]
- Lawrence, C. Advances in Zebrafish Husbandry and Management. In Methods in Cell Biology; Elsevier: Cambridge, MA, USA, 2011; Volume 104, pp. 429–451. ISBN 978-0-12-374814-0. [Google Scholar]
- Norton, A.; Franse, K.F.; Daw, T.; Gordon, L.; Vitiello, P.F.; Kinkel, M.D. Larval Rearing Methods for Small-Scale Production of Healthy Zebrafish. East. Biol. 2019, 2019, 21. [Google Scholar]
- Kawakami, K.; Patton, E.E.; Orger, M. Zebrafish: Methods and Protocols. In Methods in Molecular Biology; Springer: New York, NY, USA, 2016; Volume 1451, ISBN 978-1-4939-3769-1. [Google Scholar]
- Kawakami, K. Tol2: A Versatile Gene Transfer Vector in Vertebrates. Genome Biol. 2007, 8, S7. [Google Scholar] [CrossRef] [PubMed]
- Sheets, L.; Trapani, J.G.; Mo, W.; Obholzer, N.; Nicolson, T. Ribeye Is Required for Presynaptic CaV1.3a Channel Localization and Afferent Innervation of Sensory Hair Cells. Development 2011, 138, 1309–1319. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Kaslin, J.; Brand, M. The Zebrafish Cerebellum. In Essentials of Cerebellum and Cerebellar Disoders; Springer International Publishing: Cham, Switzerland, 2016; Volume 56, pp. 411–421. ISBN 978-3-319-24551-5. [Google Scholar]
- Miyazawa, H.; Okumura, K.; Hiyoshi, K.; Maruyama, K.; Kakinuma, H.; Amo, R.; Okamoto, H.; Yamasu, K.; Tsuda, S. Optical Interrogation of Neuronal Circuitry in Zebrafish Using Genetically Encoded Voltage Indicators. Sci. Rep. 2018, 8, 6048. [Google Scholar] [CrossRef]
- Parrock, S.; Hussain, S.; Issler, N.; Differ, A.-M.; Lench, N.; Guarino, S.; Oosterveld, M.J.S.; Keijzer-Veen, M.; Brilstra, E.; van Wieringen, H.; et al. KCNJ10 Mutations Display Differential Sensitivity to Heteromerisation with KCNJ16. Nephron Physiol. 2013, 123, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, A.; Mickoleit, M.; Weber, M.; Huisken, J. Multilayer Mounting Enables Long-Term Imaging of Zebrafish Development in a Light Sheet Microscope. Development 2012, 139, 3242–3247. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; Mickoleit, M.; Huisken, J. Multilayer Mounting for Long-Term Light Sheet Microscopy of Zebrafish. J. Vis. Exp. 2014, 51119. [Google Scholar] [CrossRef] [PubMed]
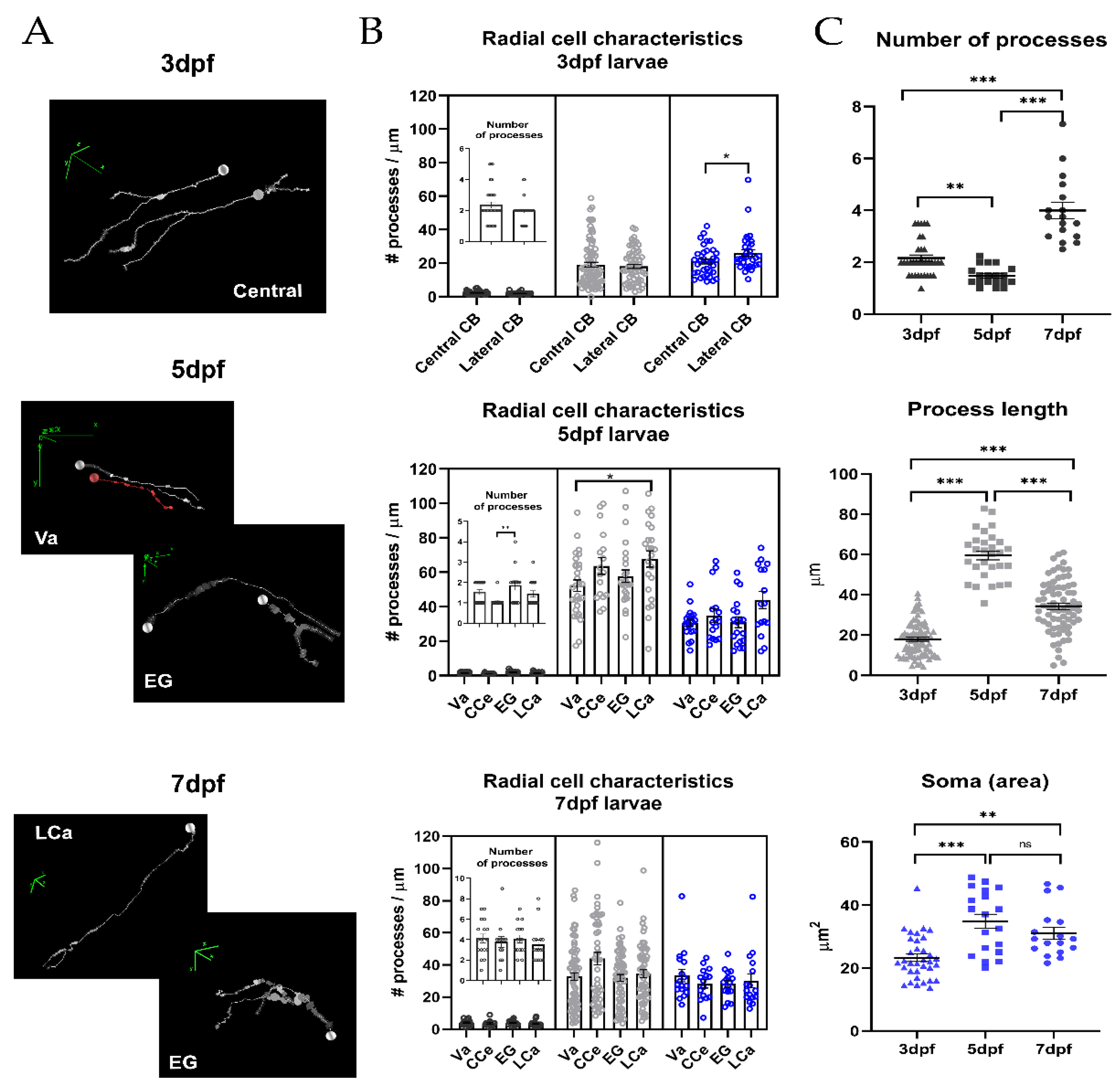
| Age | Number of Processes | Process Length (µm) | Area of Soma (µm2) |
|---|---|---|---|
| 3 dpf | 2.16 ± 0.12 | 17.89 ± 0.97 | 23.28 ± 1.19 |
| 5 dpf | 1.48 ± 0.09 | 59.53 ± 2.17 | 34.87 ± 2.27 |
| 7 dpf | 3.99 ± 0.31 | 34.37 ± 1.56 | 31.13 ± 1.90 |
| Age | Number of Processes | Process Length (µm) | Area of Soma (µm2) | |||
|---|---|---|---|---|---|---|
| 3 dpf | Lateral 1 | Central 1 | Lateral 1 | Central 1 | Lateral 1 | Central 1 |
| 2.35 ±0.20 | 1.97 ± 0.12 | 19.10 ± 1.50 | 18.10 ± 1.24 | 21.00 ± 1.51 | 26.20 ± 2.03 | |
| 5 dpf | Va 1 | CCe 1 | Va 1 | CCe 1 | Va 1 | CCe 1 |
| 52.2 ± 0.12 | 63.6 ± 0.06 | 30.7 ± 3.3 | 34.8 ± 4.9 | 21.2 ± 3.0 | 22.7 ± 4.1 | |
| EG 1 | LCa 1 | EG 1 | LCa 1 | EG 1 | LCa 1 | |
| 57.70 ± 0.23 | 67.7 ± 0.16 | 31.0 ± 3.7 | 43.9 ± 4.7 | 21.7 ± 3.4 | 28.9 ± 5.2 | |
| 7 dpf | Va 1 | CCe 1 | Va 1 | CCe 1 | Va 1 | CCe 1 |
| 4.12 ± 0.43 | 3.77 ± 0.53 | 32.9 ± 2.4 | 44.1 ± 3.9 | 33.5 ± 3.8 | 28.4 ± 2.6 | |
| EG 1 | LCa 1 | EG 1 | LCa 1 | EG 1 | LCa 1 | |
| 4.06 ± 0.37 | 3.50 ± 0.44 | 32.1 ± 2.1 | 34.7 ± 2.5 | 28.4 ± 2.2 | 30.3 ± 4.4 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pereida-Jaramillo, E.; Gómez-González, G.B.; Espino-Saldaña, A.E.; Martínez-Torres, A. Calcium Signaling in the Cerebellar Radial Glia and Its Association with Morphological Changes during Zebrafish Development. Int. J. Mol. Sci. 2021, 22, 13509. https://doi.org/10.3390/ijms222413509
Pereida-Jaramillo E, Gómez-González GB, Espino-Saldaña AE, Martínez-Torres A. Calcium Signaling in the Cerebellar Radial Glia and Its Association with Morphological Changes during Zebrafish Development. International Journal of Molecular Sciences. 2021; 22(24):13509. https://doi.org/10.3390/ijms222413509
Chicago/Turabian StylePereida-Jaramillo, Elizabeth, Gabriela B. Gómez-González, Angeles Edith Espino-Saldaña, and Ataúlfo Martínez-Torres. 2021. "Calcium Signaling in the Cerebellar Radial Glia and Its Association with Morphological Changes during Zebrafish Development" International Journal of Molecular Sciences 22, no. 24: 13509. https://doi.org/10.3390/ijms222413509
APA StylePereida-Jaramillo, E., Gómez-González, G. B., Espino-Saldaña, A. E., & Martínez-Torres, A. (2021). Calcium Signaling in the Cerebellar Radial Glia and Its Association with Morphological Changes during Zebrafish Development. International Journal of Molecular Sciences, 22(24), 13509. https://doi.org/10.3390/ijms222413509






