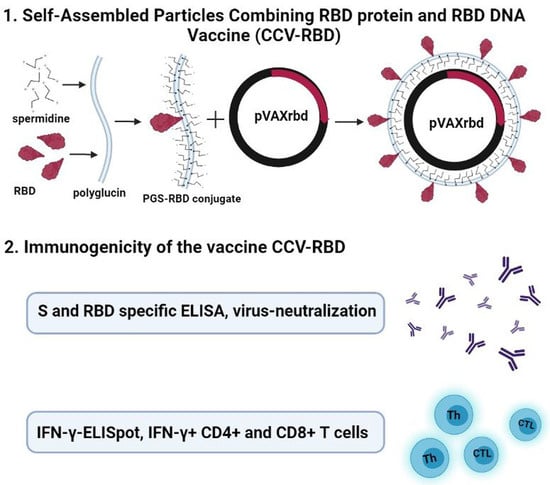
Self-Assembled Particles Combining SARS-CoV-2 RBD Protein and RBD DNA Vaccine Induce Synergistic Enhancement of the Humoral Response in Mice
Abstract
:1. Introduction
2. Results
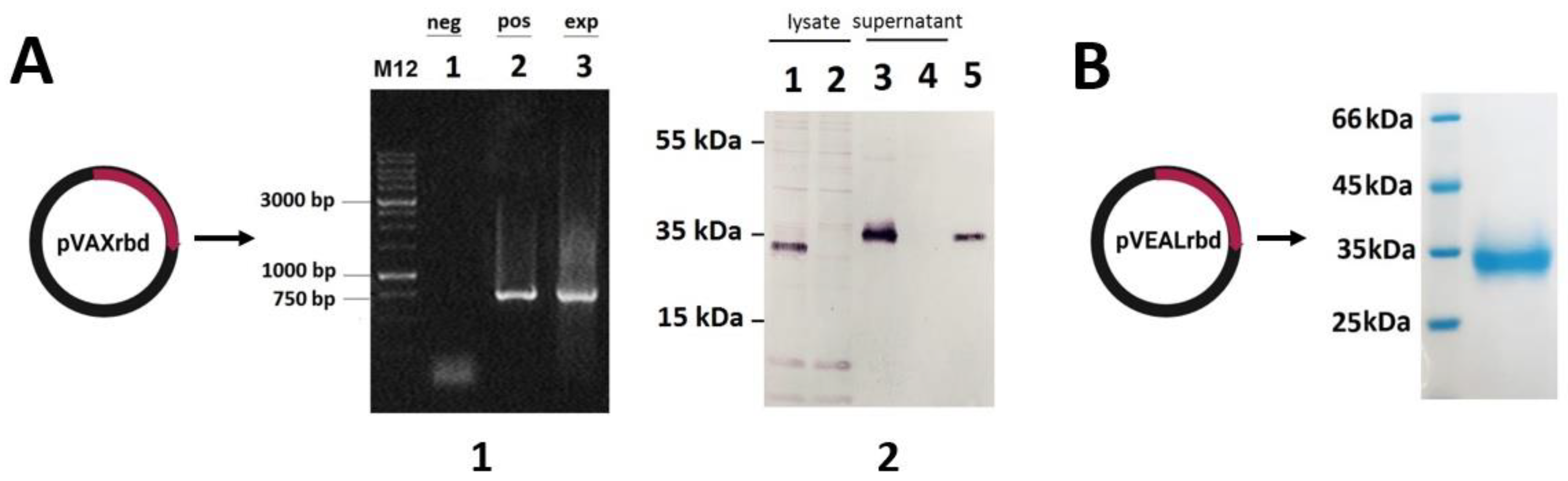
2.1. Verification of RBD Gene Transcription in Transfected HEK293T Cells
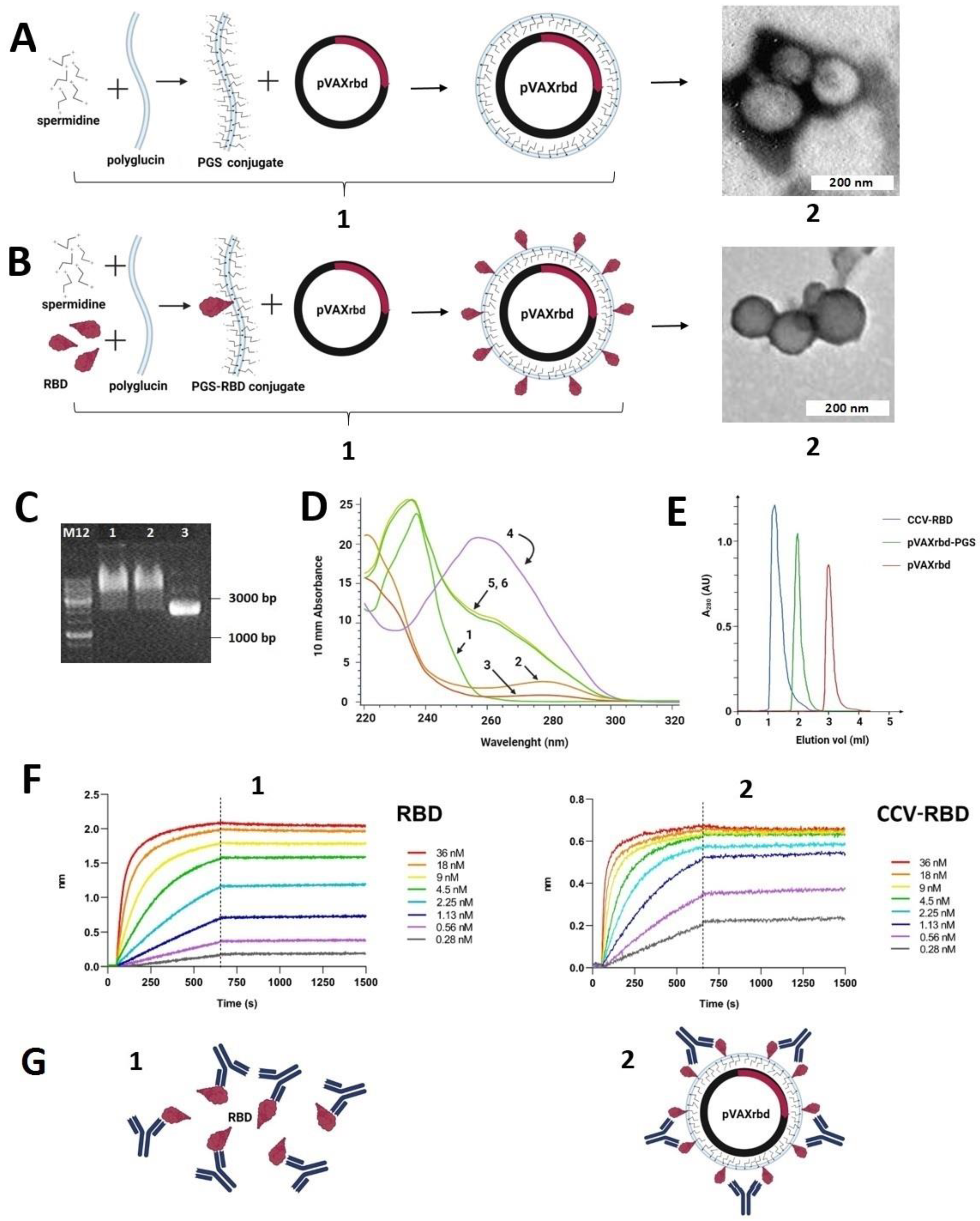
2.2. Preparation and Characterization of the Particles
2.3. Humoral Immune Response
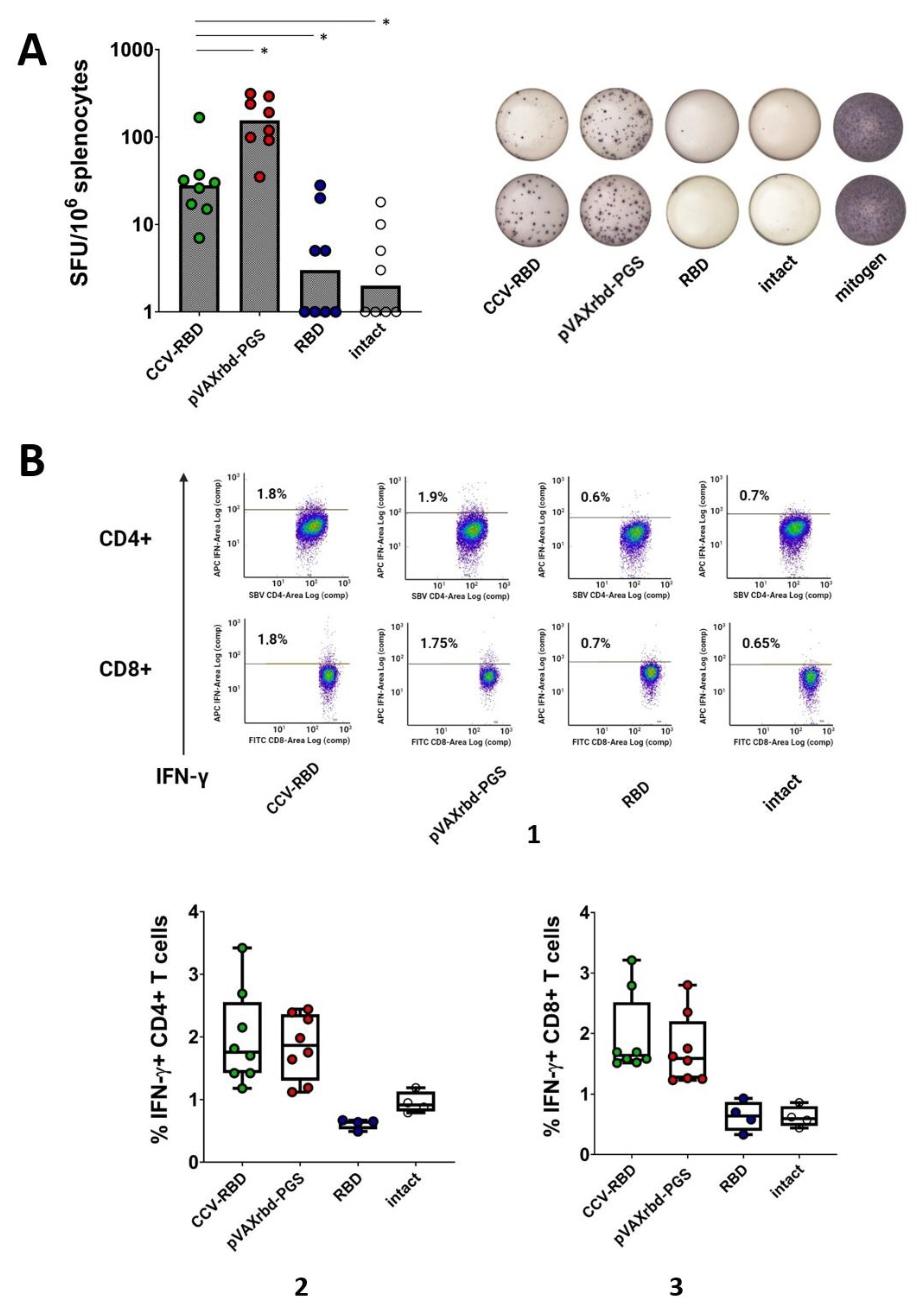
2.4. Cellular Immune Response
3. Discussion
4. Materials and Methods
4.1. Viruses and Cell Cultures
4.2. DNA Vaccine Construction and Protein Production
4.3. In Vitro Investigation of Transgene Expression
4.4. Purification of the Recombinant RBD Protein
4.5. Synthesis of PGS and PGS-RBD Conjugates
4.6. DNA Packaging into PGS and PGS-RBD Conjugates
4.7. Biolayer Interferometry
4.8. Mouse Immunization
4.9. ELISA
4.10. Virus-Neutralizing Assay
4.11. IFN-γ ELISpot
4.12. ICS
4.13. Software and Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Morens, D.M.; Taubenberger, J.K.; Fauci, A.S. Universal Coronavirus Vaccines—An Urgent Need. N. Engl. J. Med. 2022, 386, 297–299. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Zhang, Y.; Wang, Y.; Wang, H.; Yang, Y.; Gao, G.F.; Tan, W.; Wu, G.; Xu, M.; Lou, Z.; et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: A randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect. Dis. 2021, 21, 39–51. [Google Scholar] [CrossRef]
- Folegatti, P.M.; Ewer, K.J.; Aley, P.K.; Angus, B.; Becker, S.; Belij-Rammerstorfer, S.; Bellamy, D.; Bibi, S.; Bittaye, M.; Clutterbuck, E.A.; et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: A preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 2020, 396, 467–478. [Google Scholar] [CrossRef]
- Logunov, D.Y.; Dolzhikova, I.V.; Zubkova, O.V.; Tukhvatullin, A.I.; Shcheblyakov, D.V.; Dzharullaeva, A.S.; Grousova, D.M.; Erokhova, A.S.; Kovyrshina, A.V.; Botikov, A.G.; et al. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: Two open, non-randomised phase 1/2 studies from Russia. Lancet 2020, 396, 887–897. [Google Scholar] [CrossRef]
- COVID-19 Vaccine Tracker and Landscape on WHO | World Health Organization Site. Available online: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed on 21 January 2022).
- Momin, T.; Kansagra, K.; Patel, H.; Sharma, S.; Sharma, B.; Patel, J.; Mittal, R.; Sanmukhani, J.; Maithal, K.; Dey, A.; et al. Safety and Immunogenicity of a DNA SARS-CoV-2 vaccine (ZyCoV-D): Results of an open-label, non-randomized phase I part of phase I/II clinical study by intradermal route in healthy subjects in India. EClinicalMedicine 2021, 38, 101020. [Google Scholar] [CrossRef]
- Mallapaty, S. India’s DNA COVID vaccine is a world first—More are coming. Nature 2021, 597, 161–162. [Google Scholar] [CrossRef]
- Tejeda-Mansir, A.; García-Rendón, A.; Guerrero-Germán, P. Plasmid-DNA lipid and polymeric nanovaccines: A new strategic in vaccines development. Biotechnol. Genet. Eng. Rev. 2019, 35, 46–68. [Google Scholar] [CrossRef]
- Chavda, V.P.; Pandya, R.; Apostolopoulos, V. DNA vaccines for SARS-CoV-2: Toward third-generation vaccination era. Expert Rev. Vaccines 2021, 20, 1549–1560. [Google Scholar] [CrossRef]
- Tebas, P.; Kraynyak, K.A.; Patel, A.; Maslow, J.N.; Morrow, M.P.; Sylvester, A.J.; Knoblock, D.; Gillespie, E.; Amante, D.; Racine, T.; et al. Intradermal SynCon® Ebola GP DNA Vaccine Is Temperature Stable and Safely Demonstrates Cellular and Humoral Immunogenicity Advantages in Healthy Volunteers. J. Infect. Dis. 2019, 220, 400–410. [Google Scholar] [CrossRef]
- Kumru, O.S.; Joshi, S.B.; Smith, D.E.; Middaugh, C.R.; Prusik, T.; Volkin, D.B. Vaccine instability in the cold chain: Mechanisms, analysis and formulation strategies. Biologicals 2014, 42, 237–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.A. A Comparison of Plasmid DNA and mRNA as Vaccine Technologies. Vaccines 2019, 7, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hobernik, D.; Bros, M. DNA Vaccines-How Far From Clinical Use? Int. J. Mol. Sci. 2018, 19, 3605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kutzler, M.A.; Weiner, D.B. DNA vaccines: Ready for prime time? Nat. Rev. Genet. 2008, 9, 776–788. [Google Scholar] [CrossRef]
- Chang, C.; Sun, J.; Hayashi, H.; Suzuki, A.; Sakaguchi, Y.; Miyazaki, H.; Nishikawa, T.; Nakagami, H.; Yamashita, K.; Kaneda, Y. Stable Immune Response Induced by Intradermal DNA Vaccination by a Novel Needleless Pyro-Drive Jet Injector. AAPS PharmSciTech 2019, 21, 19. [Google Scholar] [CrossRef] [Green Version]
- Conforti, A.; Marra, E.; Palombo, F.; Roscilli, G.; Ravà, M.; Fumagalli, V.; Muzi, A.; Maffei, M.; Luberto, L.; Lione, L.; et al. COVID-eVax, an electroporated DNA vaccine candidate encoding the SARS-CoV-2 RBD, elicits protective responses in animal models. Mol. Ther. 2022, 30, 311–326. [Google Scholar] [CrossRef]
- Peletta, A.; Prompetchara, E.; Tharakhet, K.; Kaewpang, P.; Buranapraditkun, S.; Techawiwattanaboon, T.; Jbilou, T.; Krangvichian, P.; Sirivichayakul, S.; Manopwisedjaroen, S.; et al. DNA Vaccine Administered by Cationic Lipoplexes or by In Vivo Electroporation Induces Comparable Antibody Responses against SARS-CoV-2 in Mice. Vaccines 2021, 9, 874. [Google Scholar] [CrossRef]
- Tran, T.N.M.; May, B.P.; Ung, T.T.; Nguyen, M.K.; Nguyen, T.T.T.; Dinh, V.L.; Doan, C.C.; Tran, T.V.; Khong, H.; Nguyen, T.T.T.; et al. Preclinical Immune Response and Safety Evaluation of the Protein Subunit Vaccine Nanocovax for COVID-19. Front. Immunol. 2021, 12, 766112. [Google Scholar] [CrossRef]
- Liu, H.; Zhou, C.; An, J.; Song, Y.; Yu, P.; Li, J.; Gu, C.; Hu, D.; Jiang, Y.; Zhang, L.; et al. Development of recombinant COVID-19 vaccine based on CHO-produced, prefusion spike trimer and alum/CpG adjuvants. Vaccine 2021, 39, 7001–7011. [Google Scholar] [CrossRef]
- Guo, Y.; He, W.; Mou, H.; Zhang, L.; Chang, J.; Peng, S.; Ojha, A.; Tavora, R.; Parcells, M.S.; Luo, G.; et al. An Engineered Receptor-Binding Domain Improves the Immunogenicity of Multivalent SARS-CoV-2 Vaccines. mBio 2021, 12, e00930-21. [Google Scholar] [CrossRef] [PubMed]
- Richmond, P.; Hatchuel, L.; Dong, M.; Ma, B.; Hu, B.; Smolenov, I.; Li, P.; Liang, P.; Han, H.H.; Liang, J.; et al. Safety and immunogenicity of S-Trimer (SCB-2019), a protein subunit vaccine candidate for COVID-19 in healthy adults: A phase 1, randomised, double-blind, placebo-controlled trial. Lancet 2021, 397, 682–694. [Google Scholar] [CrossRef]
- Hsieh, S.M.; Liu, W.D.; Huang, Y.S.; Lin, Y.J.; Hsieh, E.F.; Lian, W.C.; Chen, C.; Janssen, R.; Shih, S.R.; Huang, C.G.; et al. Safety and immunogenicity of a Recombinant Stabilized Prefusion SARS-CoV-2 Spike Protein Vaccine (MVC-COV1901) Adjuvanted with CpG 1018 and Aluminum Hydroxide in healthy adults: A Phase 1, dose-escalation study. EClinicalMedicine 2021, 38, 100989. [Google Scholar] [CrossRef] [PubMed]
- DeFrancesco, L. Whither COVID-19 vaccines? Nat. Biotechnol. 2020, 38, 1132–1145. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Lin, X.; Wang, Y.; Abraham, C.; Sou, C.; Ngo, T.; Zhang, Y.; Wilson, I.A.; Zhu, J. Single-component, self-assembling, protein nanoparticles presenting the receptor binding domain and stabilized spike as SARS-CoV-2 vaccine candidates. Sci. Adv. 2021, 7, eabf1591. [Google Scholar] [CrossRef]
- Kalathiya, U.; Padariya, M.; Fahraeus, R.; Chakraborti, S.; Hupp, T.R. Multivalent Display of SARS-CoV-2 Spike (RBD Domain) of COVID-19 to Nanomaterial, Protein Ferritin Nanocages. Biomolecules 2021, 11, 297. [Google Scholar] [CrossRef]
- Almansour, I.; Chen, H.; Wang, S.; Lu, S. Cross reactivity of serum antibody responses elicited by DNA vaccines expressing HA antigens from H1N1 subtype influenza vaccines in the past 30 years. Hum. Vaccines Immunother. 2013, 9, 2049–2059. [Google Scholar] [CrossRef] [Green Version]
- Vaine, M.; Wang, S.; Hackett, A.; Arthos, J.; Lu, S. Antibody responses elicited through homologous or heterologous prime-boost DNA and protein vaccinations differ in functional activity and avidity. Vaccine 2010, 28, 2999–3007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Arthos, J.; Lawrence, J.M.; Van Ryk, D.; Mboudjeka, I.; Shen, S.; Chou, T.H.; Montefiori, D.C.; Lu, S. Enhanced immunogenicity of gp120 protein when combined with recombinant DNA priming to generate antibodies that neutralize the JR-FL primary isolate of human immunodeficiency virus type 1. J. Virol. 2005, 79, 7933–7937. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Bi, Y.; Xiao, H.; Yao, Y.; Liu, X.; Hu, Z.; Duan, J.; Yang, Y.; Li, Z.; Li, Y. A novel DNA and protein combination COVID-19 vaccine formulation provides full protection against SARS-CoV-2 in rhesus macaques. Emerg. Microbes Infect. 2021, 10, 342–355. [Google Scholar] [CrossRef]
- Felber, B.K.; Lu, Z.; Hu, X.; Valentin, A.; Rosati, M.; Remmel, C.A.L.; Weiner, J.A.; Carpenter, M.C.; Faircloth, K.; Stanfield-Oakley, S.; et al. Co-immunization of DNA and Protein in the Same Anatomical Sites Induces Superior Protective Immune Responses against SHIV Challenge. Cell Rep. 2020, 31, 107624. [Google Scholar] [CrossRef] [PubMed]
- Karpenko, L.I.; Ilyichev, A.A.; Eroshkin, A.M.; Lebedev, L.R.; Uzhachenko, R.V.; Nekrasova, N.A.; Plyasunova, O.A.; Belavin, P.A.; Seregin, S.V.; Danilyuk, N.K.; et al. Combined virus-like particle-based polyepitope DNA/protein HIV-1 vaccine design, immunogenicity and toxicity studies. Vaccine 2007, 25, 4312–4323. [Google Scholar] [CrossRef] [PubMed]
- Merkuleva, I.A.; Shcherbakov, D.N.; Borgoyakova, M.B.; Shanshin, D.V.; Rudometov, A.P.; Karpenko, L.I.; Belenkaya, S.V.; Isaeva, A.A.; Nesmeyanova, V.S.; Kazachinskaia, E.I.; et al. Comparative Immunogenicity of the Recombinant Receptor-Binding Domain of Protein S SARS-CoV-2 Obtained in Prokaryotic and Mammalian Expression Systems. Vaccines 2022, 10, 96. [Google Scholar] [CrossRef] [PubMed]
- Gorchakov, A.A.; Kulemzin, S.V.; Guselnikov, S.V.; Baranov, K.O.; Belovezhets, T.N.; Mechetina, L.V.; Volkova, O.Y.; Najakshin, A.M.; Chikaev, N.A.; Chikaev, A.N.; et al. Isolation of a panel of ultra-potent human antibodies neutralizing SARS-CoV-2 and viral variants of concern. Cell Discov. 2021, 7, 96. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, F.; Shen, C.; Peng, W.; Li, D.; Zhao, C.; Li, Z.; Li, S.; Bi, Y.; Yang, Y.; et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 2020, 368, 1274–1278. [Google Scholar] [CrossRef] [PubMed]
- Barnes, C.O.; West, A.P., Jr.; Huey-Tubman, K.E.; Hoffmann, M.A.G.; Sharaf, N.G.; Hoffman, P.R.; Koranda, N.; Gristick, H.B.; Gaebler, C.; Muecksch, F.; et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell 2020, 182, 828–842.e16. [Google Scholar] [CrossRef]
- Brouwer, P.J.M.; Caniels, T.G.; van der Straten, K.; Snitselaar, J.L.; Aldon, Y.; Bangaru, S.; Torres, J.L.; Okba, N.M.A.; Claireaux, M.; Kerster, G.; et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 2020, 369, 643–650. [Google Scholar] [CrossRef]
- Wang, C.; Li, W.; Drabek, D.; Okba, N.M.A.; van Haperen, R.; Osterhaus, A.D.M.E.; van Kuppeveld, F.J.M.; Haagmans, B.L.; Grosveld, F.; Bosch, B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020, 11, 2251. [Google Scholar] [CrossRef]
- Kam, Y.W.; Kien, F.; Roberts, A.; Cheung, Y.C.; Lamirande, E.W.; Vogel, L.; Chu, S.L.; Tse, J.; Guarner, J.; Zaki, S.R.; et al. Antibodies against trimeric S glycoprotein protect hamsters against SARS-CoV challenge despite their capacity to mediate FcgammaRII-dependent entry into B cells in vitro. Vaccine 2007, 25, 729–740. [Google Scholar] [CrossRef]
- Tseng, C.T.; Sbrana, E.; Iwata-Yoshikawa, N.; Newman, P.C.; Garron, T.; Atmar, R.L.; Peters, C.J.; Couch, R.B. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS ONE 2012, 7, e35421. [Google Scholar] [CrossRef]
- Karpenko, L.I.; Apartsin, E.K.; Dudko, S.G.; Starostina, E.V.; Kaplina, O.N.; Antonets, D.V.; Volosnikova, E.A.; Zaitsev, B.N.; Bakulina, A.Y.; Venyaminova, A.G.; et al. Cationic Polymers for the Delivery of the Ebola DNA Vaccine Encoding Artificial T-Cell Immunogen. Vaccines 2020, 8, 718. [Google Scholar] [CrossRef] [PubMed]
- Borgoyakova, M.B.; Karpenko, L.I.; Rudometov, A.P.; Shanshin, D.V.; Isaeva, A.A.; Nesmeyanova, V.S.; Volkova, N.V.; Belenkaya, S.V.; Murashkin, D.E.; Shcherbakov, D.N.; et al. Immunogenic Properties of the DNA Construct Encoding the Receptor-Binding Domain of the SARS-CoV-2 Spike Protein. Mol. Biol. 2021, 55, 889–898. [Google Scholar] [CrossRef] [PubMed]
- Karpenko, L.I.; Bazhan, S.I.; Bogryantseva, M.P.; Ryndyuk, N.N.; Ginko, Z.I.; Kuzubov, V.I.; Lebedev, L.R.; Kaplina, O.N.; Reguzova, A.Y.; Ryzhikov, A.B.; et al. Results of phase I clinical trials of a combined vaccine against HIV-1 based on synthetic polyepitope immunogens. Russ. J. Bioorg. Chem. 2016, 42, 170–182. [Google Scholar] [CrossRef]
- Bolhassani, A.; Javanzad, S.; Saleh, T.; Hashemi, M.; Aghasadeghi, M.R.; Sadat, S.M. Polymeric nanoparticles: Potent vectors for vaccine delivery targeting cancer and infectious diseases. Hum. Vaccines Immunother. 2014, 10, 321–332. [Google Scholar] [CrossRef] [Green Version]
- Singh, A. Eliciting B cell immunity against infectious diseases using nanovaccines. Nat. Nanotechnol. 2021, 16, 16–24. [Google Scholar] [CrossRef]
- Khudyakov, Y.; Pumpens, P. Viral Nanotechnology; CRC Press: Boca Raton, FL, USA, 2015; 524p. [Google Scholar] [CrossRef]
- Bazhan, S.I.; Karpenko, L.I.; Lebedev, L.R.; Uzhachenko, R.V.; Belavin, P.A.; Eroshkin, A.M.; Ilyichev, A.A. A synergistic effect of a combined bivalent DNA-protein anti-HIV-1 vaccine containing multiple T- and B-cell epitopes of HIV-1 proteins. Mol. Immunol. 2008, 45, 661–669. [Google Scholar] [CrossRef]
- Karpenko, L.I.; Rudometov, A.P.; Sharabrin, S.V.; Shcherbakov, D.N.; Borgoyakova, M.B.; Bazhan, S.I.; Volosnikova, E.A.; Rudometova, N.B.; Orlova, L.A.; Pyshnaya, I.A.; et al. Delivery of mRNA Vaccine against SARS-CoV-2 Using a Polyglucin:Spermidine Conjugate. Vaccines 2021, 9, 76. [Google Scholar] [CrossRef]

| Number | Sequence | MHC Restriction |
|---|---|---|
| 1 | VYAWNRKRI | H2-Kd |
| 2 | FERDISTEI | H2-Ld |
| 3 | CGPKKSTNL | H2-Dd |
| 4 | KNIDGYFKIYSKHTP | H2-IEd |
| 5 | RFASVYAWNRKRISN | H2-IEd, H2-IAd |
| 6 | VGGNYNYLYRLFRKS | H2-IEd |
| 7 | GGNYNYLYRLFRKSN | H2-IEd |
| 8 | YNYKLPDDFTGCVIA | H2-IEd |
| 9 | NATRFASVYAWNRKR | H2-IEd, H2-IAd |
| 10 | KNKCVNFNFNGLTGT | H2-IEd |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borgoyakova, M.B.; Karpenko, L.I.; Rudometov, A.P.; Volosnikova, E.A.; Merkuleva, I.A.; Starostina, E.V.; Zadorozhny, A.M.; Isaeva, A.A.; Nesmeyanova, V.S.; Shanshin, D.V.; et al. Self-Assembled Particles Combining SARS-CoV-2 RBD Protein and RBD DNA Vaccine Induce Synergistic Enhancement of the Humoral Response in Mice. Int. J. Mol. Sci. 2022, 23, 2188. https://doi.org/10.3390/ijms23042188
Borgoyakova MB, Karpenko LI, Rudometov AP, Volosnikova EA, Merkuleva IA, Starostina EV, Zadorozhny AM, Isaeva AA, Nesmeyanova VS, Shanshin DV, et al. Self-Assembled Particles Combining SARS-CoV-2 RBD Protein and RBD DNA Vaccine Induce Synergistic Enhancement of the Humoral Response in Mice. International Journal of Molecular Sciences. 2022; 23(4):2188. https://doi.org/10.3390/ijms23042188
Chicago/Turabian StyleBorgoyakova, Mariya B., Larisa I. Karpenko, Andrey P. Rudometov, Ekaterina A. Volosnikova, Iuliia A. Merkuleva, Ekaterina V. Starostina, Alexey M. Zadorozhny, Anastasiya A. Isaeva, Valentina S. Nesmeyanova, Daniil V. Shanshin, and et al. 2022. "Self-Assembled Particles Combining SARS-CoV-2 RBD Protein and RBD DNA Vaccine Induce Synergistic Enhancement of the Humoral Response in Mice" International Journal of Molecular Sciences 23, no. 4: 2188. https://doi.org/10.3390/ijms23042188
APA StyleBorgoyakova, M. B., Karpenko, L. I., Rudometov, A. P., Volosnikova, E. A., Merkuleva, I. A., Starostina, E. V., Zadorozhny, A. M., Isaeva, A. A., Nesmeyanova, V. S., Shanshin, D. V., Baranov, K. O., Volkova, N. V., Zaitsev, B. N., Orlova, L. A., Zaykovskaya, A. V., Pyankov, O. V., Danilenko, E. D., Bazhan, S. I., Shcherbakov, D. N., ... Ilyichev, A. A. (2022). Self-Assembled Particles Combining SARS-CoV-2 RBD Protein and RBD DNA Vaccine Induce Synergistic Enhancement of the Humoral Response in Mice. International Journal of Molecular Sciences, 23(4), 2188. https://doi.org/10.3390/ijms23042188