Tceal5 and Tceal7 Function in C2C12 Myogenic Differentiation via Exosomes in Fetal Bovine Serum
Abstract
1. Introduction
2. Results
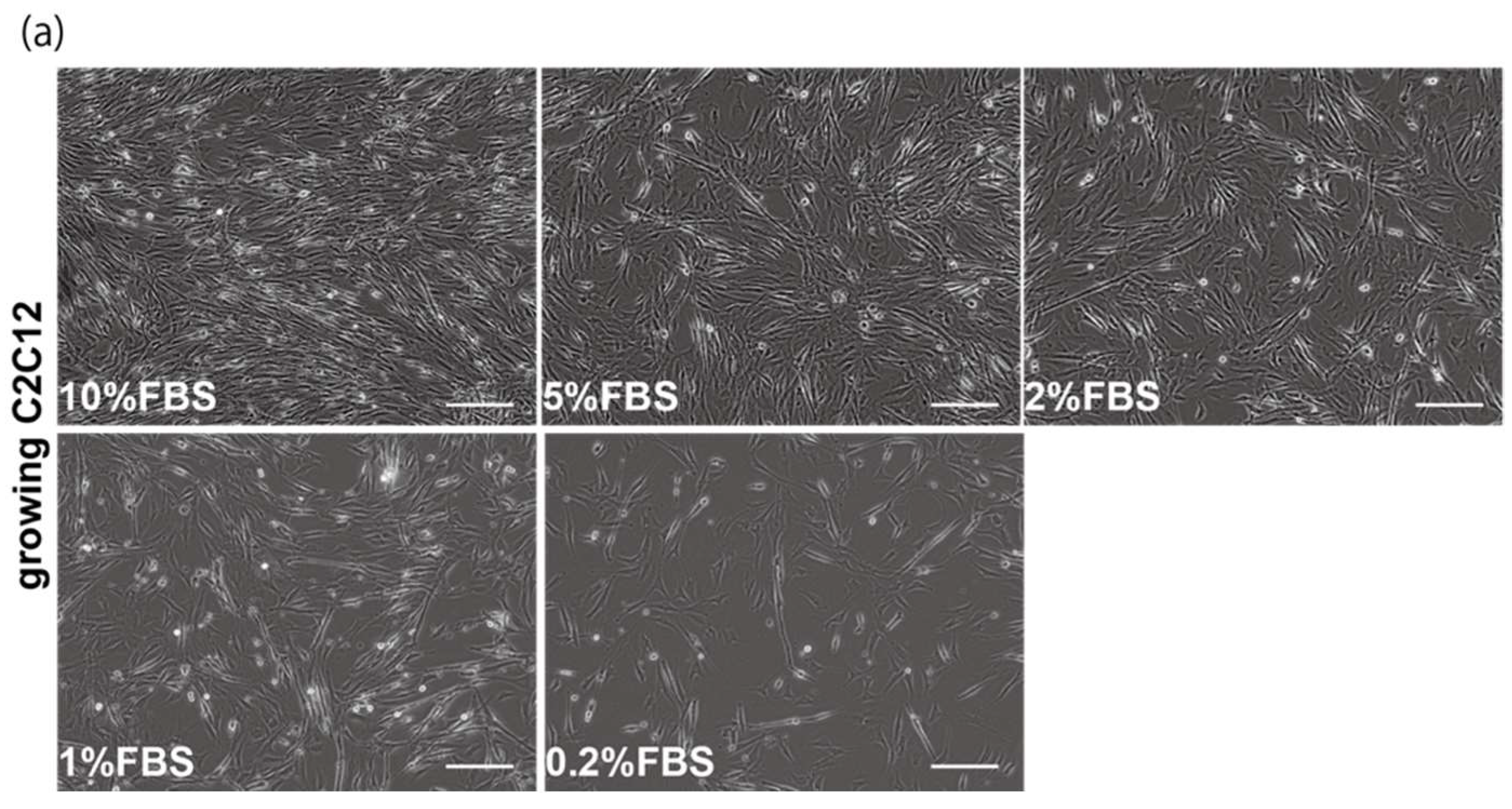
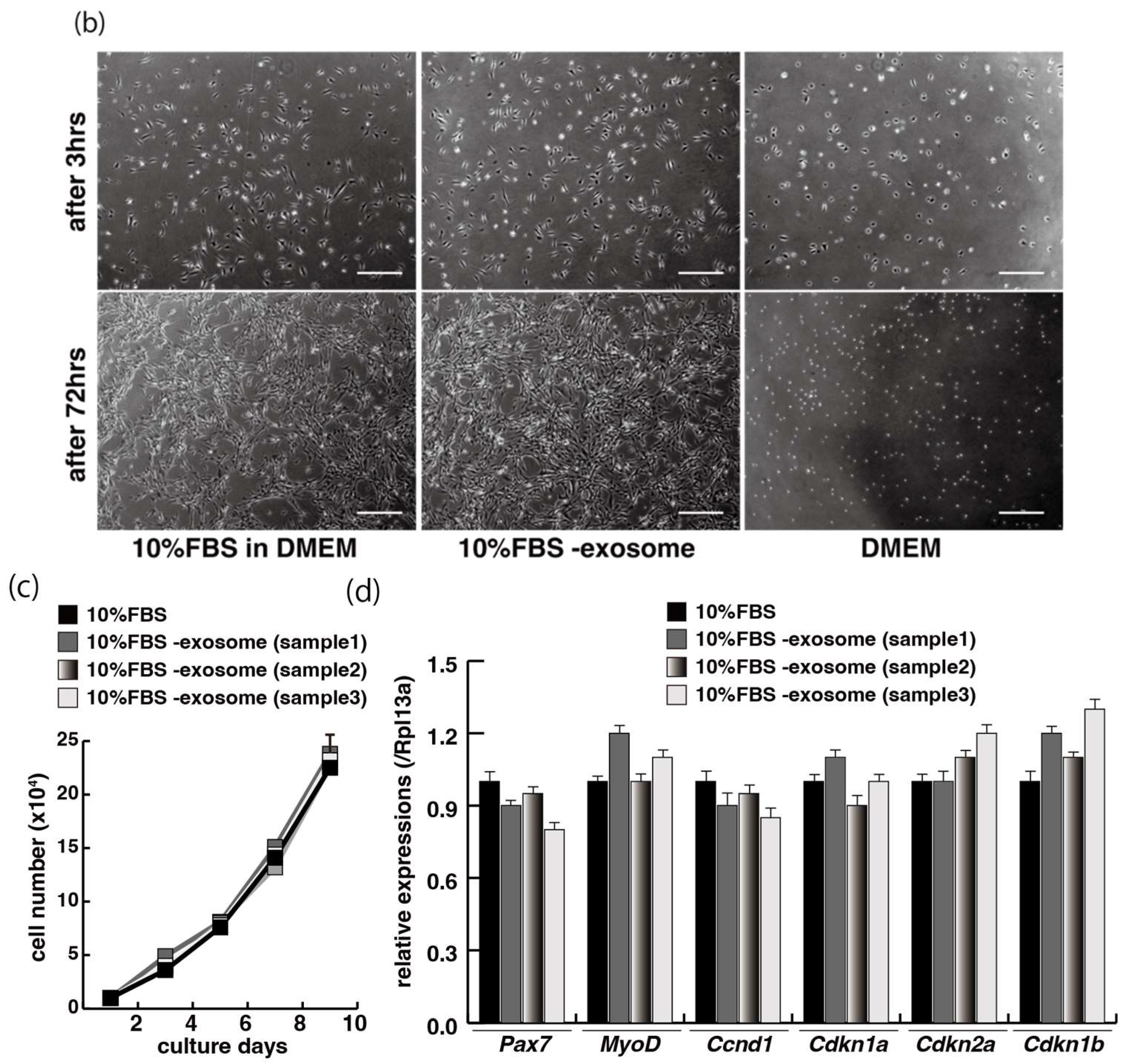
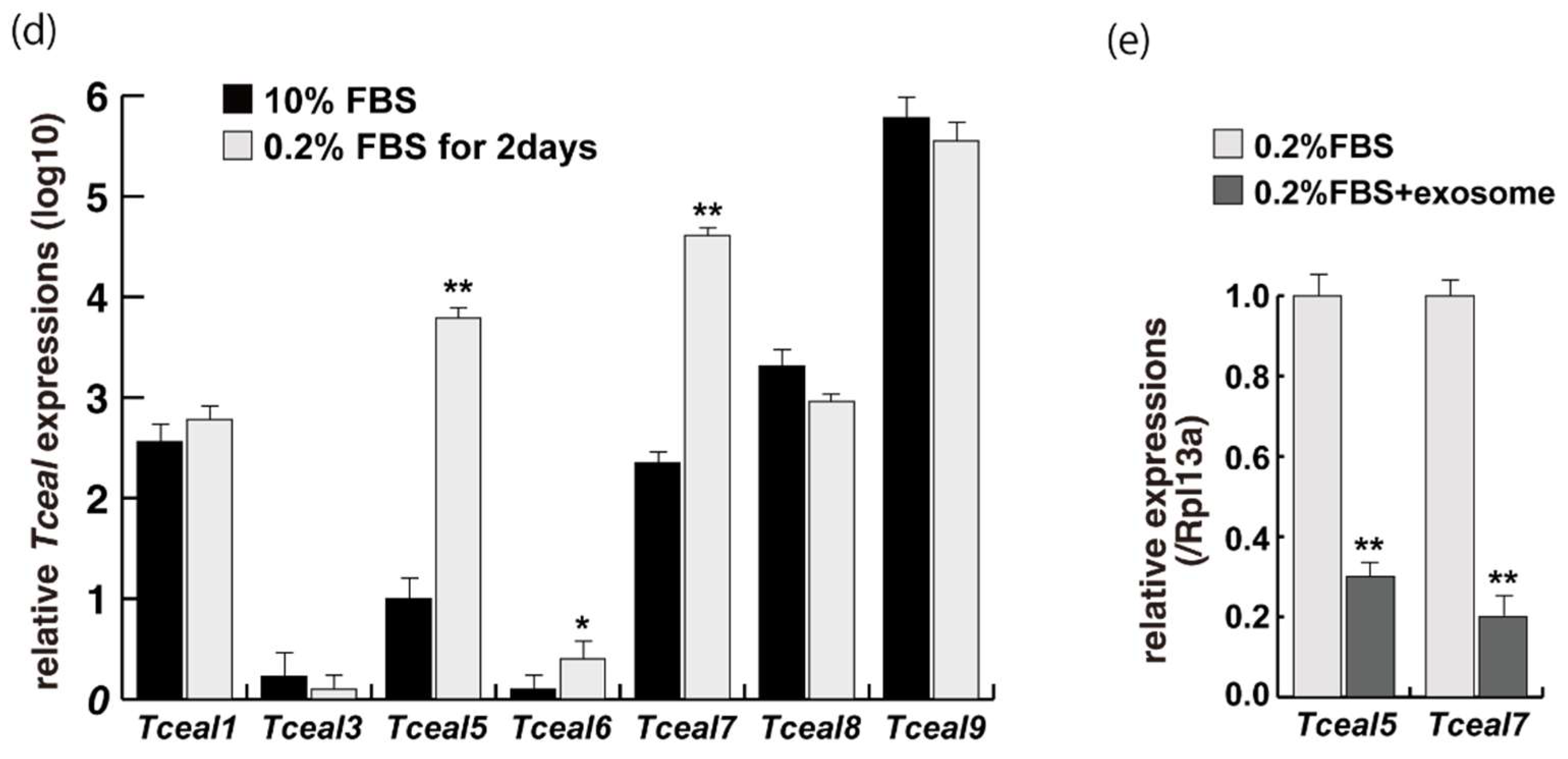
2.1. Myogenic Cell Growth without Exosomes in Serum
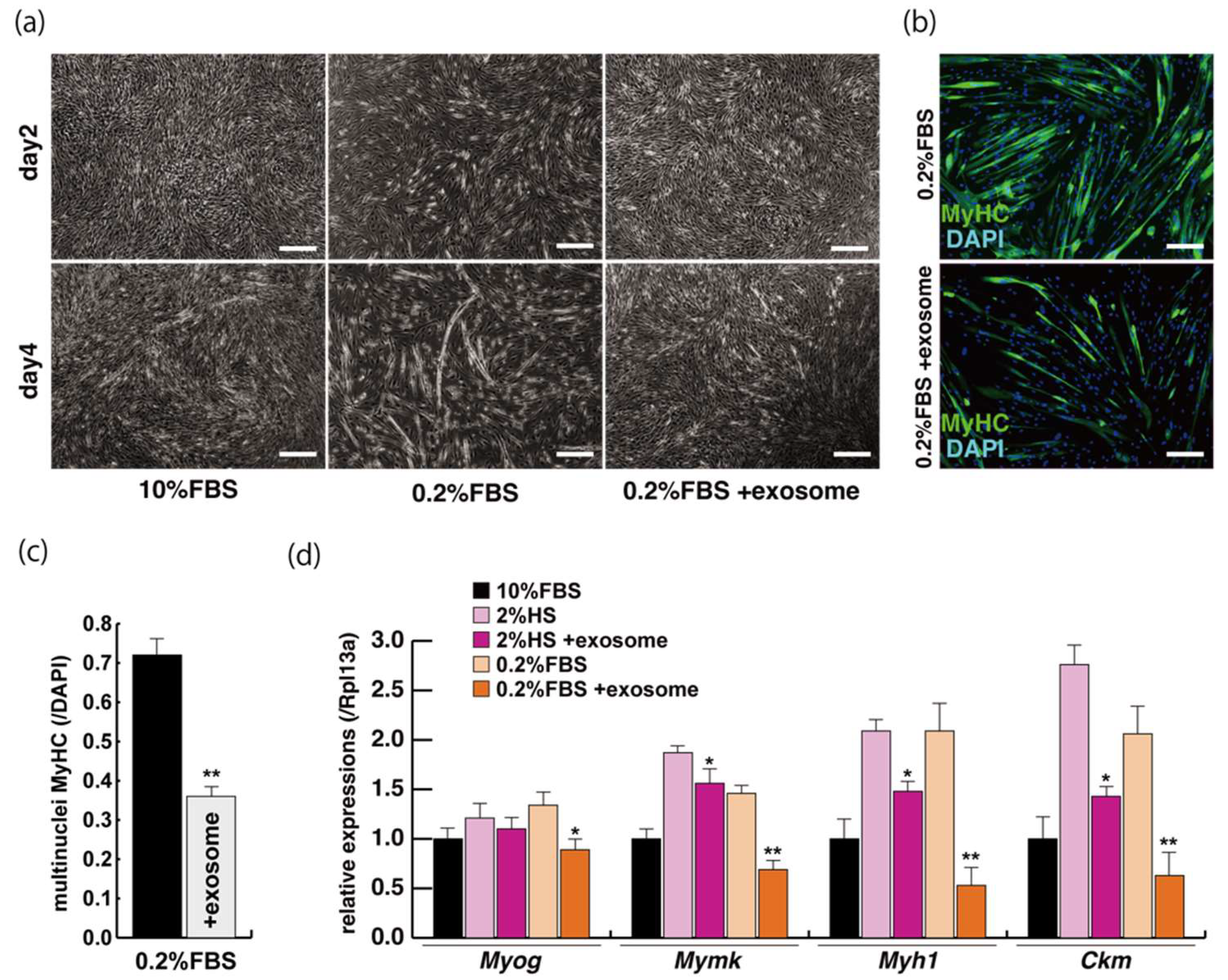
2.2. Myogenic Differentiation with Exosomes from Serum
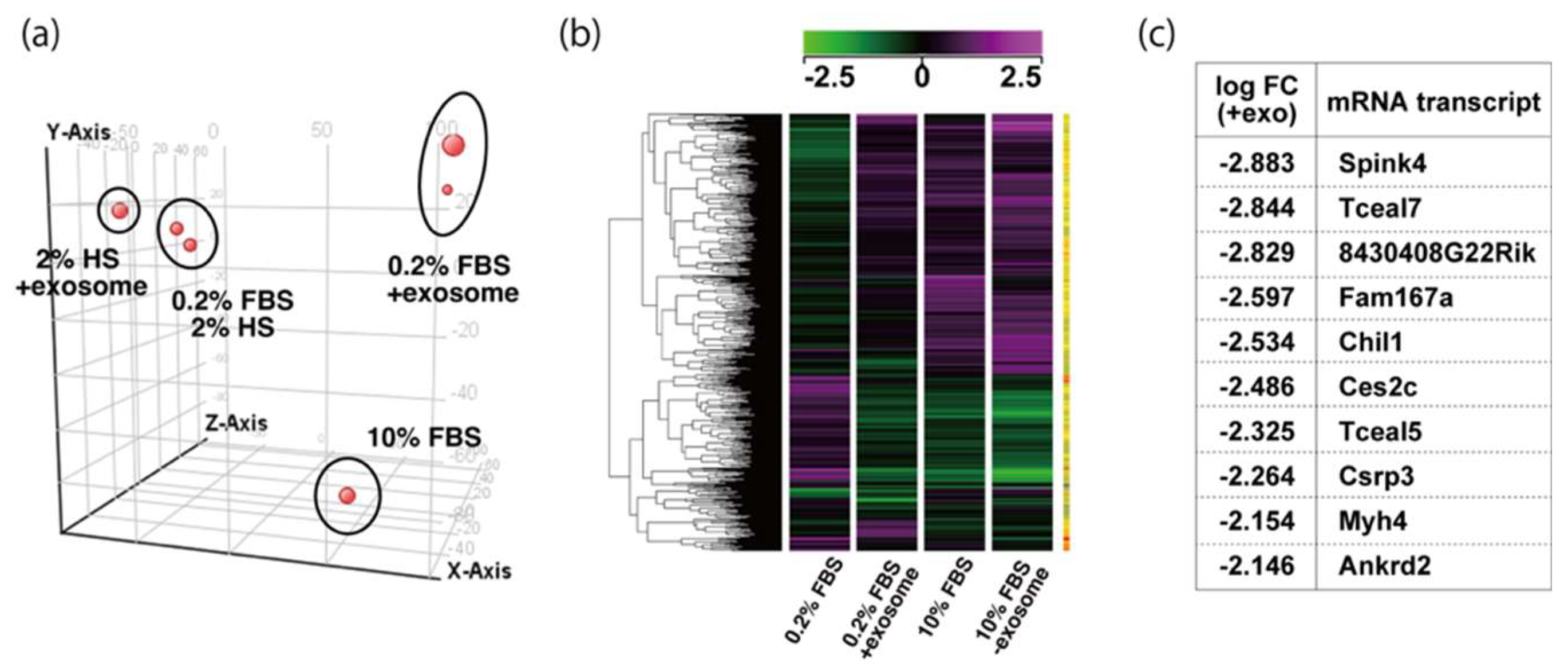
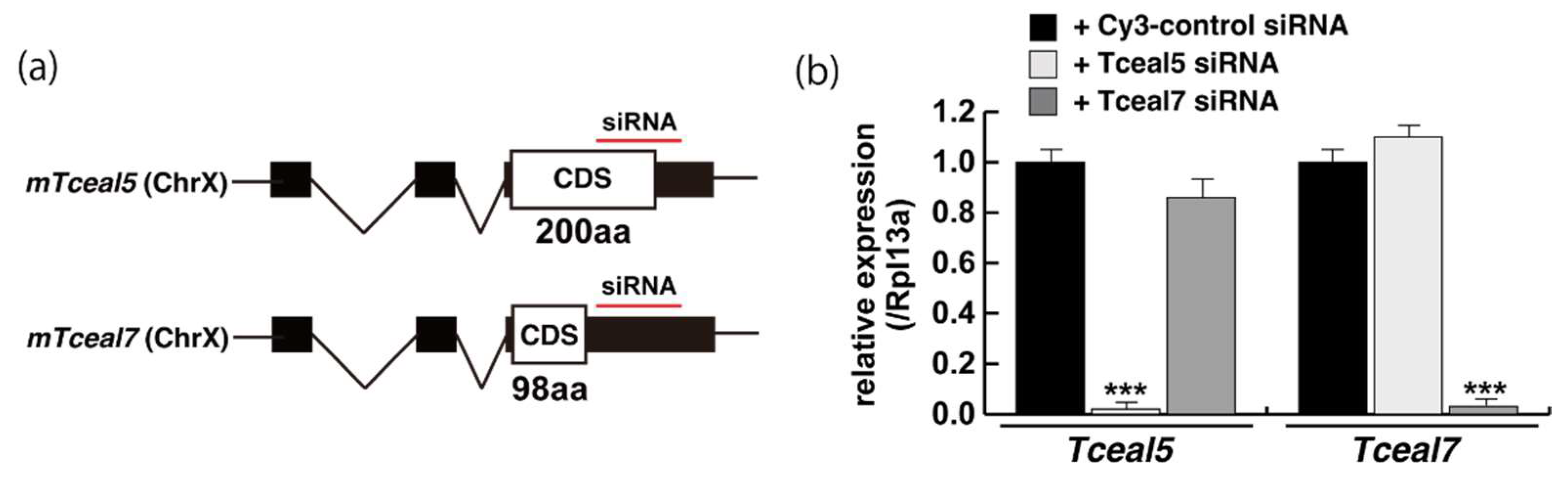
2.3. Tceal5 and Tceal7 in Differentiated C2C12 Cells
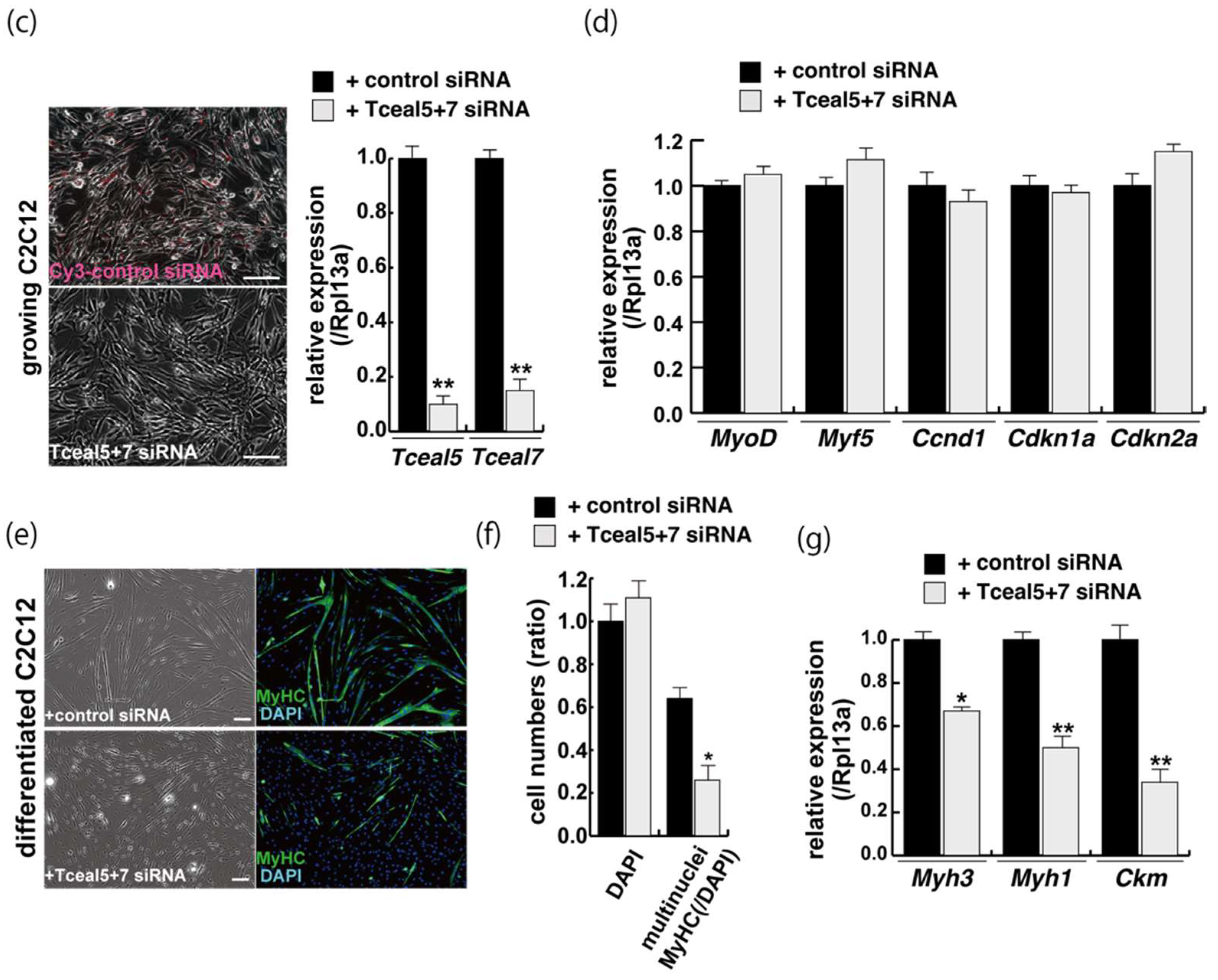
2.4. Attenuated Tceal5 and Tceal7 Expression Effects to Myogenic Differentiation
2.5. Increased Expression of Tceal5 and Tceal7 Rescues Myogenic Differentiation from the Negative Effects of Exosomes
3. Discussions
4. Materials and Methods
4.1. Isolation of Exosomes from FBS and C2C12 Myogenic Cell Cultures
4.2. Next-Generation Sequencing and RT-qPCR Analyses
4.3. Immunofluorescence
4.4. Western Blot
4.5. Tceal Overexpression and siRNA Constructs
4.6. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schneider, M.D.; Olson, E.N. Control of myogenic differentiation by cellular oncogenes. Mol. Neurobiol. 1988, 2, 1–39. [Google Scholar] [CrossRef] [PubMed]
- Clegg, C.H.; Linkhart, T.A.; Olwin, B.B.; Hauschka, S.D. Growth factor control of skeletal muscle differentiation: Commitment to terminal differentiation occurs in G1 phase and is repressed by fibroblast growth factor. J. Cell Biol. 1987, 105, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Magri, K.A.; Ewton, D.Z.; Florini, J.R. The role of the IGFs in myogenesis differentiation. Adv. Exp. Med. Biol. 1991, 293, 57–76. [Google Scholar] [PubMed]
- Jin, P.; Sejersen, T.; Ringertz, N.R. Recombinant platelet-derived growth factor-BB stimulates growth and inhibits differentiation of rat L6 myoblasts. J. Biol. Chem. 1991, 266, 1245–1249. [Google Scholar] [CrossRef]
- Zentella, A.; Massague, J. Transforming growth factor beta induces myoblast differentiation in the presence of mitogens. Proc. Natl. Acad. Sci. USA 1992, 89, 5176–5180. [Google Scholar] [CrossRef]
- Yoshiko, Y.; Hirao, K.; Sakabe, K.; Seiki, K.; Takezawa, J.; Maeda, N. Autonomous control of expression of genes for insulin-like growth factors during the proliferation and differentiation of C2C12 mouse myoblasts in serum-free culture. Life Sci. 1996, 59, 1961–1968. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Schmid, M.; Jensen, T.H. The exosome: A multipurpose RNA-decay machine. Trends Biochem. Sci. 2008, 33, 501–510. [Google Scholar] [CrossRef]
- Rome, S.; Forterre, A.; Mizgier, M.L.; Bouzakri, K. Skeletal Muscle-Released Extracellular Vesicles: State of the Art. Front. Physiol. 2019, 10, 929. [Google Scholar] [CrossRef]
- Mytidou, C.; Koutsoulidou, A.; Katsioloudi, A.; Prokopi, M.; Kapnisis, K.; Michailidou, K.; Anayiotos, A.; Phylactou, L.A. Muscle-derived exosomes encapsulate myomiRs and are involved in local skeletal muscle tissue communication. FASEB J. 2021, 35, e21279. [Google Scholar] [CrossRef] [PubMed]
- Gergics, P.; Christian, H.C.; Choo, M.S.; Ajmal, A.; Camper, S.A. Gene Expression in Mouse Thyrotrope Adenoma: Transcription Elongation Factor Stimulates Proliferation. Endocrinology 2016, 157, 3631–3646. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chien, J.; Staub, J.; Avula, R.; Zhang, H.; Liu, W.; Hartmann, L.C.; Kaufmann, S.H.; Smith, D.I.; Shridhar, V. Epigenetic silencing of TCEAL7 (Bex4) in ovarian cancer. Oncogene 2005, 24, 5089–5100. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Rai, A.J.; DeCastro, G.J.; Zeringer, E.; Barta, T.; Magdaleno, S.; Setterquist, R.; Vlassov, A.V. An optimized procedure for exosome isolation and analysis using serum samples: Application to cancer biomarker discovery. Methods 2015, 87, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.H.; Shatkin, A.J. Down-regulation of Rous sarcoma virus long terminal repeat promoter activity by a HeLa cell basic protein. Proc. Natl. Acad. Sci. USA 1994, 91, 11002–11006. [Google Scholar] [CrossRef] [PubMed]
- Rattan, R.; Narita, K.; Chien, J.; Maguire, J.L.; Shridhar, R.; Giri, S.; Shridhar, V. TCEAL7, a putative tumor suppressor gene, negatively regulates NF-kappaB pathway. Oncogene 2010, 29, 1362–1373. [Google Scholar] [CrossRef]
- Guo, Y.; Liao, Y.; Jia, C.; Ren, J.; Wang, J.; Li, T. MicroRNA-182 promotes tumor cell growth by targeting transcription elongation factor A-like 7 in endometrial carcinoma. Cell Physiol. Biochem. 2013, 32, 581–590. [Google Scholar] [CrossRef]
- Shi, X.; Garry, D.J. Myogenic regulatory factors transactivate the Tceal7 gene and modulate muscle differentiation. Biochem. J. 2010, 428, 213–221. [Google Scholar] [CrossRef][Green Version]
- Rana, K.; Lee, N.K.; Zajac, J.D.; MacLean, H.E. Expression of androgen receptor target genes in skeletal muscle. Asian J. Androl. 2014, 16, 675–683. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Horikiri, T.; Ohi, H.; Shibata, M.; Ikeya, M.; Ueno, M.; Sotozono, C.; Kinoshita, S.; Sato, T. SOX10-Nano-Lantern Reporter Human iPS Cells; A Versatile Tool for Neural Crest Research. PLoS ONE 2017, 12, e0170342. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sawada, A.; Yamamoto, T.; Sato, T. Tceal5 and Tceal7 Function in C2C12 Myogenic Differentiation via Exosomes in Fetal Bovine Serum. Int. J. Mol. Sci. 2022, 23, 2036. https://doi.org/10.3390/ijms23042036
Sawada A, Yamamoto T, Sato T. Tceal5 and Tceal7 Function in C2C12 Myogenic Differentiation via Exosomes in Fetal Bovine Serum. International Journal of Molecular Sciences. 2022; 23(4):2036. https://doi.org/10.3390/ijms23042036
Chicago/Turabian StyleSawada, Aika, Takuya Yamamoto, and Takahiko Sato. 2022. "Tceal5 and Tceal7 Function in C2C12 Myogenic Differentiation via Exosomes in Fetal Bovine Serum" International Journal of Molecular Sciences 23, no. 4: 2036. https://doi.org/10.3390/ijms23042036
APA StyleSawada, A., Yamamoto, T., & Sato, T. (2022). Tceal5 and Tceal7 Function in C2C12 Myogenic Differentiation via Exosomes in Fetal Bovine Serum. International Journal of Molecular Sciences, 23(4), 2036. https://doi.org/10.3390/ijms23042036





