Identification of the Regulatory Genes of UV-B-Induced Anthocyanin Biosynthesis in Pepper Fruit
Abstract
1. Introduction
2. Results
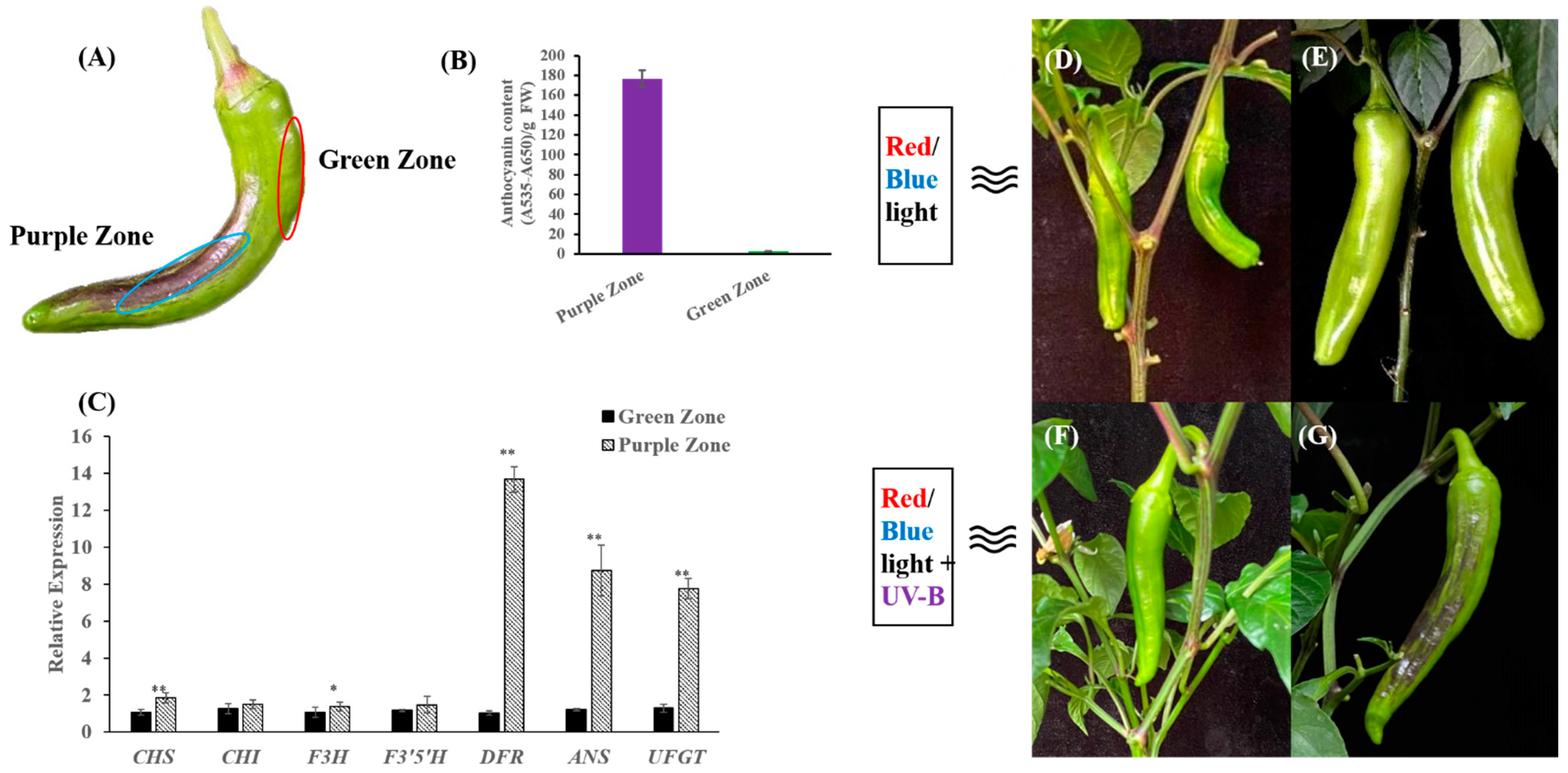
2.1. Anthocyanin Content and Expression of Anthocyanin Biosynthesis Genes Were Induced by UV-B in the Peel of 19Q6100
2.2. Transcriptome Analysis of Purple and Green Peels of UV-B-Treated Fruits
2.3. Transcription Factors and Structural Genes Involved in UV-B-Induced Anthocyanin Biosynthesis
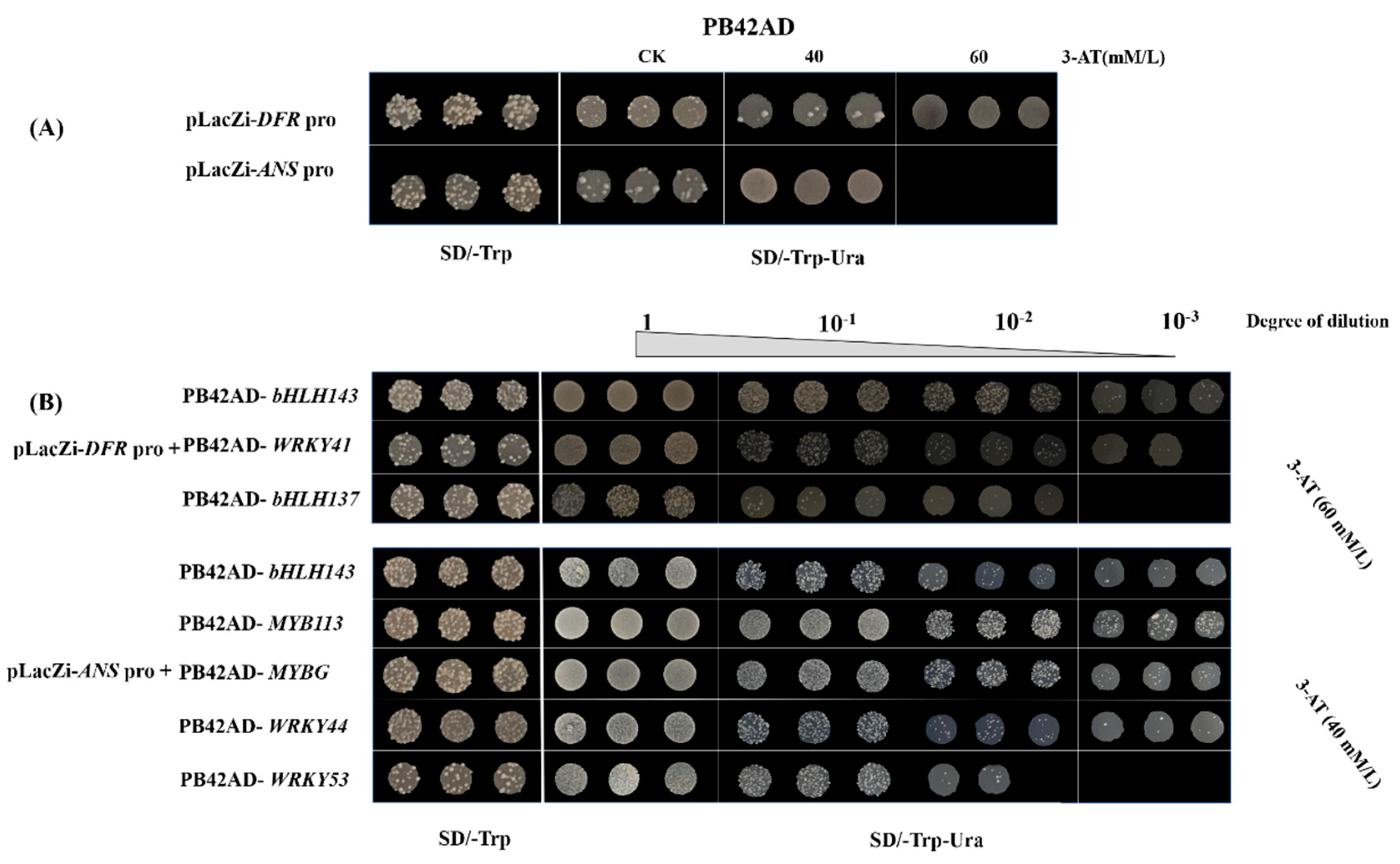
2.4. Interactions between TFs and Anthocyanin Biosynthesis Structural Genes
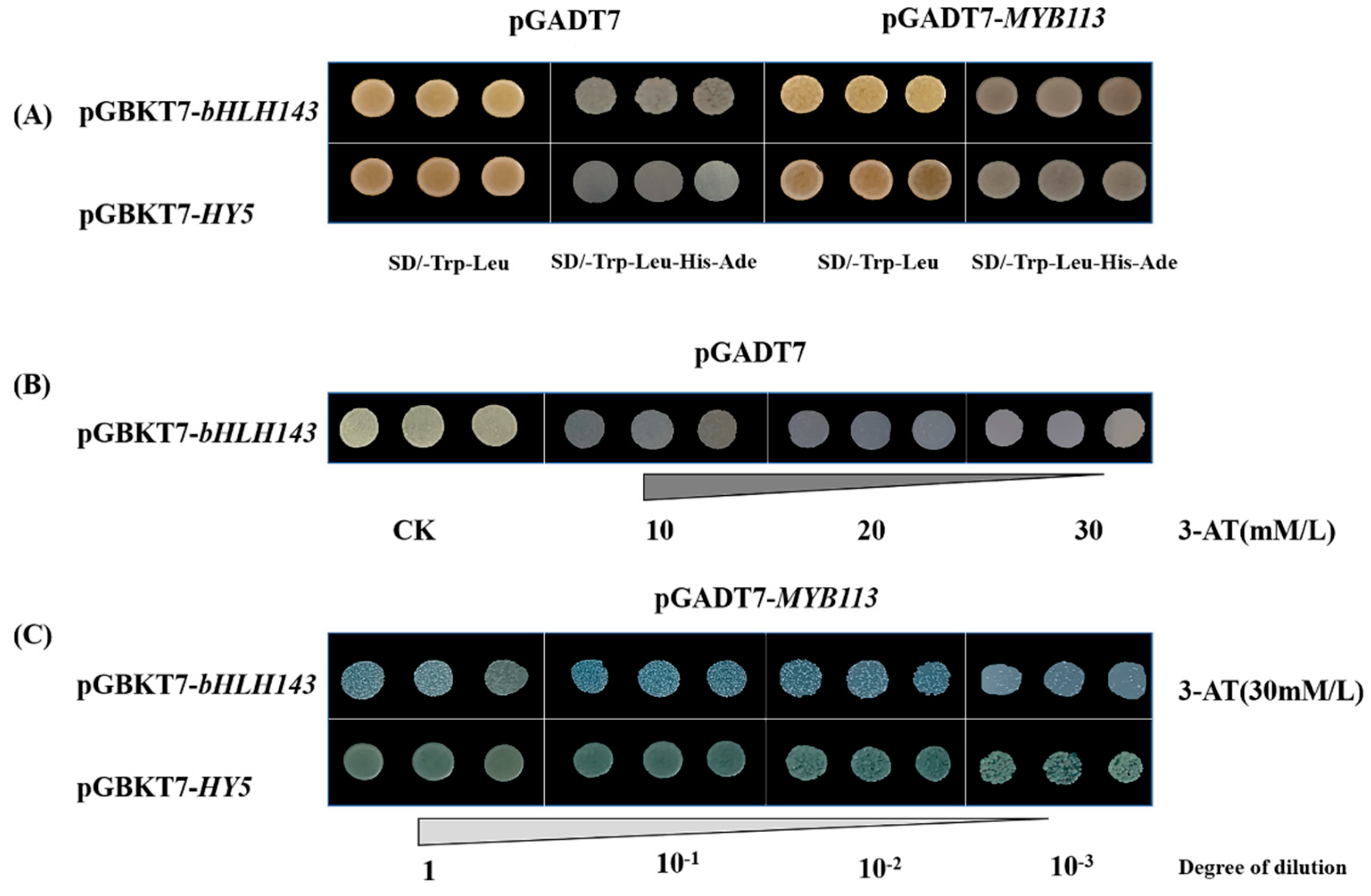
2.5. CaMYB113 Directly Bound to CabHLH143 and CaHY5
2.6. Function Analysis of CaMYB113 by VIGS
3. Discussion
3.1. UV-B Effectively Induced Anthocyanin Biosynthesis in the Peel of 19Q6100 Fruit
3.2. Upstream Regulators May Be Involved in UV-B-Induced Anthocyanin Accumulation in 19Q6100 Fruit
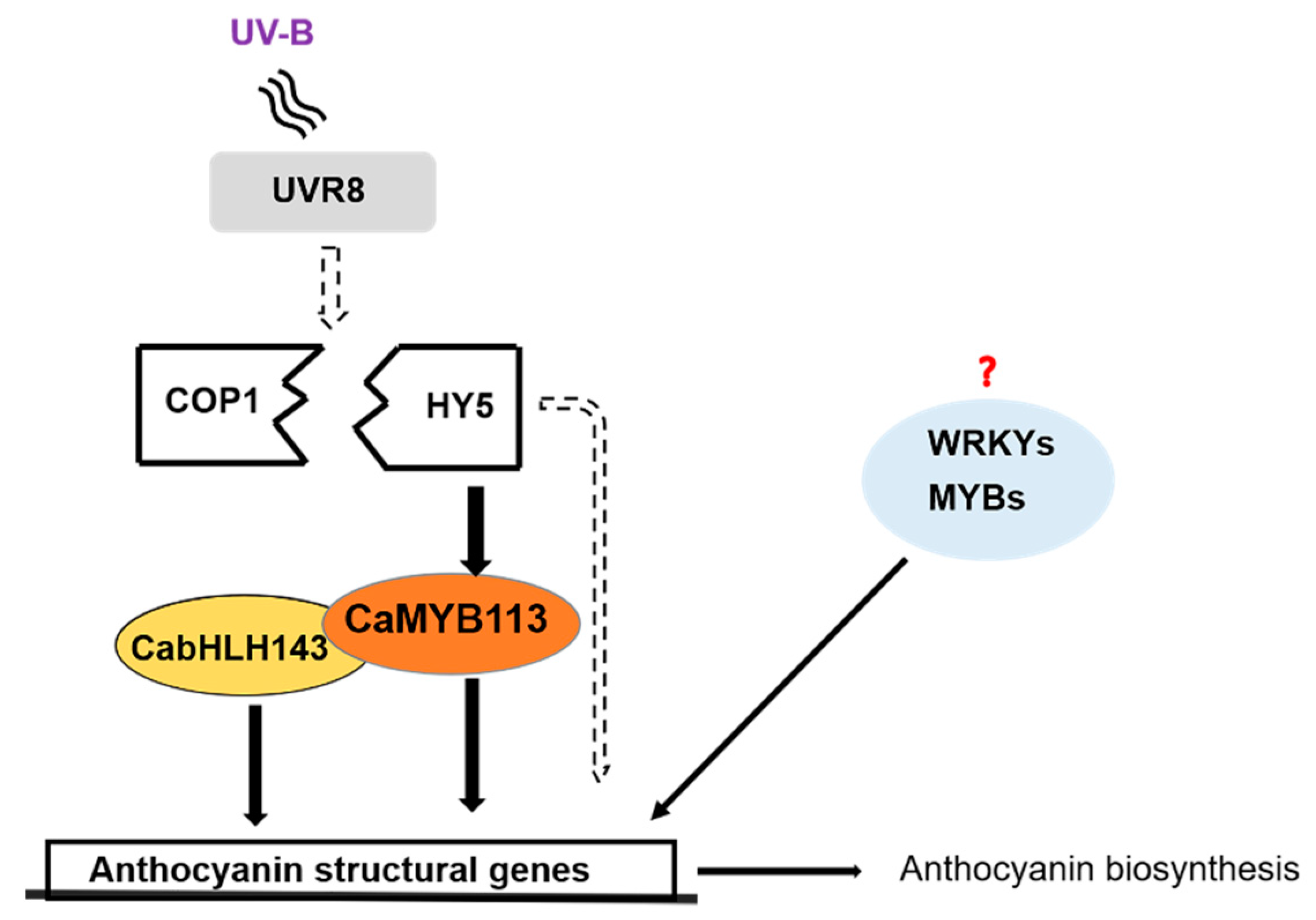
3.3. The HY5-CaMYB113-CabHLH143 Pathway
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Anthocyanin Extraction and Determination
4.3. RNA Extraction, cDNA Synthesis and Real-Time PCR Analysis
4.4. RNA-Seq Analysis
4.5. Yeast One-Hybrid Assay
4.6. Yeast Two-Hybrid Assay
4.7. VIGS
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Gould, K.S.; Dudle, D.A.; Neufeld, H.S. Why some stems are red: Cauline anthocyanins shield photosystem II against high light stress. J. Exp. Bot. 2010, 61, 2707–2717. [Google Scholar] [CrossRef] [PubMed]
- Olsen, K.; Slimestad, R.; Lea, U.; Brede, C.; Løvdal, T.; Ruoff, P.; Verheul, M.; Lillo, C. Temperature and nitrogen effects on regulators and products of the flavonoid pathway: Experimental and kinetic model studies. Plant Cell Envion. 2009, 32, 286–299. [Google Scholar] [CrossRef] [PubMed]
- Howe, G.; Jander, G. Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 2008, 59, 41–66. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Venail, J.; Mackay, S.; Bailey, P.C.; Schwinn, K.E.; Jameson, P.E.; Cathie, R.M.; Davies, K.M. The molecular basis for venation patterning of pigmentation and its effect on pollinator attraction in flowers of Antirrhinum. New Phytol. 2011, 189, 602–615. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Hichri, I.; Barrieu, F.; Bogs, J.; Kappel, C.; Delrot, S.; Lauvergeat, V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011, 62, 2465–2483. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.; Lloyd, A. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Albert, N.; Davies, K.; Lewis, D.; Zhang, H.; Montefiori, M.; Brendolise, C.; Boase, M.; Ngo, H.; Jameson, P.; Schwinn, K. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell. 2014, 26, 962–980. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lin-Wang, K.; Espley, R.; Wang, L.; Yang, H.; Yu, B.; Dare, A.; Varkonyi-Gasic, E.; Wang, J.; Zhang, J.; et al. Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 2016, 67, 2159–2176. [Google Scholar] [CrossRef]
- Spelt, C.; Quattrocchio, F.; Mol, J.; Koes, R. anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell. 2000, 12, 1619–1632. [Google Scholar] [CrossRef]
- Quattrocchio, F.; Verweij, W.; Kroon, A.; Spelt, C.; Mol, J.; Koes, R. PH4 of Petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell. 2006, 18, 1274–1291. [Google Scholar] [CrossRef] [PubMed]
- Stommel, J.R.; Dumm, J.M. Coordinated Regulation of Biosynthetic and Regulatory Genes Coincides with Anthocyanin Accumulation in Developing Eggplant Fruit. J. Am. Soc. Horitc. Sci. 2015, 140, 129–135. [Google Scholar] [CrossRef]
- Ravaglia, D.; Espley, R.V.; Henry-Kirk, R.A.; Andreotti, C.; Ziosi, V.; Hellens, R.P.; Costa, G.; Allan, A.C. Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol. 2013, 13, 68. [Google Scholar] [CrossRef]
- Hu, J.; Fang, H.; Wang, J.; Yue, X.; Su, M.; Mao, Z.; Zou, Q.; Jiang, H.; Guo, Z.; Yu, L.; et al. Ultraviolet B-induced MdWRKY72 expression promotes anthocyanin synthesis in apple. Plant Sci. 2020, 292, 110377. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, K.; Xu, Z.; El-Kereamy, A.; Casaretto, J.; Rothstein, S. The Arabidopsis Transcription Factor ANAC032 Represses Anthocyanin Biosynthesis in Response to High Sucrose and Oxidative and Abiotic Stresses. Front. Plant Sci. 2016, 7, 1548. [Google Scholar] [CrossRef]
- Shi, H.; Liu, G.; Wei, Y.; Chan, Z. The zinc-finger transcription factor ZAT6 is essential for hydrogen peroxide induction of anthocyanin synthesis in Arabidopsis. Plant Mol. Biol. 2018, 97, 165–176. [Google Scholar] [CrossRef]
- Guo, J.; Han, W.; Wang, M. Ultraviolet and environmental stresses involved in the induction and regulation of anthocyanin biosynthesis: A review. Afr. J. Biotechnol. 2008, 7, 1–4. [Google Scholar] [CrossRef][Green Version]
- van Gelderen, K.; Kang, C.; Pierik, R. Light Signaling, Root Development, and Plasticity. Plant Physiol. 2018, 176, 1049–1060. [Google Scholar] [CrossRef] [PubMed]
- Zoratti, L.; Karppinen, K.; Luengo Escobar, A.; Häggman, H.; Jaakola, L. Light-controlled flavonoid biosynthesis in fruits. Front. Plant Sci. 2014, 5, 534. [Google Scholar] [CrossRef]
- Zhao, Y.; Min, T.; Chen, M.; Wang, H.; Zhu, C.; Jin, R.; Allan, A.; Lin-Wang, K.; Xu, C. The Photomorphogenic Transcription Factor PpHY5 Regulates Anthocyanin Accumulation in Response to UVA and UVB Irradiation. Front. Plant Sci. 2020, 11, 603178. [Google Scholar] [CrossRef]
- Li, D.; Luo, Z.; Mou, W.; Wang, Y.; Ying, T.; Mao, L. ABA and UV-C effects on quality, antioxidant capacity and anthocyanin contents of strawberry fruit (Fragaria ananassa Duch.). Postharvest Biol. Technol. 2014, 90, 56–62. [Google Scholar] [CrossRef]
- Henry-Kirk, R.; Plunkett, B.; Hall, M.; McGhie, T.; Allan, A.; Wargent, J.; Espley, R. Solar UV light regulates flavonoid metabolism in apple (Malus x domestica). Plant Cell Envion. 2018, 41, 675–688. [Google Scholar] [CrossRef]
- Qian, M.; Zhang, D.; Yue, X.; Wang, S.; Li, X.; Teng, Y. Analysis of different pigmentation patterns in ‘Mantianhong’ (Pyrus pyrifolia Nakai) and ‘Cascade’ (Pyrus communis L.) under bagging treatment and postharvest UV-B/visible irradiation conditions. Sci. Hortic. 2013, 151, 75–82. [Google Scholar] [CrossRef]
- Catola, S.; Castagna, A.; Santin, M.; Calvenzani, V.; Petroni, K.; Mazzucato, A.; Ranieri, A. The dominant allele Aft induces a shift from flavonol to anthocyanin production in response to UV-B radiation in tomato fruit. Planta 2017, 246, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Chen, Z.; Liu, Q.; Mao, W.; Chen, Y.; Tian, W.; Liu, Y.; Han, J.; Ouyang, X.; Huang, X. Coordinated Transcriptional Regulation by the UV-B Photoreceptor and Multiple Transcription Factors for Plant UV-B Responses. Mol. Plant 2020, 13, 777–792. [Google Scholar] [CrossRef] [PubMed]
- Podolec, R.; Demarsy, E.; Ulm, R. Perception and Signaling of Ultraviolet-B Radiation in Plants. Annu. Rev. Plant Biol. 2021, 72, 793–822. [Google Scholar] [CrossRef]
- Qiu, Z.; Wang, H.; Li, D.; Yu, B.; Hui, Q.; Yan, S.; Huang, Z.; Cui, X.; Cao, B. Identification of Candidate HY5-Dependent and -Independent Regulators of Anthocyanin Biosynthesis in Tomato. Plant Cell Physiol. 2019, 60, 643–656. [Google Scholar] [CrossRef]
- An, J.; Qu, F.; Yao, J.; Wang, X.; You, C.; Wang, X.; Hao, Y. Erratum: The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 2017, 4, 17056. [Google Scholar] [CrossRef] [PubMed]
- Lian, H.; Xu, P.; He, S.; Wu, J.; Pan, J.; Wang, W.; Xu, F.; Wang, S.; Pan, J.; Huang, J.; et al. Photoexcited CRYPTOCHROME 1 Interacts Directly with G-Protein β Subunit AGB1 to Regulate the DNA-Binding Activity of HY5 and Photomorphogenesis in Arabidopsis. Mol. Plant 2018, 11, 1248–1263. [Google Scholar] [CrossRef]
- Liu, C.; Chi, C.; Jin, L.; Zhu, J.; Yu, J.; Zhou, Y. The bZip transcription factor HY5 mediates CRY1a-induced anthocyanin biosynthesis in tomato. Plant Cell Envion. 2018, 41, 1762–1775. [Google Scholar] [CrossRef] [PubMed]
- Bursch, K.; Niemann, E.; Nelson, D.; Johansson, H. Karrikins control seedling photomorphogenesis and anthocyanin biosynthesis through a HY5-BBX transcriptional module. Plant J. 2021, 107, 1346–1362. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.; Choi, M.; Kim, K.; Bang, G.; Cho, M.; Choi, S.; Choi, G.; Park, Y. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013, 587, 1543–1547. [Google Scholar] [CrossRef]
- Stracke, R.; Favory, J.; Gruber, H.; Bartelniewoehner, L.; Bartels, S.; Binkert, M.; Funk, M.; Weisshaar, B.; Ulm, R. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 2010, 33, 88–103. [Google Scholar] [CrossRef]
- Aguilar-Barragán, A.; Ochoa-Alejo, N. Virus-induced silencing of MYB and WD40 transcription factor genes affects the accumulation of anthocyanins in chilli pepper fruit. Bio. Plant. 2014, 58, 567–574. [Google Scholar] [CrossRef]
- Zhen, Z.; Li, D.W.; Jin, J.H.; Yin, Y.X.; Zhang, H.X.; Chai, W.G.; Gong, Z.H. VIGS approach reveals the modulation of anthocyanin biosynthetic genes by CaMYB in chili pepper leaves. Front. Plant Sci. 2015, 6, 500. [Google Scholar] [CrossRef]
- Borovsky, Y.; Oren-Shamir, M.; Ovadia, R.; Jong, W.D.; Paran, I. The A locus that controls anthocyanin accumulation in pepper encodes a MYB transcription factor homologous to Anthocyanin2 of Petunia. Theor. Appl. Genet. 2004, 109, 23–29. [Google Scholar] [CrossRef]
- Tang, B.; Li, L.; Hu, Z.; Chen, Y.; Tan, T.; Jia, Y.; Xie, Q.; Chen, G. Anthocyanin Accumulation and Transcriptional Regulation of Anthocyanin Biosynthesis in Purple Pepper. J. Agric. Food. Chem. 2020, 68, 12152–12163. [Google Scholar] [CrossRef]
- Liu, J.; Ai, X.; Wang, Y.; Lu, Q.; Li, T.; Wu, L.; Sun, L.; Shen, H. Fine mapping of the Ca3GT gene controlling anthocyanin biosynthesis in mature unripe fruit of Capsicum annuum L. Theor. Appl. Genet. 2020, 133, 2729–2742. [Google Scholar] [CrossRef] [PubMed]
- Castagna, A.; Dall’Asta, C.; Chiavaro, E.; Galaverna, G.; Ranieri, A. Effect of Post-harvest UV-B Irradiation on Polyphenol Profile and Antioxidant Activity in Flesh and Peel of Tomato Fruits. Food Bioprocess Tech. 2014, 7, 2241–2250. [Google Scholar] [CrossRef]
- Lau, O.; Deng, X. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012, 17, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Kubasek, W.; Shirley, B.; McKillop, A.; Goodman, H.; Briggs, W.; Ausubel, F. Regulation of Flavonoid Biosynthetic Genes in Germinating Arabidopsis Seedlings. Plant Cell. 1992, 4, 1229–1236. [Google Scholar] [CrossRef]
- Wang, L.; Lu, W.; Ran, L.; Dou, L.; Yao, S.; Hu, J.; Fan, D.; Li, C.; Luo, K. R2R3-MYB transcription factor MYB6 promotes anthocyanin and proanthocyanidin biosynthesis but inhibits secondary cell wall formation in Populus tomentosa. Plant J. 2019, 99, 733–751. [Google Scholar] [CrossRef]
- Huang, D.; Tang, Z.; Fu, J.; Yuan, Y.; Deng, X.; Xu, Q. CsMYB3 and CsRuby1 form an ‘Activator-and-Repressor’ Loop for the Regulation of Anthocyanin Biosynthesis in Citrus. Plant Cell Physiol. 2020, 61, 318–330. [Google Scholar] [CrossRef]
- Peng, Y.; Thrimawithana, A.; Cooney, J.; Jensen, D.; Espley, R.; Allan, A. The proanthocyanin-related transcription factors MYBC1 and WRKY44 regulate branch points in the kiwifruit anthocyanin pathway. Sci. Rep. 2020, 10, 141–161. [Google Scholar] [CrossRef]
- Duan, S.; Wang, J.; Gao, C.; Jin, C.; Li, D.; Peng, D.; Du, G.; Li, Y.; Chen, M. Functional characterization of a heterologously expressed Brassica napus WRKY41-1 transcription factor in regulating anthocyanin biosynthesis in Arabidopsis thaliana. Plant Sci. 2018, 268, 47–53. [Google Scholar] [CrossRef]
- Song, X.; Yang, Q.; Liu, Y.; Li, J.; Chang, X.; Xian, L.; Zhang, J. Genome-wide identification of Pistacia R2R3-MYB gene family and function characterization of PcMYB113 during autumn leaf coloration in Pistacia chinensis. Int. J. Biol. Macromol. 2021, 192, 16–27. [Google Scholar] [CrossRef]
- Zhang, Y.; Fang, Z.; Ye, X.; Pan, S. Identification of candidate genes involved in anthocyanin accumulation in the peel of jaboticaba (Myrciaria cauliflora) fruits by transcriptomic analysis. Gene 2018, 676, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Wang, X.; Gao, J.; Guo, Y.; Huang, Z.; Du, Y. The Tomato Hoffman’s Anthocyaninless Gene Encodes a bHLH Transcription Factor Involved in Anthocyanin Biosynthesis That Is Developmentally Regulated and Induced by Low Temperatures. PLoS ONE 2016, 11, e0151067. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D.L. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S. TopHat2: Accurate alignment of transcriptomes in the p resence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.H.; Huber, W. Differential Expression of RNA-Seq Data at the Gene Level—The DESeqPackage. Available online: https://www.semanticscholar.org/paper/Differential-expression-of-RNA-Seq-data-at-the-gene-Anders-Huber/81c42bdf29edd0035b237375e2270f5a31762147 (accessed on 10 April 2019).
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.; Frank, M.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- JASPAR. Available online: https://jaspar.genereg.net/ (accessed on 20 August 2021).
- Wang, X.; An, J.; Liu, X.; Su, L.; You, C.; Hao, Y. The Nitrate-Responsive Protein MdBT2 Regulates Anthocyanin Biosynthesis by Interacting with the MdMYB1 Transcription Factor. Plant Physiol. 2018, 178, 890–906. [Google Scholar] [CrossRef]
- AraPPINet. Available online: https://netbio.sjtu.edu.cn/arappinet/ (accessed on 20 August 2021).
- SGN. Available online: https://vigs.solgenomics.net/ (accessed on 11 September 2021).


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Liu, S.; Wang, H.; Zhang, Y.; Li, W.; Liu, J.; Cheng, Q.; Sun, L.; Shen, H. Identification of the Regulatory Genes of UV-B-Induced Anthocyanin Biosynthesis in Pepper Fruit. Int. J. Mol. Sci. 2022, 23, 1960. https://doi.org/10.3390/ijms23041960
Wang Y, Liu S, Wang H, Zhang Y, Li W, Liu J, Cheng Q, Sun L, Shen H. Identification of the Regulatory Genes of UV-B-Induced Anthocyanin Biosynthesis in Pepper Fruit. International Journal of Molecular Sciences. 2022; 23(4):1960. https://doi.org/10.3390/ijms23041960
Chicago/Turabian StyleWang, Yihao, Sujun Liu, Haoran Wang, Yingxue Zhang, Wenjie Li, Jinkui Liu, Qing Cheng, Liang Sun, and Huolin Shen. 2022. "Identification of the Regulatory Genes of UV-B-Induced Anthocyanin Biosynthesis in Pepper Fruit" International Journal of Molecular Sciences 23, no. 4: 1960. https://doi.org/10.3390/ijms23041960
APA StyleWang, Y., Liu, S., Wang, H., Zhang, Y., Li, W., Liu, J., Cheng, Q., Sun, L., & Shen, H. (2022). Identification of the Regulatory Genes of UV-B-Induced Anthocyanin Biosynthesis in Pepper Fruit. International Journal of Molecular Sciences, 23(4), 1960. https://doi.org/10.3390/ijms23041960





