Control of Gene Expression via the Yeast CWI Pathway
Abstract
1. Introduction
2. Chromatin and Gene Expression in Response to Stress
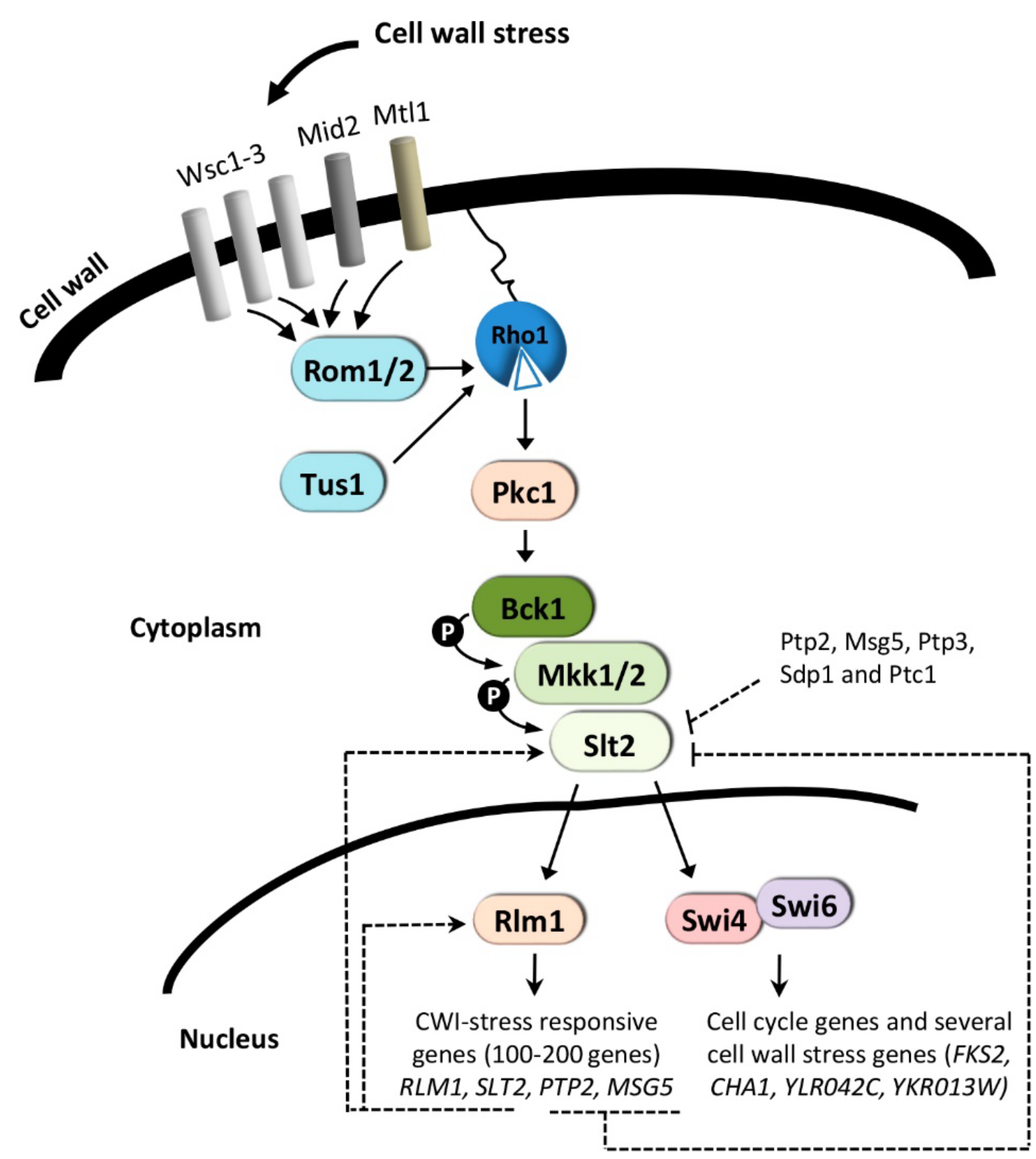
3. The CWI Pathway and Regulation of Gene Expression
3.1. Cell Wall-Stress Conditions
3.1.1. Transcriptional Activation Mechanism for SBF-Dependent Genes
3.1.2. Transcriptional Activation Mechanism for Rlm1-Dependent Genes
3.2. Other CWI-Activating Conditions
4. Conclusions and Future Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krishna, M.; Narang, H. The complexity of mitogen-activated protein kinases (MAPKs) made simple. Cell. Mol. Life Sci. 2008, 65, 3525–3544. [Google Scholar] [CrossRef] [PubMed]
- Elion, E.A.; Qi, M.; Chen, W. Signal transduction. Signaling specificity in yeast. Science 2005, 307, 687–688. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.E.; Thorner, J. Function and regulation in MAPK signaling pathways: Lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 2007, 1773, 1311–1340. [Google Scholar] [CrossRef] [PubMed]
- Engelberg, D.; Perlman, R.; Levitzki, A. Transmembrane signaling in Saccharomyces cerevisiae as a model for signaling in metazoans: State of the art after 25 years. Cell. Signal. 2014, 26, 2865–2878. [Google Scholar] [CrossRef] [PubMed]
- Levin, D.E. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Gutierrez, E.; Alegria-Carrasco, E.; Sellers-Moya, A.; Molina, M.; Martin, H. Not just the wall: The other ways to turn the yeast CWI pathway on. Int. Microbiol. 2020, 23, 107–119. [Google Scholar] [CrossRef]
- Berry, D.B.; Gasch, A.P. Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol. Biol. Cell 2008, 19, 4580–4587. [Google Scholar] [CrossRef]
- Chen, F.; Evans, A.; Pham, J.; PLoSky, B. Cellular stress responses: A balancing act. Mol. Cell 2010, 40, 175. [Google Scholar] [CrossRef]
- Martin, H.; Flandez, M.; Nombela, C.; Molina, M. Protein phosphatases in MAPK signalling: We keep learning from yeast. Mol. Microbiol. 2005, 58, 6–16. [Google Scholar] [CrossRef]
- Gonzalez-Rubio, G.; Fernandez-Acero, T.; Martin, H.; Molina, M. Mitogen-Activated Protein Kinase Phosphatases (MKPs) in Fungal Signaling: Conservation, Function, and Regulation. Int. J. Mol. Sci. 2019, 20, 1709. [Google Scholar] [CrossRef]
- Molina, M.; Cid, V.J.; Martin, H. Fine regulation of Saccharomyces cerevisiae MAPK pathways by post-translational modifications. Yeast 2010, 27, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Ibarra, A.; Rodriguez-Martinez, G.; Guerrero-Serrano, G.; Kawasaki, L.; Ongay-Larios, L.; Coria, R. Negative feedback-loop mechanisms regulating HOG- and pheromone-MAPK signaling in yeast. Curr. Genet. 2020, 66, 867–880. [Google Scholar] [CrossRef] [PubMed]
- Atay, O.; Skotheim, J.M. Spatial and temporal signal processing and decision making by MAPK pathways. J. Cell Biol. 2017, 216, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Cairns, B.R. The logic of chromatin architecture and remodelling at promoters. Nature 2009, 461, 193–198. [Google Scholar] [CrossRef]
- Singh, A.K.; Mueller-Planitz, F. Nucleosome Positioning and Spacing: From Mechanism to Function. J. Mol. Biol. 2021, 433, 166847. [Google Scholar] [CrossRef]
- Cutter, A.R.; Hayes, J.J. A brief review of nucleosome structure. FEBS Lett. 2015, 589, 2914–2922. [Google Scholar] [CrossRef]
- Kornberg, R.D. Chromatin structure: A repeating unit of histones and DNA. Science 1974, 184, 868–871. [Google Scholar] [CrossRef]
- Luger, K.; Mader, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- Whitlock, J.P., Jr.; Simpson, R.T. Localization of the sites along nucleosome DNA which interact with NH2-terminal histone regions. J. Biol. Chem. 1977, 252, 6516–6520. [Google Scholar] [CrossRef]
- Ausio, J.; Dong, F.; van Holde, K.E. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone "tails" in the stabilization of the nucleosome. J. Mol. Biol. 1989, 206, 451–463. [Google Scholar] [CrossRef]
- Zheng, C.; Hayes, J.J. Structures and interactions of the core histone tail domains. Biopolymers 2003, 68, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Carey, M.; Workman, J.L. The role of chromatin during transcription. Cell 2007, 128, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.D.; Widom, J. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol. 2000, 296, 979–987. [Google Scholar] [CrossRef]
- Jansen, A.; Verstrepen, K.J. Nucleosome positioning in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2011, 75, 301–320. [Google Scholar] [CrossRef]
- Field, Y.; Kaplan, N.; Fondufe-Mittendorf, Y.; Moore, I.K.; Sharon, E.; Lubling, Y.; Widom, J.; Segal, E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput. Biol. 2008, 4, e1000216. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Pugh, B.F. Nucleosome positioning and gene regulation: Advances through genomics. Nat. Rev. Genet. 2009, 10, 161–172. [Google Scholar] [CrossRef]
- Lee, W.; Tillo, D.; Bray, N.; Morse, R.H.; Davis, R.W.; Hughes, T.R.; Nislow, C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007, 39, 1235–1244. [Google Scholar] [CrossRef]
- Lorch, Y.; Kornberg, R.D. Chromatin-remodeling and the initiation of transcription. Q. Rev. Biophys. 2015, 48, 465–470. [Google Scholar] [CrossRef]
- Bowman, G.D.; McKnight, J.N. Sequence-specific targeting of chromatin remodelers organizes precisely positioned nucleosomes throughout the genome. Bioessays 2017, 39, 1–8. [Google Scholar] [CrossRef][Green Version]
- Kornberg, R.D.; Lorch, Y. Primary Role of the Nucleosome. Mol. Cell 2020, 79, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Shivaswamy, S.; Iyer, V.R. Stress-dependent dynamics of global chromatin remodeling in yeast: Dual role for SWI/SNF in the heat shock stress response. Mol. Cell Biol. 2008, 28, 2221–2234. [Google Scholar] [CrossRef] [PubMed]
- Shivaswamy, S.; Bhinge, A.; Zhao, Y.; Jones, S.; Hirst, M.; Iyer, V.R. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008, 6, e65. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wysocka, J.; Perlin, J.R.; Leonelli, L.; Allis, C.D.; Coonrod, S.A. Linking covalent histone modifications to epigenetics: The rigidity and plasticity of the marks. Cold Spring Harb. Symp. Quant. Biol. 2004, 69, 161–169. [Google Scholar] [CrossRef]
- Zhang, T.; Cooper, S.; Brockdorff, N. The interplay of histone modifications—Writers that read. EMBO Rep. 2015, 16, 1467–1481. [Google Scholar] [CrossRef]
- Clapier, C.R.; Cairns, B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef]
- Narlikar, G.J.; Fan, H.Y.; Kingston, R.E. Cooperation between complexes that regulate chromatin structure and transcription. Cell 2002, 108, 475–487. [Google Scholar] [CrossRef]
- Lin, A.; Du, Y.; Xiao, W. Yeast chromatin remodeling complexes and their roles in transcription. Curr. Genet. 2020, 66, 657–670. [Google Scholar] [CrossRef]
- Lorch, Y.; Kornberg, R.D. Chromatin-remodeling for transcription. Q. Rev. Biophys. 2017, 50, e5. [Google Scholar] [CrossRef]
- Villaseñor, R.; Baubec, T. Regulatory mechanisms governing chromatin organization and function. Curr. Opin. Cell Biol. 2020, 70, 10–17. [Google Scholar] [CrossRef]
- Weiner, A.; Chen, H.V.; Liu, C.L.; Rahat, A.; Klien, A.; Soares, L.; Gudipati, M.; Pfeffner, J.; Regev, A.; Buratowski, S.; et al. Systematic dissection of roles for chromatin regulators in a yeast stress response. PLoS Biol. 2012, 10, e1001369. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.T.; Workman, J.L. Chromatin proteins: Key responders to stress. PLoS Biol. 2012, 10, e1001371. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Al Aboud, N.M.; Tupper, C.; Jialal, I. Genetics, Epigenetic Mechanism; StatPearls: Treasure Island, FL, USA, 2021. [Google Scholar]
- Freitag, M. Histone Methylation by SET Domain Proteins in Fungi. Annu. Rev. Microbiol. 2017, 71, 413–439. [Google Scholar] [CrossRef]
- Separovich, R.J.; Wong, M.W.M.; Chapman, T.R.; Slavich, E.; Hamey, J.J.; Wilkins, M.R. Post-translational modification analysis of Saccharomyces cerevisiae histone methylation enzymes reveals phosphorylation sites of regulatory potential. J. Biol. Chem. 2021, 296, 100192. [Google Scholar] [CrossRef]
- Wilson, N.R.; Hochstrasser, M. The Regulation of Chromatin by Dynamic SUMO Modifications. Methods Mol. Biol. 2016, 1475, 23–38. [Google Scholar]
- Lawrence, M.; Daujat, S.; Schneider, R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet. 2016, 32, 42–56. [Google Scholar] [CrossRef]
- Lee, K.K.; Workman, J.L. Histone acetyltransferase complexes: One size doesn’t fit all. Nat. Rev. Mol. Cell Biol. 2007, 8, 284–295. [Google Scholar] [CrossRef]
- Cheon, Y.; Kim, H.; Park, K.; Kim, M.; Lee, D. Dynamic modules of the coactivator SAGA in eukaryotic transcription. Exp. Mol. Med. 2020, 52, 991–1003. [Google Scholar] [CrossRef]
- Nuño-Cabanes, C.; Rodriguez-Navarro, S. The promiscuity of the SAGA complex subunits: Multifunctional or moonlighting proteins? Biochim. Biophys. Acta Gene Regul. Mech. 2021, 1864, 194607. [Google Scholar] [CrossRef]
- Kuo, Y.M.; Andrews, A.J. Quantitating the specificity and selectivity of Gcn5-mediated acetylation of histone H3. PLoS ONE 2013, 8, e54896. [Google Scholar] [CrossRef]
- Pokholok, D.K.; Harbison, C.T.; Levine, S.; Cole, M.; Hannett, N.M.; Lee, T.I.; Bell, G.W.; Walker, K.; Rolfe, P.A.; Herbolsheimer, E.; et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 2005, 122, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.L.; Kaplan, T.; Kim, M.; Buratowski, S.; Schreiber, S.L.; Friedman, N.; Rando, O.J. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005, 3, e328. [Google Scholar] [CrossRef]
- Robert, F.; Pokholok, D.K.; Hannett, N.M.; Rinaldi, N.J.; Chandy, M.; Rolfe, A.; Workman, J.L.; Gifford, D.K.; Young, R.A. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell 2004, 16, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.J. Lysine acetylation and the bromodomain: A new partnership for signaling. Bioessays 2004, 26, 1076–1087. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Zhou, M.M. Bromodomain: An acetyl-lysine binding domain. FEBS Lett. 2002, 513, 124–128. [Google Scholar] [CrossRef]
- Winston, F.; Allis, C.D. The bromodomain: A chromatin-targeting module? Nat. Struct. Biol. 1999, 6, 601–604. [Google Scholar] [CrossRef]
- Mohibullah, N.; Hahn, S. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev. 2008, 22, 2994–3006. [Google Scholar] [CrossRef]
- Bhaumik, S.R.; Green, M.R. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 2002, 22, 7365–7371. [Google Scholar] [CrossRef]
- Dudley, A.M.; Rougeulle, C.; Winston, F. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 1999, 13, 2940–2945. [Google Scholar] [CrossRef]
- Huisinga, K.L.; Pugh, B.F. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 2004, 13, 573–585. [Google Scholar] [CrossRef]
- Lee, T.I.; Causton, H.C.; Holstege, F.C.; Shen, W.C.; Hannett, N.; Jennings, E.G.; Winston, F.; Green, M.R.; Young, R.A. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 2000, 405, 701–704. [Google Scholar] [CrossRef] [PubMed]
- Henry, K.W.; Wyce, A.; Lo, W.S.; Duggan, L.J.; Emre, N.C.; Kao, C.F.; Pillus, L.; Shilatifard, A.; Osley, M.A.; Berger, S.L. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003, 17, 2648–2663. [Google Scholar] [CrossRef] [PubMed]
- Daniel, J.A.; Torok, M.S.; Sun, Z.W.; Schieltz, D.; Allis, C.D.; Yates, J.R., 3rd; Grant, P.A. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J. Biol. Chem. 2004, 279, 1867–1871. [Google Scholar] [CrossRef]
- Prajapati, H.K.; Ocampo, J.; Clark, D.J. Interplay among ATP-Dependent Chromatin Remodelers Determines Chromatin Organisation in Yeast. Biology 2020, 9, 190. [Google Scholar] [CrossRef]
- Flaus, A.; Martin, D.M.; Barton, G.J.; Owen-Hughes, T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006, 34, 2887–2905. [Google Scholar] [CrossRef]
- Kasten, M.M.; Clapier, C.R.; Cairns, B.R. SnapShot: Chromatin remodeling: SWI/SNF. Cell 2011, 144, 310. [Google Scholar] [CrossRef]
- Cote, J.; Quinn, J.; Workman, J.L.; Peterson, C.L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 1994, 265, 53–60. [Google Scholar] [CrossRef]
- Smith, C.L.; Horowitz-Scherer, R.; Flanagan, J.F.; Woodcock, C.L.; Peterson, C.L. Structural analysis of the yeast SWI/SNF chromatin remodeling complex. Nat. Struct. Biol. 2003, 10, 141–145. [Google Scholar] [CrossRef]
- Cosma, M.P.; Tanaka, T.; Nasmyth, K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 1999, 97, 299–311. [Google Scholar] [CrossRef]
- Neely, K.E.; Hassan, A.H.; Wallberg, A.E.; Steger, D.J.; Cairns, B.R.; Wright, A.P.; Workman, J.L. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol. Cell 1999, 4, 649–655. [Google Scholar] [CrossRef]
- Yudkovsky, N.; Logie, C.; Hahn, S.; Peterson, C.L. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 1999, 13, 2369–2374. [Google Scholar] [CrossRef] [PubMed]
- Peterson, C.L.; Workman, J.L. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 2000, 10, 187–192. [Google Scholar] [CrossRef]
- Hahn, S.; Young, E.T. Transcriptional regulation in Saccharomyces cerevisiae: Transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics 2011, 189, 705–736. [Google Scholar] [CrossRef] [PubMed]
- De Nadal, E.; Zapater, M.; Alepuz, P.M.; Sumoy, L.; Mas, G.; Posas, F. The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature 2004, 427, 370–374. [Google Scholar] [CrossRef]
- Mas, G.; de Nadal, E.; Dechant, R.; Rodríguez de la Concepcion, M.L.; Logie, C.; Jimeno-Gonzalez, S.; Chavez, S.; Ammerer, G.; Posas, F. Recruitment of a chromatin remodelling complex by the Hog1 MAP kinase to stress genes. EMBO J. 2009, 28, 326–336. [Google Scholar] [CrossRef]
- Klopf, E.; Paskova, L.; Sole, C.; Mas, G.; Petryshyn, A.; Posas, F.; Wintersberger, U.; Ammerer, G.; Schuller, C. Cooperation between the INO80 complex and histone chaperones determines adaptation of stress gene transcription in the yeast Saccharomyces cerevisiae. Mol. Cell Biol. 2009, 29, 4994–5007. [Google Scholar] [CrossRef]
- Zapater, M.; Sohrmann, M.; Peter, M.; Posas, F.; de Nadal, E. Selective requirement for SAGA in Hog1-mediated gene expression depending on the severity of the external osmostress conditions. Mol. Cell. Biol. 2007, 27, 3900–3910. [Google Scholar] [CrossRef]
- Solé, C.; Nadal-Ribelles, M.; Kraft, C.; Peter, M.; Posas, F.; de Nadal, E. Control of Ubp3 ubiquitin protease activity by the Hog1 SAPK modulates transcription upon osmostress. EMBO J. 2011, 30, 3274–3284. [Google Scholar] [CrossRef]
- Bradley, A.I.; Marsh, N.M.; Borror, H.R.; Mostoller, K.E.; Gama, A.I.; Gardner, R.G. Acute ethanol stress induces sumoylation of conserved chromatin structural proteins in Saccharomyces cerevisiae. Mol. Biol. Cell 2021, 32, 1121–1133. [Google Scholar] [CrossRef]
- Sanz, A.B.; García, R.; Rodríguez-Peña, J.M.; Nombela, C.; Arroyo, J. Cooperation between SAGA and SWI/SNF complexes is required for efficient transcriptional responses regulated by the yeast MAPK Slt2. Nucleic Acids Res. 2016, 44, 7159–7172. [Google Scholar] [CrossRef] [PubMed]
- Sanz, A.B.; García, R.; Rodríguez-Peña, J.M.; Díez-Muñiz, S.; Nombela, C.; Peterson, C.L.; Arroyo, J. Chromatin remodeling by the SWI/SNF complex is essential for transcription mediated by the yeast cell wall integrity MAPK pathway. Mol. Biol. Cell 2012, 23, 2805–2817. [Google Scholar] [CrossRef] [PubMed]
- Nadal-Ribelles, M.; Mas, G.; Millan-Zambrano, G.; Sole, C.; Ammerer, G.; Chavez, S.; Posas, F.; de Nadal, E. H3K4 monomethylation dictates nucleosome dynamics and chromatin remodeling at stress-responsive genes. Nucleic Acids Res. 2015, 43, 4937–4949. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Liu, Y.; Xu, C.; Wang, Y.; Gong, J.; Shen, Y.; Wu, Q.; Boeke, J.D.; Dai, J. Dissecting Nucleosome Function with a Comprehensive Histone H2A and H2B Mutant Library. G3 2017, 7, 3857–3866. [Google Scholar] [CrossRef]
- Williams, S.K.; Truong, D.; Tyler, J.K. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc. Natl. Acad. Sci. USA 2008, 105, 9000–9005. [Google Scholar] [CrossRef]
- Weiner, A.; Hsieh, T.H.; Appleboim, A.; Chen, H.V.; Rahat, A.; Amit, I.; Rando, O.J.; Friedman, N. High-resolution chromatin dynamics during a yeast stress response. Mol. Cell 2015, 58, 371–386. [Google Scholar] [CrossRef]
- Vieitez, C.; Martinez-Cebrian, G.; Sole, C.; Bottcher, R.; Potel, C.M.; Savitski, M.M.; Onnebo, S.; Fabregat, M.; Shilatifard, A.; Posas, F.; et al. A genetic analysis reveals novel histone residues required for transcriptional reprogramming upon stress. Nucleic Acids Res. 2020, 48, 3455–3475. [Google Scholar] [CrossRef]
- Yang, S.H.; Sharrocks, A.D.; Whitmarsh, A.J. Transcriptional regulation by the MAP kinase signaling cascades. Gene 2003, 320, 3–21. [Google Scholar] [CrossRef]
- Nadal-Ribelles, M.; Solé, M.; Martínez-Cebrián, G.; Posas, F.; de Nadal, E. Shaping the Transcriptional Landscape through MAPK Signaling. In Gene Expression and Control; Uchiumi, F., Ed.; IntechOpen: London, UK, 2018; pp. 1–22. [Google Scholar]
- de Nadal, E.; Posas, F. Elongating under Stress. Genet. Res. Int. 2011, 2011, 326286. [Google Scholar] [CrossRef]
- Ferreiro, I.; Barragan, M.; Gubern, A.; Ballestar, E.; Joaquin, M.; Posas, F. The p38 SAPK is recruited to chromatin via its interaction with transcription factors. J. Biol. Chem. 2010, 285, 31819–31828. [Google Scholar] [CrossRef]
- Lawrence, M.C.; Shao, C.; McGlynn, K.; Naziruddin, B.; Levy, M.F.; Cobb, M.H. Multiple chromatin-bound protein kinases assemble factors that regulate insulin gene transcription. Proc. Natl. Acad. Sci. USA 2009, 106, 22181–22186. [Google Scholar] [CrossRef] [PubMed]
- Pokholok, D.K.; Zeitlinger, J.; Hannett, N.M.; Reynolds, D.B.; Young, R.A. Activated signal transduction kinases frequently occupy target genes. Science 2006, 313, 533–536. [Google Scholar] [CrossRef] [PubMed]
- Chow, C.W.; Davis, R.J. Proteins kinases: Chromatin-associated enzymes? Cell 2006, 127, 887–890. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Reiter, W.; Watt, S.; Dawson, K.; Lawrence, C.L.; Bahler, J.; Jones, N.; Wilkinson, C.R. Fission yeast MAP kinase Sty1 is recruited to stress-induced genes. J. Biol. Chem. 2008, 283, 9945–9956. [Google Scholar] [CrossRef] [PubMed]
- Sanso, M.; Vargas-Pérez, I.; Quintales, L.; Antequera, F.; Ayte, J.; Hidalgo, E. Gcn5 facilitates Pol II progression, rather than recruitment to nucleosome-depleted stress promoters, in Schizosaccharomyces pombe. Nucleic Acids Res. 2011, 39, 6369–6379. [Google Scholar] [CrossRef]
- Kim, K.Y.; Truman, A.W.; Levin, D.E. Yeast Mpk1 mitogen-activated protein kinase activates transcription through Swi4/Swi6 by a noncatalytic mechanism that requires upstream signal. Mol. Cell Biol. 2008, 28, 2579–2589. [Google Scholar] [CrossRef]
- Kim, K.Y.; Levin, D.E. Mpk1 MAPK association with the paf1 complex blocks sen1-mediated premature transcription termination. Cell 2011, 144, 745–756. [Google Scholar] [CrossRef]
- Truman, A.W.; Kim, K.Y.; Levin, D.E. Mechanism of Mpk1 mitogen-activated protein kinase binding to the Swi4 transcription factor and its regulation by a novel caffeine-induced phosphorylation. Mol. Cell Biol. 2009, 29, 6449–6461. [Google Scholar] [CrossRef]
- Sanz, A.B.; García, R.; Rodríguez-Peña, J.M.; Nombela, C.; Arroyo, J. Slt2 MAPK association with chromatin is required for transcriptional activation of Rlm1 dependent genes upon cell wall stress. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 1029–1039. [Google Scholar] [CrossRef]
- Sanz, A.B.; García, R.; Rodríguez-Peña, J.M.; Arroyo, J. The CWI Pathway: Regulation of the Transcriptional Adaptive Response to Cell Wall Stress in Yeast. J. Fungi 2017, 4, 1. [Google Scholar] [CrossRef]
- Gonzalez-Rubio, G.; Sellers-Moya, A.; Martin, H.; Molina, M. A walk-through MAPK structure and functionality with the 30-year-old yeast MAPK Slt2. Int. Microbiol. 2021, 24, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Quilis, I.; Gomar-Alba, M.; Igual, J.C. The CWI Pathway: A Versatile Toolbox to Arrest Cell-Cycle Progression. J. Fungi 2021, 7, 1041. [Google Scholar] [CrossRef] [PubMed]
- Pujol-Carrion, N.; Pavon-Verges, M.; Arroyo, J.; de la Torre-Ruiz, M.A. The MAPK Slt2/Mpk1 plays a role in iron homeostasis through direct regulation of the transcription factor Aft1. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118974. [Google Scholar] [CrossRef] [PubMed]
- Baetz, K.; Moffat, J.; Haynes, J.; Chang, M.; Andrews, B. Transcriptional coregulation by the cell integrity mitogen-activated protein kinase Slt2 and the cell cycle regulator Swi4. Mol. Cell Biol. 2001, 21, 6515–6528. [Google Scholar] [CrossRef]
- Watanabe, Y.; Takaesu, G.; Hagiwara, M.; Irie, K.; Matsumoto, K. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell Biol. 1997, 17, 2615–2623. [Google Scholar] [CrossRef]
- García, R.; Bermejo, C.; Grau, C.; Pérez, R.; Rodríguez-Peña, J.M.; Francois, J.; Nombela, C.; Arroyo, J. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J. Biol. Chem. 2004, 279, 15183–15195. [Google Scholar] [CrossRef]
- Boorsma, A.; De Nobel, H.; ter Riet, B.; Bargmann, B.; Brul, S.; Hellingwerf, K.J.; Klis, F.M. Characterization of the transcriptional response to cell wall stress in Saccharomyces cerevisiae. Yeast 2004, 21, 413–427. [Google Scholar] [CrossRef]
- García, R.; Itto-Nakama, K.; Rodríguez-Peña, J.M.; Chen, X.; Sanz, A.B.; de Lorenzo, A.; Pavon-Verges, M.; Kubo, K.; Ohnuki, S.; Nombela, C.; et al. Poacic acid, a beta-1,3-glucan-binding antifungal agent, inhibits cell-wall remodeling and activates transcriptional responses regulated by the cell-wall integrity and high-osmolarity glycerol pathways in yeast. FASEB J. 2021, 35, e21778. [Google Scholar] [CrossRef]
- García, R.; Rodríguez-Peña, J.M.; Bermejo, C.; Nombela, C.; Arroyo, J. The high osmotic response and cell wall integrity pathways cooperate to regulate transcriptional responses to zymolyase-induced cell wall stress in Saccharomyces cerevisiae. J. Biol. Chem. 2009, 284, 10901–10911. [Google Scholar] [CrossRef]
- Reinoso-Martin, C.; Schuller, C.; Schuetzer-Muehlbauer, M.; Kuchler, K. The yeast protein kinase C cell integrity pathway mediates tolerance to the antifungal drug caspofungin through activation of Slt2p mitogen-activated protein kinase signaling. Eukaryot. Cell 2003, 2, 1200–1210. [Google Scholar] [CrossRef]
- Agarwal, A.K.; Rogers, P.D.; Baerson, S.R.; Jacob, M.R.; Barker, K.S.; Cleary, J.D.; Walker, L.A.; Nagle, D.G.; Clark, A.M. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J. Biol. Chem. 2003, 278, 34998–35015. [Google Scholar] [CrossRef]
- Bermejo, C.; García, R.; Straede, A.; Rodríguez-Peña, J.M.; Nombela, C.; Heinisch, J.J.; Arroyo, J. Characterization of sensor-specific stress response by transcriptional profiling of wsc1 and mid2 deletion strains and chimeric sensors in Saccharomyces cerevisiae. OMICS 2010, 14, 679–688. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.; Bermejo, C.; García, R.; Rodríguez-Peña, J.M. Genomics in the detection of damage in microbial systems: Cell wall stress in yeast. Clin. Microbiol. Infect. 2009, 15, 44–46. [Google Scholar] [CrossRef] [PubMed]
- Bermejo, C.; Rodríguez, E.; García, R.; Rodríguez-Peña, J.M.; Rodríguez de la Concepcion, M.L.; Rivas, C.; Arias, P.; Nombela, C.; Posas, F.; Arroyo, J. The sequential activation of the yeast HOG and SLT2 pathways is required for cell survival to cell wall stress. Mol. Biol. Cell. 2008, 19, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Laz, E.V.; Lee, J.; Levin, D.E. Crosstalk between Saccharomyces cerevisiae SAPKs Hog1 and Mpk1 is mediated by glycerol accumulation. Fungal Biol. 2020, 124, 361–367. [Google Scholar] [CrossRef]
- Dunayevich, P.; Baltanas, R.; Clemente, J.A.; Couto, A.; Sapochnik, D.; Vasen, G.; Colman-Lerner, A. Heat-stress triggers MAPK crosstalk to turn on the hyperosmotic response pathway. Sci. Rep. 2018, 8, 15168. [Google Scholar] [CrossRef]
- Cañonero, L.; Pautasso, C.; Galello, F.; Sigaut, L.; Pietrasanta, L.; Arroyo, J.; Bermudez-Moretti, M.; Portela, P.; Rossi, S. Heat stress regulates the expression of TPK1 gene at transcriptional and post-transcriptional levels in Saccharomyces cerevisiae. Biochim. Biophys. Acta Mol. Cell Res. 2022, 1869, 119209. [Google Scholar] [CrossRef]
- García, R.; Bravo, E.; Díez-Muñiz, S.; Nombela, C.; Rodríguez-Peña, J.M.; Arroyo, J. A novel connection between the Cell Wall Integrity and the PKA pathways regulates cell wall stress response in yeast. Sci. Rep. 2017, 7, 5703. [Google Scholar] [CrossRef]
- Roberts, C.J.; Nelson, B.; Marton, M.J.; Stoughton, R.; Meyer, M.R.; Bennett, H.A.; He, Y.D.; Dai, H.; Walker, W.L.; Hughes, T.R.; et al. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 2000, 287, 873–880. [Google Scholar] [CrossRef]
- García, R.; Sanz, A.B.; Rodríguez-Peña, J.M.; Nombela, C.; Arroyo, J. Rlm1 mediates positive autoregulatory transcriptional feedback that is essential for Slt2-dependent gene expression. J. Cell Sci. 2016, 129, 1649–1660. [Google Scholar] [CrossRef]
- Taylor, I.A.; McIntosh, P.B.; Pala, P.; Treiber, M.K.; Howell, S.; Lane, A.N.; Smerdon, S.J. Characterization of the DNA-binding domains from the yeast cell-cycle transcription factors Mbp1 and Swi4. Biochemistry 2000, 39, 3943–3954. [Google Scholar] [CrossRef] [PubMed]
- Sedgwick, S.G.; Taylor, I.A.; Adam, A.C.; Spanos, A.; Howell, S.; Morgan, B.A.; Treiber, M.K.; Kanuga, N.; Banks, G.R.; Foord, R.; et al. Structural and functional architecture of the yeast cell-cycle transcription factor Swi6. J. Mol. Biol. 1998, 281, 763–775. [Google Scholar] [CrossRef]
- Breeden, L.L. Periodic transcription: A cycle within a cycle. Curr.Biol. 2003, 13, R31–R38. [Google Scholar] [CrossRef]
- Hendler, A.; Medina, E.M.; Buchler, N.E.; de Bruin, R.A.M.; Aharoni, A. The evolution of a G1/S transcriptional network in yeasts. Curr. Genet. 2018, 64, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.Y.; Levin, D.E. Transcriptional reporters for genes activated by cell wall stress through a non-catalytic mechanism involving Mpk1 and SBF. Yeast 2010, 27, 541–548. [Google Scholar] [CrossRef]
- Yurko, N.; Liu, X.; Yamazaki, T.; Hoque, M.; Tian, B.; Manley, J.L. MPK1/SLT2 Links Multiple Stress Responses with Gene Expression in Budding Yeast by Phosphorylating Tyr1 of the RNAP II CTD. Mol. Cell 2017, 68, 913–925. [Google Scholar] [CrossRef]
- Mayer, A.; Heidemann, M.; Lidschreiber, M.; Schreieck, A.; Sun, M.; Hintermair, C.; Kremmer, E.; Eick, D.; Cramer, P. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science 2012, 336, 1723–1725. [Google Scholar] [CrossRef]
- Jung, U.S.; Levin, D.E. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 1999, 34, 1049–1057. [Google Scholar] [CrossRef]
- Watanabe, Y.; Irie, K.; Matsumoto, K. Yeast RLM1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell Biol. 1995, 15, 5740–5749. [Google Scholar] [CrossRef]
- Dodou, E.; Treisman, R. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol. Cell Biol. 1997, 17, 1848–1859. [Google Scholar] [CrossRef]
- Jung, U.S.; Sobering, A.K.; Romeo, M.J.; Levin, D.E. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol. Microbiol. 2002, 46, 781–789. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Pugh, B.F. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 2009, 10, R109. [Google Scholar] [CrossRef] [PubMed]
- Albert, I.; Mavrich, T.N.; Tomsho, L.P.; Qi, J.; Zanton, S.J.; Schuster, S.C.; Pugh, B.F. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 2007, 446, 572–576. [Google Scholar] [CrossRef] [PubMed]
- Tirosh, I.; Barkai, N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008, 18, 1084–1091. [Google Scholar] [CrossRef]
- Rando, O.J.; Winston, F. Chromatin and transcription in yeast. Genetics 2012, 190, 351–387. [Google Scholar] [CrossRef]
- Basehoar, A.D.; Zanton, S.J.; Pugh, B.F. Identification and distinct regulation of yeast TATA box-containing genes. Cell 2004, 116, 699–709. [Google Scholar] [CrossRef]
- De Nadal, E.; Posas, F. Regulation of gene expression in response to osmostress by the yeast stress-activated protein kinase Hog1. In Topics in Current Genetics; Posas, F., Nebreda, A.R., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; Volume 20, pp. 81–97. [Google Scholar]
- Proft, M.; Struhl, K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell. 2002, 9, 1307–1317. [Google Scholar] [CrossRef]
- de Nadal, E.; Posas, F. Multilayered control of gene expression by stress-activated protein kinases. EMBO J. 2010, 29, 4–13. [Google Scholar] [CrossRef]
- Alepuz, P.M.; de Nadal, E.; Zapater, M.; Ammerer, G.; Posas, F. Osmostress-induced transcription by Hot1 depends on a Hog1-mediated recruitment of the RNA Pol II. EMBO J. 2003, 22, 2433–2442. [Google Scholar] [CrossRef]
- Alepuz, P.M.; Jovanovic, A.; Reiser, V.; Ammerer, G. Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol. Cell. 2001, 7, 767–777. [Google Scholar] [CrossRef]
- Ruiz-Roig, C.; Vieitez, C.; Posas, F.; de Nadal, E. The Rpd3L HDAC complex is essential for the heat stress response in yeast. Mol. Microbiol. 2010, 76, 1049–1062. [Google Scholar] [CrossRef]
- Alejandro-Osorio, A.L.; Huebert, D.J.; Porcaro, D.T.; Sonntag, M.E.; Nillasithanukroh, S.; Will, J.L.; Gasch, A.P. The histone deacetylase Rpd3p is required for transient changes in genomic expression in response to stress. Genome Biol. 2009, 10, R57. [Google Scholar] [CrossRef]
- Geng, F.; Laurent, B.C. Roles of SWI/SNF and HATs throughout the dynamic transcription of a yeast glucose-repressible gene. EMBO J. 2004, 23, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Erkina, T.Y.; Zou, Y.; Freeling, S.; Vorobyev, V.I.; Erkine, A.M. Functional interplay between chromatin remodeling complexes RSC, SWI/SNF and ISWI in regulation of yeast heat shock genes. Nucleic Acids Res. 2010, 38, 1441–1449. [Google Scholar] [CrossRef] [PubMed]
- Angus-Hill, M.L.; Schlichter, A.; Roberts, D.; Erdjument-Bromage, H.; Tempst, P.; Cairns, B.R. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell 2001, 7, 741–751. [Google Scholar] [CrossRef]
- García, R.; Botet, J.; Rodríguez-Peña, J.M.; Bermejo, C.; Ribas, J.C.; Revuelta, J.L.; Nombela, C.; Arroyo, J. Genomic profiling of fungal cell wall-interfering compounds: Identification of a common gene signature. BMC Genomics 2015, 16, 683. [Google Scholar] [CrossRef]
- Wilson, B.; Erdjument-Bromage, H.; Tempst, P.; Cairns, B.R. The RSC chromatin remodeling complex bears an essential fungal-specific protein module with broad functional roles. Genetics. 2006, 172, 795–809. [Google Scholar] [CrossRef]
- Kasler, H.G.; Victoria, J.; Duramad, O.; Winoto, A. ERK5 is a novel type of mitogen-activated protein kinase containing a transcriptional activation domain. Mol. Cell. Biol. 2000, 20, 8382–8389. [Google Scholar] [CrossRef]
- Soler, M.; Plovins, A.; Martin, H.; Molina, M.; Nombela, C. Characterization of domains in the yeast MAP kinase Slt2 (Mpk1) required for functional activity and in vivo interaction with protein kinases Mkk1 and Mkk2. Mol. Microbiol. 1995, 17, 833–842. [Google Scholar] [CrossRef]
- Truman, A.W.; Millson, S.H.; Nuttall, J.M.; King, V.; Mollapour, M.; Prodromou, C.; Pearl, L.H.; Piper, P.W. Expressed in the yeast Saccharomyces cerevisiae, human ERK5 is a client of the Hsp90 chaperone that complements loss of the Slt2p (Mpk1p) cell integrity stress-activated protein kinase. Eukaryot. Cell 2006, 5, 1914–1924. [Google Scholar] [CrossRef]
- Morimoto, H.; Kondoh, K.; Nishimoto, S.; Terasawa, K.; Nishida, E. Activation of a C-terminal transcriptional activation domain of ERK5 by autophosphorylation. J. Biol. Chem. 2007, 282, 35449–35456. [Google Scholar] [CrossRef] [PubMed]
- Sohn, S.J.; Li, D.; Lee, L.K.; Winoto, A. Transcriptional regulation of tissue-specific genes by the ERK5 mitogen-activated protein kinase. Mol. Cell. Biol. 2005, 25, 8553–8566. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.Y.; Cosano, I.C.; Levin, D.E.; Molina, M.; Martin, H. Dissecting the transcriptional activation function of the cell wall integrity MAP kinase. Yeast 2007, 24, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Sariki, S.K.; Kumawat, R.; Singh, V.; Tomar, R.S. Flocculation of Saccharomyces cerevisiae is dependent on activation of Slt2 and Rlm1 regulated by the cell wall integrity pathway. Mol. Microbiol. 2019, 112, 1350–1369. [Google Scholar] [CrossRef]
- Proft, M.; Mas, G.; de Nadal, E.; Vendrell, A.; Noriega, N.; Struhl, K.; Posas, F. The stress-activated Hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Mol. Cell 2006, 23, 241–250. [Google Scholar] [CrossRef]
- Klein, A.M.; Zaganjor, E.; Cobb, M.H. Chromatin-tethered MAPKs. Curr. Opin. Cell Biol. 2013, 25, 272–277. [Google Scholar] [CrossRef]
- Silva, A.; Cavero, S.; Begley, V.; Sole, C.; Bottcher, R.; Chavez, S.; Posas, F.; de Nadal, E. Regulation of transcription elongation in response to osmostress. PLoS Genet. 2017, 13, e1007090. [Google Scholar] [CrossRef]
- Wright, D.E.; Wang, C.Y.; Kao, C.F. Histone ubiquitylation and chromatin dynamics. Frontiers in bioscience (Landmark edition) 2012, 17, 1051–1078. [Google Scholar] [CrossRef]
- Workman, J.L. CHROMATIN. It takes teamwork to modify chromatin. Science 2016, 351, 667. [Google Scholar] [CrossRef]
- Batta, K.; Zhang, Z.; Yen, K.; Goffman, D.B.; Pugh, B.F. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 2011, 25, 2254–2265. [Google Scholar] [CrossRef]
- Fierz, B.; Chatterjee, C.; McGinty, R.K.; Bar-Dagan, M.; Raleigh, D.P.; Muir, T.W. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat. Chem. Biol. 2011, 7, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekharan, M.B.; Huang, F.; Sun, Z.W. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc. Natl. Acad. Sci. USA 2009, 106, 16686–16691. [Google Scholar] [CrossRef] [PubMed]
- García, R.; Mascaraque, V.; Pavón-Vergés, M.; Sanz, A.B.; Rodríguez-Peña, J.M.; Nombela, C.; Arroyo, J. Manuscript in preparation 2022.
- Lee, J.; Liu, L.; Levin, D.E. Stressing out or stressing in: Intracellular pathways for SAPK activation. Curr. Genet. 2019, 65, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Levin, D.E. Intracellular mechanism by which arsenite activates the yeast stress MAPK Hog1. Mol. Biol. Cell 2018, 29, 1904–1915. [Google Scholar] [CrossRef]
- Liu, L.; Levin, D.E. Intracellular mechanism by which genotoxic stress activates yeast SAPK Mpk1. Mol. Biol. Cell 2018, 29, 2898–2909. [Google Scholar] [CrossRef]
- Sotelo, J.; Rodriguez-Gabriel, M.A. Mitogen-activated protein kinase Hog1 is essential for the response to arsenite in Saccharomyces cerevisiae. Eukaryot. Cell 2006, 5, 1826–1830. [Google Scholar] [CrossRef][Green Version]
- Liu, L.; Veis, J.; Reiter, W.; Motari, E.; Costello, C.E.; Samuelson, J.C.; Ammerer, G.; Levin, D.E. Regulation of Pkc1 Hyper-Phosphorylation by Genotoxic Stress. J. Fungi 2021, 7, 874. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, S.; Wang, J.; Liu, Y.; Deng, Y. Roles of High Osmolarity Glycerol and Cell Wall Integrity Pathways in Cadmium Toxicity in Saccharomyces cerevisiae. Int. J. Mol. Sci. 2021, 22, 6169. [Google Scholar] [CrossRef]
- Techo, T.; Charoenpuntaweesin, S.; Auesukaree, C. Involvement of the Cell Wall Integrity Pathway of Saccharomyces cerevisiae in Protection against Cadmium and Arsenate Stresses. Appl. Environ. Microbiol. 2020, 86, e01339-20. [Google Scholar] [CrossRef]
- Wosika, V.; Pelet, S. Single-particle imaging of stress-promoters induction reveals the interplay between MAPK signaling, chromatin and transcription factors. Nat. Commun. 2020, 11, 3171. [Google Scholar] [CrossRef]
- Adamson, B.; Norman, T.M.; Jost, M.; Cho, M.Y.; Nunez, J.K.; Chen, Y.W.; Villalta, J.E.; Gilbert, L.A.; Horlbeck, M.A.; Hein, M.Y.; et al. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell 2016, 167, 1867–1882. [Google Scholar] [CrossRef] [PubMed]
- Vihervaara, A.; Duarte, F.M.; Lis, J.T. Molecular mechanisms driving transcriptional stress responses. Nat. Rev. Genet. 2018, 19, 385–397. [Google Scholar] [CrossRef] [PubMed]
- García, R.; Pulido, V.; Orellana-Munoz, S.; Nombela, C.; Vazquez de Aldana, C.R.; Rodríguez-Peña, J.M.; Arroyo, J. Signalling through the yeast MAPK Cell Wall Integrity pathway controls P-body assembly upon cell wall stress. Sci. Rep. 2019, 9, 3186. [Google Scholar] [CrossRef]
- Nithianandarajah-Jones, G.N.; Wilm, B.; Goldring, C.E.; Muller, J.; Cross, M.J. ERK5: Structure, regulation and function. Cell. Signal. 2012, 24, 2187–2196. [Google Scholar] [CrossRef] [PubMed]
- Pereira, D.M.; Rodrigues, C.M.P. Targeted Avenues for Cancer Treatment: The MEK5-ERK5 Signaling Pathway. Trends Mol. Med. 2020, 26, 394–407. [Google Scholar] [CrossRef] [PubMed]
- Simoes, A.E.; Rodrigues, C.M.; Borralho, P.M. The MEK5/ERK5 signalling pathway in cancer: A promising novel therapeutic target. Drug Discov. Today 2016, 21, 1654–1663. [Google Scholar] [CrossRef]
- Hoang, V.T.; Yan, T.J.; Cavanaugh, J.E.; Flaherty, P.T.; Beckman, B.S.; Burow, M.E. Oncogenic signaling of MEK5-ERK5. Cancer Lett. 2017, 392, 51–59. [Google Scholar] [CrossRef]
- Walker, L.A.; Gow, N.A.; Munro, C.A. Fungal echinocandin resistance. Fungal Genet. Biol. 2010, 47, 117–126. [Google Scholar] [CrossRef]
- Fortwendel, J.R.; Juvvadi, P.R.; Perfect, B.Z.; Rogg, L.E.; Perfect, J.R.; Steinbach, W.J. Transcriptional regulation of chitin synthases by calcineurin controls paradoxical growth of Aspergillus fumigatus in response to caspofungin. Antimicrob. Agents Chemother. 2010, 54, 1555–1563. [Google Scholar] [CrossRef]
- Lara-Aguilar, V.; Rueda, C.; Garcia-Barbazan, I.; Varona, S.; Monzon, S.; Jimenez, P.; Cuesta, I.; Zaballos, A.; Zaragoza, O. Adaptation of the emerging pathogenic yeast Candida auris to high caspofungin concentrations correlates with cell wall changes. Virulence 2021, 12, 1400–1417. [Google Scholar] [CrossRef]
- de Oliveira, H.C.; Rossi, S.A.; Garcia-Barbazan, I.; Zaragoza, O.; Trevijano-Contador, N. Cell Wall Integrity Pathway Involved in Morphogenesis, Virulence and Antifungal Susceptibility in Cryptococcus neoformans. J. Fungi 2021, 7, 831. [Google Scholar] [CrossRef] [PubMed]
- Ibe, C.; Munro, C.A. Fungal Cell Wall Proteins and Signaling Pathways Form a Cytoprotective Network to Combat Stresses. J. Fungi 2021, 7, 739. [Google Scholar] [CrossRef] [PubMed]
- Robbins, N.; Caplan, T.; Cowen, L.E. Molecular Evolution of Antifungal Drug Resistance. Ann. Rev. Microbiol. 2017, 71, 753–775. [Google Scholar] [CrossRef] [PubMed]
- Mota Fernandes, C.; Dasilva, D.; Haranahalli, K.; McCarthy, J.B.; Mallamo, J.; Ojima, I.; Del Poeta, M. The Future of Antifungal Drug Therapy: Novel Compounds and Targets. Antimicrob. Agents Chemother. 2021, 65, e01719-20. [Google Scholar] [CrossRef]
- Sussman, A.; Huss, K.; Chio, L.C.; Heidler, S.; Shaw, M.; Ma, D.; Zhu, G.; Campbell, R.M.; Park, T.S.; Kulanthaivel, P.; et al. Discovery of cercosporamide, a known antifungal natural product, as a selective Pkc1 kinase inhibitor through high-throughput screening. Eukaryot. Cell. 2004, 3, 932–943. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.K.; Feng, Q.; Liu, L.; Levin, D.E.; Roy, K.K.; Doerksen, R.J.; Baerson, S.R.; Shi, X.; Pan, X.; Xu, W.H.; et al. Puupehenone, a Marine-Sponge-Derived Sesquiterpene Quinone, Potentiates the Antifungal Drug Caspofungin by Disrupting Hsp90 Activity and the Cell Wall Integrity Pathway. mSphere 2020, 5, e00818-19. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.; Farkas, V.; Sanz, A.B.; Cabib, E. Strengthening the fungal cell wall through chitin-glucan cross-links: Effects on morphogenesis and cell integrity. Cell. Microbiol. 2016, 18, 1239–1250. [Google Scholar] [CrossRef]
- Cabib, E.; Arroyo, J. How carbohydrates sculpt cells: Chemical control of morphogenesis in the yeast cell wall. Nat. Rev. Microbiol. 2013, 11, 648–655. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanz, A.B.; García, R.; Pavón-Vergés, M.; Rodríguez-Peña, J.M.; Arroyo, J. Control of Gene Expression via the Yeast CWI Pathway. Int. J. Mol. Sci. 2022, 23, 1791. https://doi.org/10.3390/ijms23031791
Sanz AB, García R, Pavón-Vergés M, Rodríguez-Peña JM, Arroyo J. Control of Gene Expression via the Yeast CWI Pathway. International Journal of Molecular Sciences. 2022; 23(3):1791. https://doi.org/10.3390/ijms23031791
Chicago/Turabian StyleSanz, Ana Belén, Raúl García, Mónica Pavón-Vergés, José Manuel Rodríguez-Peña, and Javier Arroyo. 2022. "Control of Gene Expression via the Yeast CWI Pathway" International Journal of Molecular Sciences 23, no. 3: 1791. https://doi.org/10.3390/ijms23031791
APA StyleSanz, A. B., García, R., Pavón-Vergés, M., Rodríguez-Peña, J. M., & Arroyo, J. (2022). Control of Gene Expression via the Yeast CWI Pathway. International Journal of Molecular Sciences, 23(3), 1791. https://doi.org/10.3390/ijms23031791






