miR-196a Upregulation Contributes to Gefitinib Resistance through Inhibiting GLTP Expression
Abstract
1. Introduction
2. Results
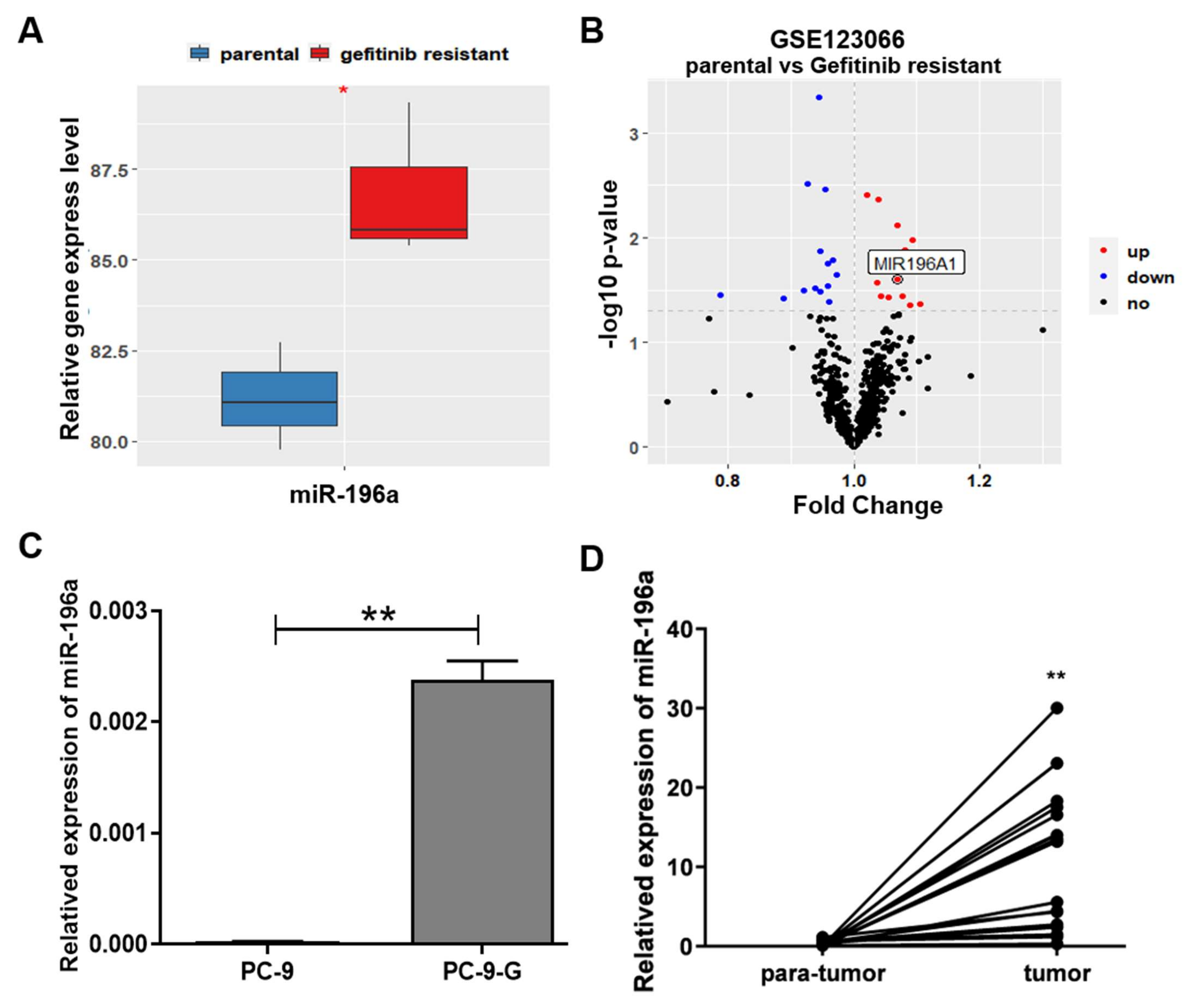
2.1. miR-196a Expression Levels Were Greatly Upregulated in Gefitinib-Resistant Lung Cancer Cells, and Its Levels Were Positively Correlated with Poor Overall Survival in Lung Cancer Patients
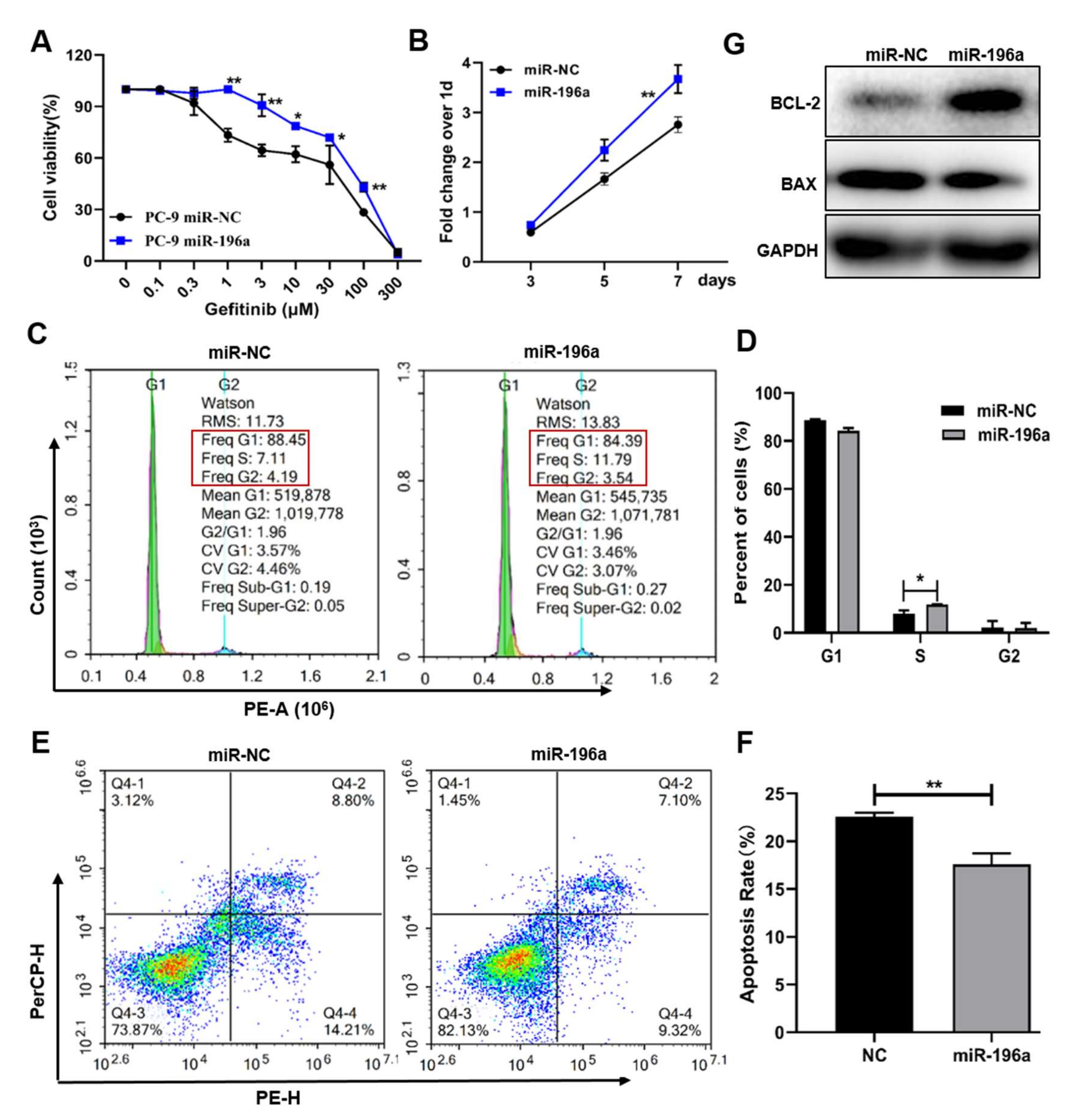
2.2. Forced Expression of miR-196a-Induced Gefitinib Resistance and Cell Proliferation, and Inhibited Cell Apoptosis That Was Important in Cell Resistance to Gefitinib Treatment
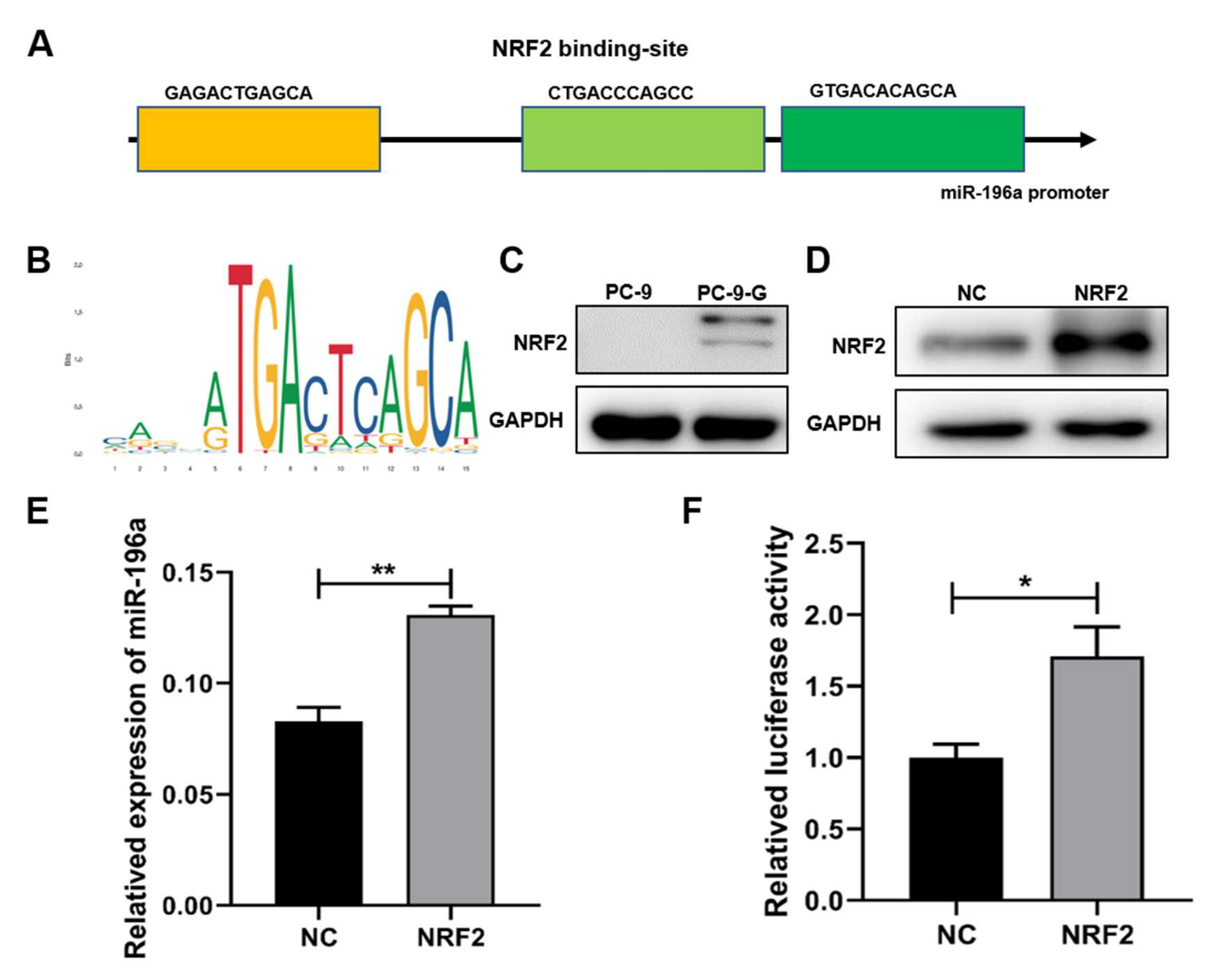
2.3. NRF2 Expression Levels Were Increased in Gefitinib-Resistant Cells and NRF2 Induced miR-196a Expression at Transcriptional Level
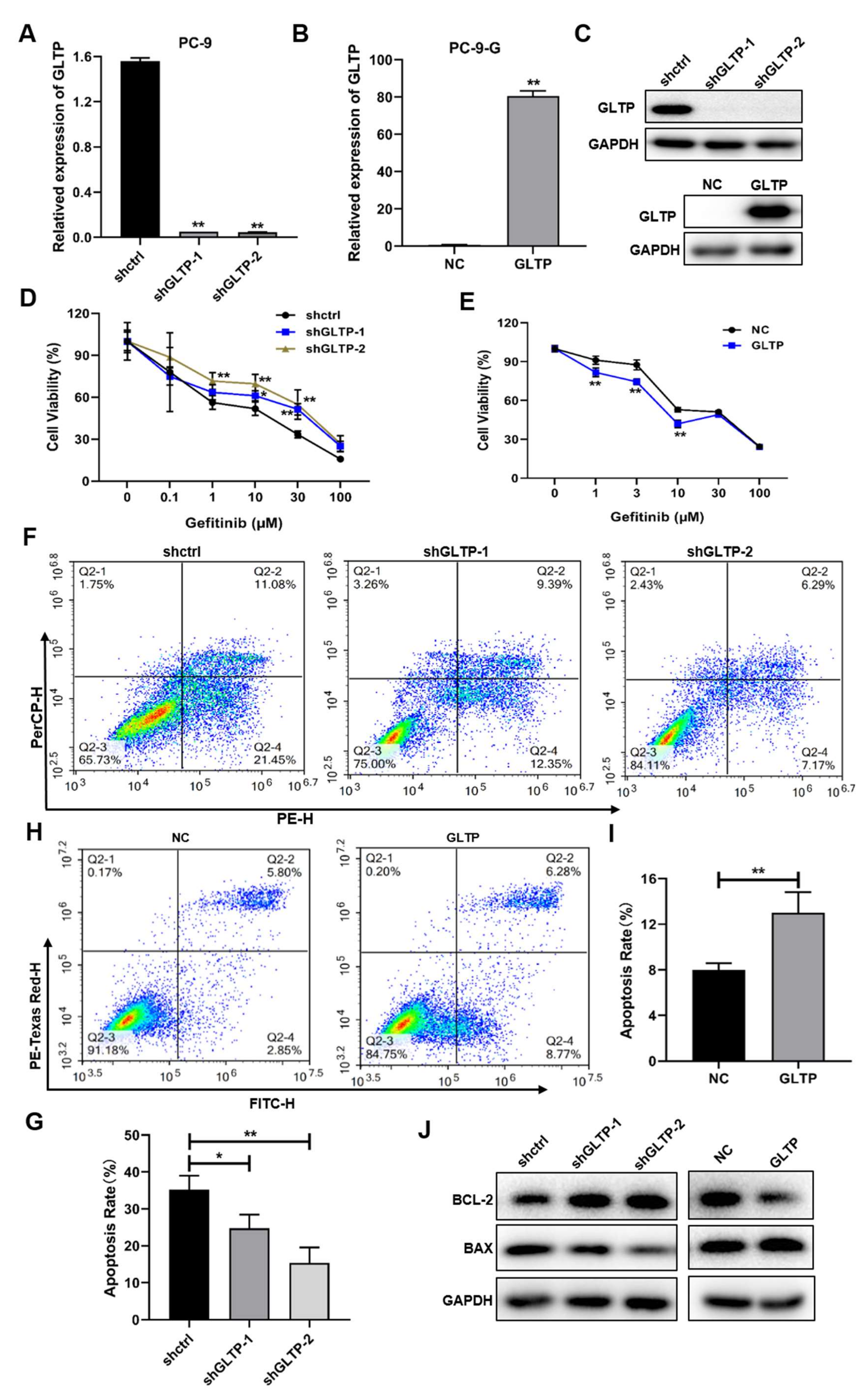
2.4. GLTP Was a Direct Functional Target of miR-196a, and GLTP Suppression Was Important for the Resistance of Cells to Gefitinib Treatment
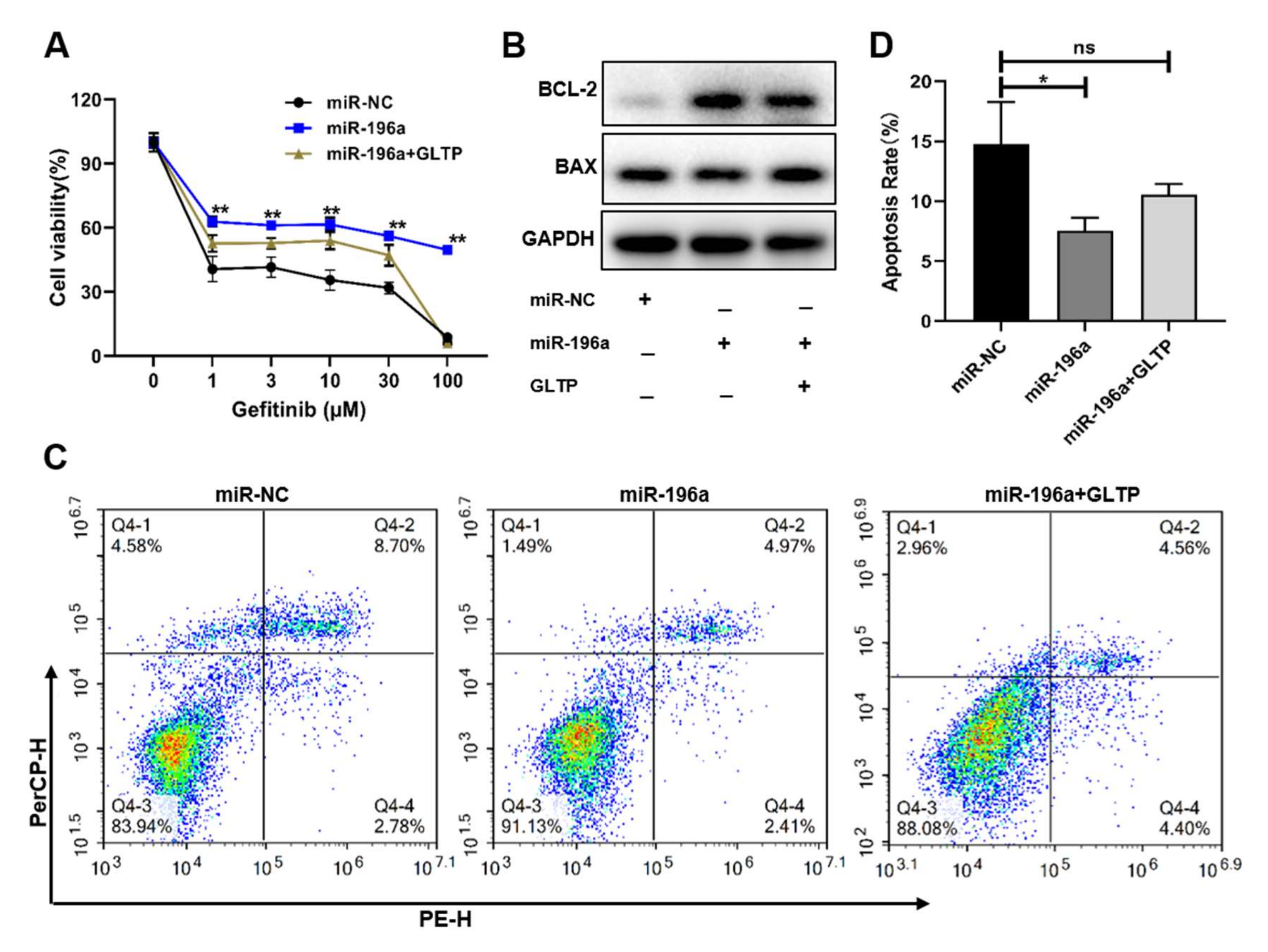
2.5. GLTP-Forced Expression Inhibited miR-196a-Inducing Gefitinib Resistance
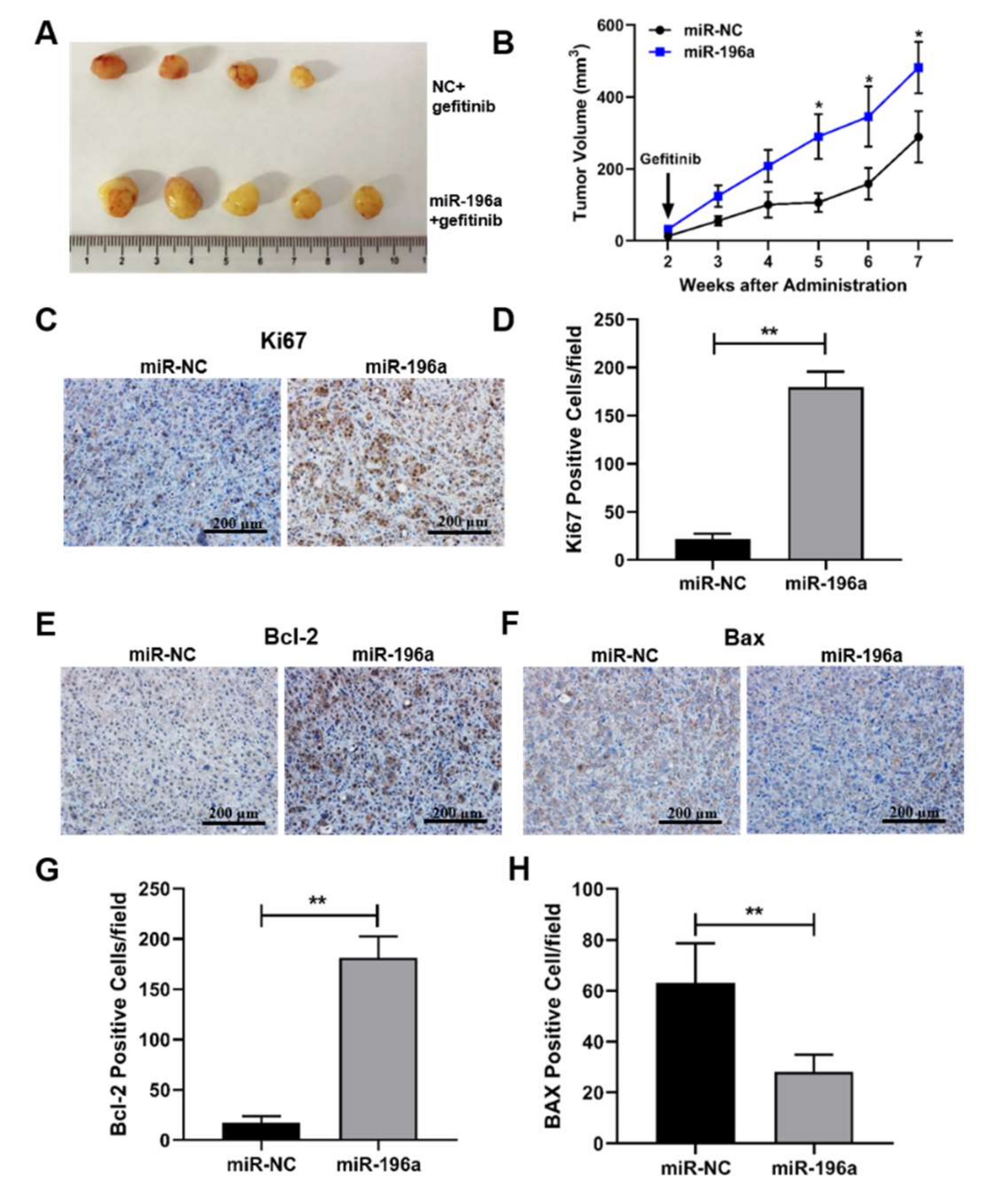
2.6. miR-196a-Forced Expression Induced Tumor Growth and Inhibited Cell Apoptosis In Vivo, Which May Contribute to Gefitinib Resistance
3. Discussion
4. Materials and Methods
4.1. Human Lung Cancer Specimens
4.2. Cell Culture and Reagents
4.3. RNA Extraction and Quantitative Real-Time PCR Analysis
4.4. Cell Viability Assay
4.5. Cell Apoptosis Assay
4.6. Western Blot Analysis
4.7. Transfection
4.8. Luciferase Reporter Assay
4.9. Tumor Growth Assay In Vivo
4.10. Immunohistochemistry (IHC) Assay
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Thai, A.A.; Solomon, B.J.; Sequist, L.V.; Gainor, J.F.; Heist, R.S. Lung cancer. Lancet 2021, 398, 535–554. [Google Scholar] [CrossRef]
- Pelayo Alvarez, M.; Gallego Rubio, O.; Bonfill Cosp, X.; Agra Varela, Y. Chemotherapy versus best supportive care for extensive small cell lung cancer. Cochrane Database Syst. Rev. 2009, CD001990. [Google Scholar] [CrossRef]
- Miller, M.; Hanna, N. Advances in systemic therapy for non-small cell lung cancer. BMJ 2021, 375, n2363. [Google Scholar] [CrossRef]
- Schaake, E.E.; Kappers, I.; Codrington, H.E.; Valdes Olmos, R.A.; Teertstra, H.J.; van Pel, R.; Burgers, J.A.; van Tinteren, H.; Klomp, H.M. Tumor response and toxicity of neoadjuvant erlotinib in patients with early-stage non-small-cell lung cancer. J. Clin. Oncol. 2012, 30, 2731–2738. [Google Scholar] [CrossRef]
- Jankovic, R.; Goncalves, H.J.; Cavic, M.; Clemente, C.; Lind, M.; Murillo Carrasco, A.; Nadifi, S.; Khyatti, M.; Adebambo, T.; Egamberdiev, D. LungCARD—Report on worldwide research and clinical practices related to lung cancer. J. BUON 2019, 24, 11–19. [Google Scholar]
- Midha, A.; Dearden, S.; McCormack, R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: A systematic review and global map by ethnicity (mutMapII). Am. J. Cancer Res. 2015, 5, 2892–2911. [Google Scholar]
- Wang, S.; Gao, A.; Liu, J.; Sun, Y. First-Line therapy for advanced non-small cell lung cancer with activating EGFR mutation: Is combined EGFR-TKIs and chemotherapy a better choice? Cancer Chemother. Pharmacol. 2018, 81, 443–453. [Google Scholar] [CrossRef]
- Kazandjian, D.; Blumenthal, G.M.; Yuan, W.; He, K.; Keegan, P.; Pazdur, R. FDA approval of gefitinib for the treatment of patients with metastatic EGFR mutation-positive non-small cell lung cancer. Clin. Cancer Res. 2016, 22, 1307–1312. [Google Scholar] [CrossRef]
- Noronha, V.; Patil, V.M.; Joshi, A.; Menon, N.; Chougule, A.; Mahajan, A.; Janu, A.; Purandare, N.; Kumar, R.; More, S.; et al. Gefitinib versus gefitinib plus pemetrexed and carboplatin chemotherapy in EGFR-mutated lung cancer. J. Clin. Oncol. 2020, 38, 124–136. [Google Scholar] [CrossRef]
- Wang, J.; Wang, B.; Chu, H.; Yao, Y. Intrinsic resistance to EGFR tyrosine kinase inhibitors in advanced non-small-cell lung cancer with activating EGFR mutations. Onco Targets Ther. 2016, 9, 3711–3726. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Huang, Y.; Shi, J.; Dai, Y.; Wu, L.; Zhou, H. ABCC10 Plays a significant role in the transport of gefitinib and contributes to acquired resistance to gefitinib in NSCLC. Front. Pharmacol. 2018, 9, 1312. [Google Scholar] [CrossRef] [PubMed]
- Radmanesh, F.; Sadeghi Abandansari, H.; Ghanian, M.H.; Pahlavan, S.; Varzideh, F.; Yakhkeshi, S.; Alikhani, M.; Moradi, S.; Braun, T.; Baharvand, H. Hydrogel-Mediated delivery of microRNA-92a inhibitor polyplex nanoparticles induces localized angiogenesis. Angiogenesis 2021, 24, 657–676. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Xing, Z.; Guo, Z.; Qiu, Y.; Liu, Z. Hypoxia-Induced microRNA-155 overexpression in extracellular vesicles promotes renal cell carcinoma progression by targeting FOXO3. Aging 2021, 13, 9613–9626. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, Y.; Zhang, Q.; Liu, B.; Cheng, Y.; Zhang, Y.; Sun, Y.; Liu, J. Exosomal miR-590-3p derived from cancer-associated fibroblasts confers radioresistance in colorectal cancer. Mol. Ther. Nucleic Acids 2021, 24, 113–126. [Google Scholar] [CrossRef]
- Hill, M.; Tran, N. MicroRNAs regulating MicroRNAs in cancer. Trends Cancer 2018, 4, 465–468. [Google Scholar] [CrossRef]
- Li, W.; Wang, L.; Ji, X.B.; Wang, L.H.; Ge, X.; Liu, W.T.; Chen, L.; Zheng, Z.; Shi, Z.M.; Liu, L.Z.; et al. MiR-199a inhibits tumor growth and attenuates chemoresistance by targeting K-RAS via AKT and ERK signalings. Front. Oncol. 2019, 9, 1071. [Google Scholar] [CrossRef]
- Wang, L.; Ji, X.B.; Wang, L.H.; Qiu, J.G.; Zhou, F.M.; Liu, W.J.; Wan, D.D.; Lin, M.C.; Liu, L.Z.; Zhang, J.Y.; et al. Regulation of MicroRNA-497-targeting AKT2 influences tumor growth and chemoresistance to cisplatin in lung cancer. Front. Cell Dev. Biol. 2020, 8, 840. [Google Scholar] [CrossRef]
- Jiang, C.F.; Shi, Z.M.; Li, D.M.; Qian, Y.C.; Ren, Y.; Bai, X.M.; Xie, Y.X.; Wang, L.; Ge, X.; Liu, W.T.; et al. Estrogen-Induced miR-196a elevation promotes tumor growth and metastasis via targeting SPRED1 in breast cancer. Mol. Cancer 2018, 17, 83. [Google Scholar] [CrossRef]
- Sun, M.; Liu, X.H.; Li, J.H.; Yang, J.S.; Zhang, E.B.; Yin, D.D.; Liu, Z.L.; Zhou, J.; Ding, Y.; Li, S.Q.; et al. MiR-196a is upregulated in gastric cancer and promotes cell proliferation by downregulating p27(kip1). Mol. Cancer Ther. 2012, 11, 842–852. [Google Scholar] [CrossRef]
- Takkar, S.; Sharma, V.; Ghosh, S.; Suri, A.; Sarkar, C.; Kulshreshtha, R. Hypoxia-Inducible miR-196a modulates glioblastoma cell proliferation and migration through complex regulation of NRAS. Cell Oncol. 2021, 44, 433–451. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, R.H.; Pasha, H.F.; Gad, D.M.; Toam, M.M. miR-146a and miR-196a-2 genes polymorphisms and its circulating levels in lung cancer patients. J. Biochem. 2019, 166, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.H.; Lu, K.H.; Wang, K.M.; Sun, M.; Zhang, E.B.; Yang, J.S.; Yin, D.D.; Liu, Z.L.; Zhou, J.; Liu, Z.J.; et al. MicroRNA-196a promotes non-small cell lung cancer cell proliferation and invasion through targeting HOXA5. BMC Cancer 2012, 12, 348. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.F.; Chang, Y.W.; Kuo, K.T.; Shen, Y.S.; Liu, C.Y.; Yu, Y.H.; Cheng, C.C.; Lee, K.Y.; Chen, F.C.; Hsu, M.K.; et al. NF-kappaB-driven suppression of FOXO3a contributes to EGFR mutation-independent gefitinib resistance. Proc. Natl. Acad. Sci. USA 2016, 113, E2526–E2535. [Google Scholar] [CrossRef] [PubMed]
- Busser, B.; Sancey, L.; Josserand, V.; Niang, C.; Favrot, M.C.; Coll, J.L.; Hurbin, A. Amphiregulin promotes BAX inhibition and resistance to gefitinib in non-small-cell lung cancers. Mol. Ther. 2010, 18, 528–535. [Google Scholar] [CrossRef]
- Xiao, Y.; Yin, C.; Wang, Y.; Lv, H.; Wang, W.; Huang, Y.; Perez-Losada, J.; Snijders, A.M.; Mao, J.H.; Zhang, P. FBXW7 deletion contributes to lung tumor development and confers resistance to gefitinib therapy. Mol. Oncol. 2018, 12, 883–895. [Google Scholar] [CrossRef]
- Chen, R.; Qian, Z.; Xu, X.; Zhang, C.; Niu, Y.; Wang, Z.; Sun, J.; Zhang, X.; Yu, Y. Exosomes-Transmitted miR-7 reverses gefitinib resistance by targeting YAP in non-small-cell lung cancer. Pharmacol. Res. 2021, 165, 105442. [Google Scholar] [CrossRef]
- Chen, Z.; Chen, Q.; Cheng, Z.; Gu, J.; Feng, W.; Lei, T.; Huang, J.; Pu, J.; Chen, X.; Wang, Z. Long non-coding RNA CASC9 promotes gefitinib resistance in NSCLC by epigenetic repression of DUSP1. Cell Death Dis. 2020, 11, 858. [Google Scholar] [CrossRef]
- Jin, X.; Liu, X.; Zhang, Z.; Guan, Y. lncRNA CCAT1 Acts as a MicroRNA-218 sponge to increase gefitinib resistance in NSCLC by targeting HOXA1. Mol. Ther. Nucleic Acids 2020, 19, 1266–1275. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, L.; Zhang, X.; Xing, C.; Liu, R.; Zhang, F. MiR-196a promoted cell migration, invasion and the epithelial-mesenchymal transition by targeting HOXA5 in osteosarcoma. Cancer Biomark 2020, 29, 291–298. [Google Scholar] [CrossRef]
- Lee, S.; Hong, J.H.; Kim, J.S.; Yoon, J.S.; Chun, S.H.; Hong, S.A.; Kim, E.J.; Kang, K.; Lee Kang, J.; Ko, Y.H.; et al. Cancer-Associated fibroblasts activated by miR-196a promote the migration and invasion of lung cancer cells. Cancer Lett. 2021, 508, 92–103. [Google Scholar] [CrossRef]
- Jin, C.; Zhang, Y.; Li, J. Upregulation of MiR-196a promotes cell proliferation by downregulating p27(kip1) in laryngeal cancer. Biol. Res. 2016, 49, 40. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ma, Y.; Wang, B.; Guo, Y.; Zhang, Y.; Huang, S.; Bao, X.; Bai, M. Inhibition of miR-196a affects esophageal cancer cell growth in vitro. Biomed. Pharm. 2016, 84, 22–27. [Google Scholar] [CrossRef]
- Zhang, S.; Guo, J.; Liang, M.; Qi, J.; Wang, Z.; Jian, X.; Zhang, Z.; Sun, B.; Li, Z. miR-196a promotes proliferation and inhibits apoptosis of immature porcine sertoli cells. DNA Cell Biol. 2019, 38, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Samygina, V.R.; Ochoa-Lizarralde, B.; Popov, A.N.; Cabo-Bilbao, A.; Goni-de-Cerio, F.; Molotkovsky, J.G.; Patel, D.J.; Brown, R.E.; Malinina, L. Structural insights into lipid-dependent reversible dimerization of human GLTP. Acta Cryst. D Biol. Cryst. 2013, 69, 603–616. [Google Scholar] [CrossRef] [PubMed]
- Malinina, L.; Malakhova, M.L.; Kanack, A.T.; Lu, M.; Abagyan, R.; Brown, R.E.; Patel, D.J. The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure. PLoS Biol. 2006, 4, e362. [Google Scholar] [CrossRef]
- Mishra, S.K.; Stephenson, D.J.; Chalfant, C.E.; Brown, R.E. Upregulation of human glycolipid transfer protein (GLTP) induces necroptosis in colon carcinoma cells. Biochim. Biophys Acta Mol. Cell Biol. Lipids 2019, 1864, 158–167. [Google Scholar] [CrossRef]
- Mishra, S.K.; Gao, Y.G.; Zou, X.; Stephenson, D.J.; Malinina, L.; Hinchcliffe, E.H.; Chalfant, C.E.; Brown, R.E. Emerging roles for human glycolipid transfer protein superfamily members in the regulation of autophagy, inflammation, and cell death. Prog. Lipid Res. 2020, 78, 101031. [Google Scholar] [CrossRef]
- Rojo de la Vega, M.; Chapman, E.; Zhang, D.D. NRF2 and the hallmarks of cancer. Cancer Cell 2018, 34, 21–43. [Google Scholar] [CrossRef]
- Bauer, A.K.; Cho, H.Y.; Miller-Degraff, L.; Walker, C.; Helms, K.; Fostel, J.; Yamamoto, M.; Kleeberger, S.R. Targeted deletion of Nrf2 reduces urethane-induced lung tumor development in mice. PLoS ONE 2011, 6, e26590. [Google Scholar] [CrossRef]
- Long, M.; Tao, S.; Rojo de la Vega, M.; Jiang, T.; Wen, Q.; Park, S.L.; Zhang, D.D.; Wondrak, G.T. Nrf2-dependent suppression of azoxymethane/dextran sulfate sodium-induced colon carcinogenesis by the cinnamon-derived dietary factor cinnamaldehyde. Cancer Prev. Res. 2015, 8, 444–454. [Google Scholar] [CrossRef] [PubMed]
- DeNicola, G.M.; Karreth, F.A.; Humpton, T.J.; Gopinathan, A.; Wei, C.; Frese, K.; Mangal, D.; Yu, K.H.; Yeo, C.J.; Calhoun, E.S.; et al. Oncogene-Induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 2011, 475, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, X.; Long, M.; Huang, Y.; Zhang, L.; Zhang, R.; Zheng, Y.; Liao, X.; Wang, Y.; Liao, Q.; et al. NRF2 activation by antioxidant antidiabetic agents accelerates tumor metastasis. Sci. Transl. Med. 2016, 8, 334ra351. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, B.-J.; Li, F.-F.; Xie, Y.-X.; Fan, C.-Y.; Liu, W.-J.; Qiu, J.-G.; Jiang, B.-H. miR-196a Upregulation Contributes to Gefitinib Resistance through Inhibiting GLTP Expression. Int. J. Mol. Sci. 2022, 23, 1785. https://doi.org/10.3390/ijms23031785
Liu B-J, Li F-F, Xie Y-X, Fan C-Y, Liu W-J, Qiu J-G, Jiang B-H. miR-196a Upregulation Contributes to Gefitinib Resistance through Inhibiting GLTP Expression. International Journal of Molecular Sciences. 2022; 23(3):1785. https://doi.org/10.3390/ijms23031785
Chicago/Turabian StyleLiu, Bing-Jie, Fang-Fang Li, Yun-Xia Xie, Chong-Yuan Fan, Wen-Jing Liu, Jian-Ge Qiu, and Bing-Hua Jiang. 2022. "miR-196a Upregulation Contributes to Gefitinib Resistance through Inhibiting GLTP Expression" International Journal of Molecular Sciences 23, no. 3: 1785. https://doi.org/10.3390/ijms23031785
APA StyleLiu, B.-J., Li, F.-F., Xie, Y.-X., Fan, C.-Y., Liu, W.-J., Qiu, J.-G., & Jiang, B.-H. (2022). miR-196a Upregulation Contributes to Gefitinib Resistance through Inhibiting GLTP Expression. International Journal of Molecular Sciences, 23(3), 1785. https://doi.org/10.3390/ijms23031785






