Characterization of Five Meloidogyne incognita Effectors Associated with PsoRPM3
Abstract
:1. Introduction
2. Results
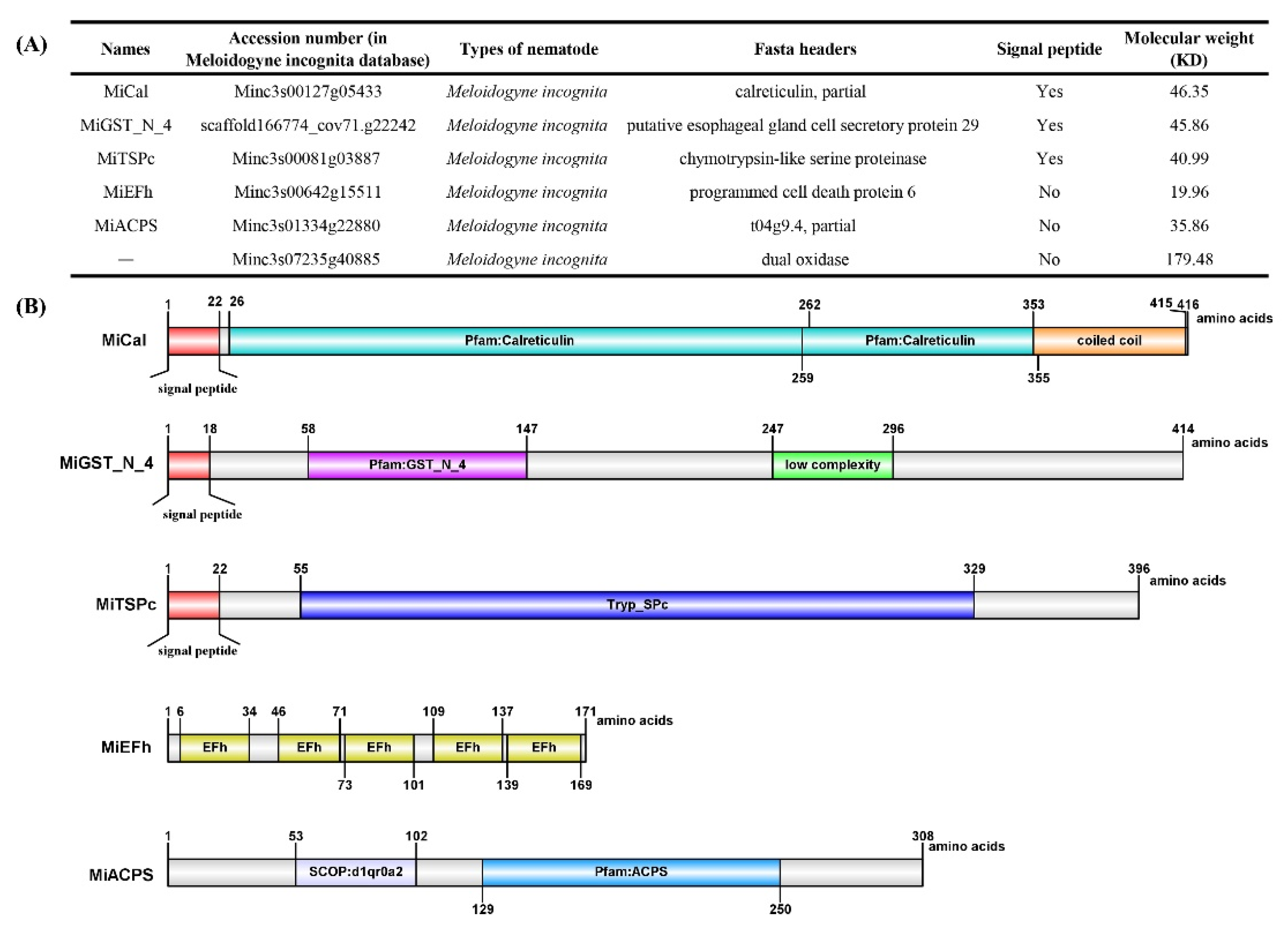
2.1. Five Candidate Effectors: Gene Amplification and Sequence Analysis
2.2. In Situ Hybridization in Pre-Parasitic Second-Stage Juveniles (Pre-J2) and Localization Analysis of Effectors
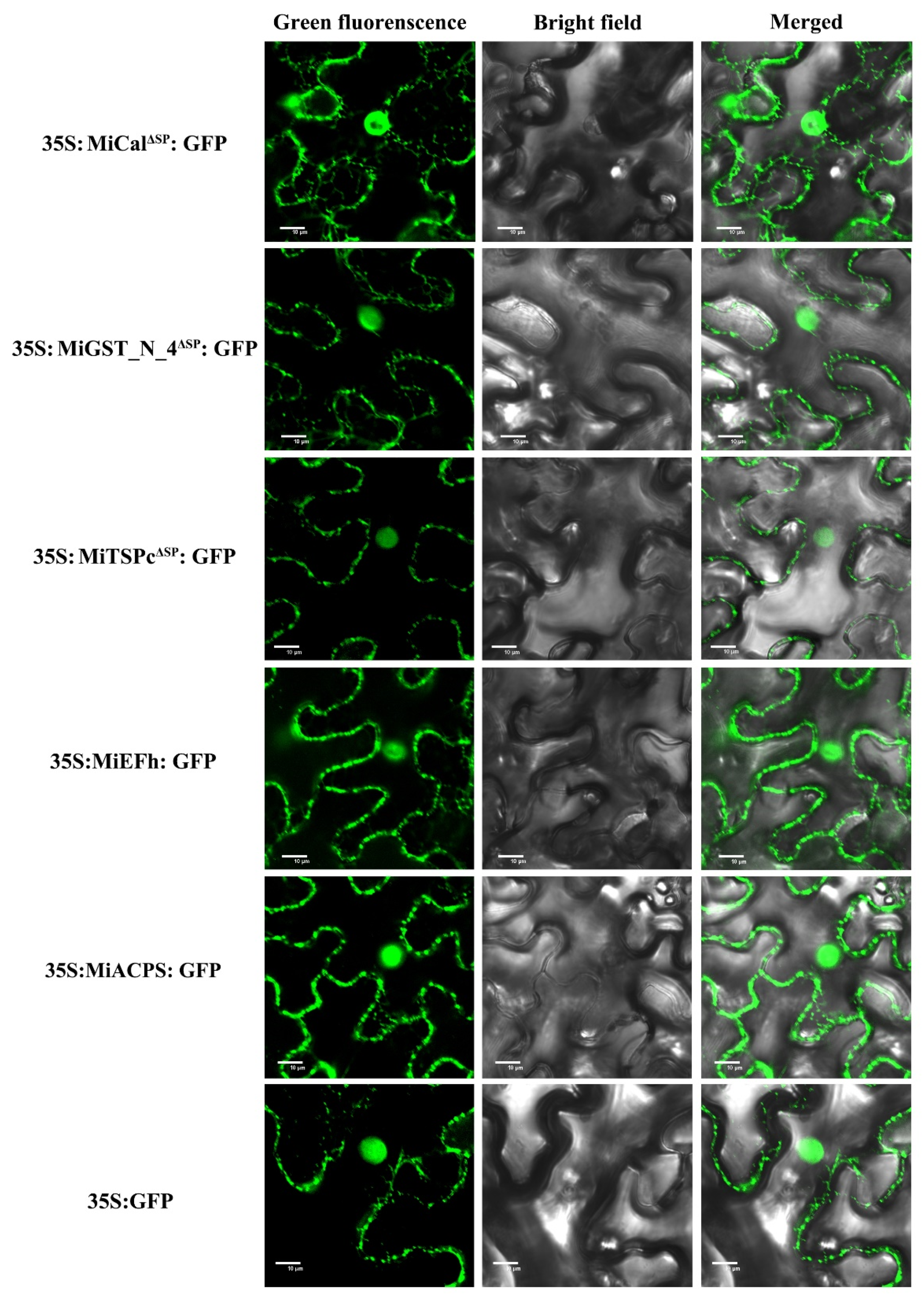
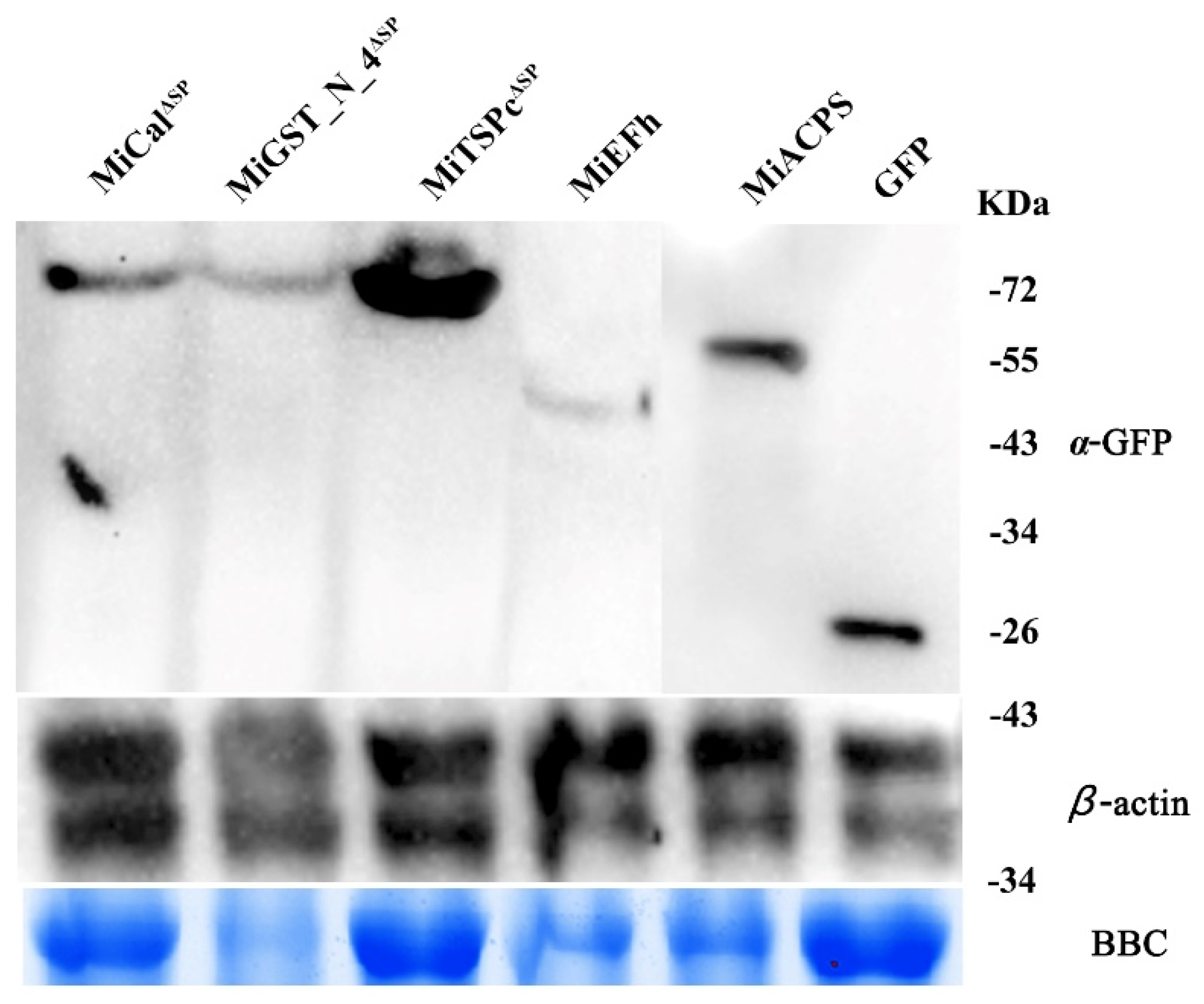
2.3. All Effectors Localize in the Nucleus and Cytoplasm when Expressed in Nicotiana benthamiana Cells
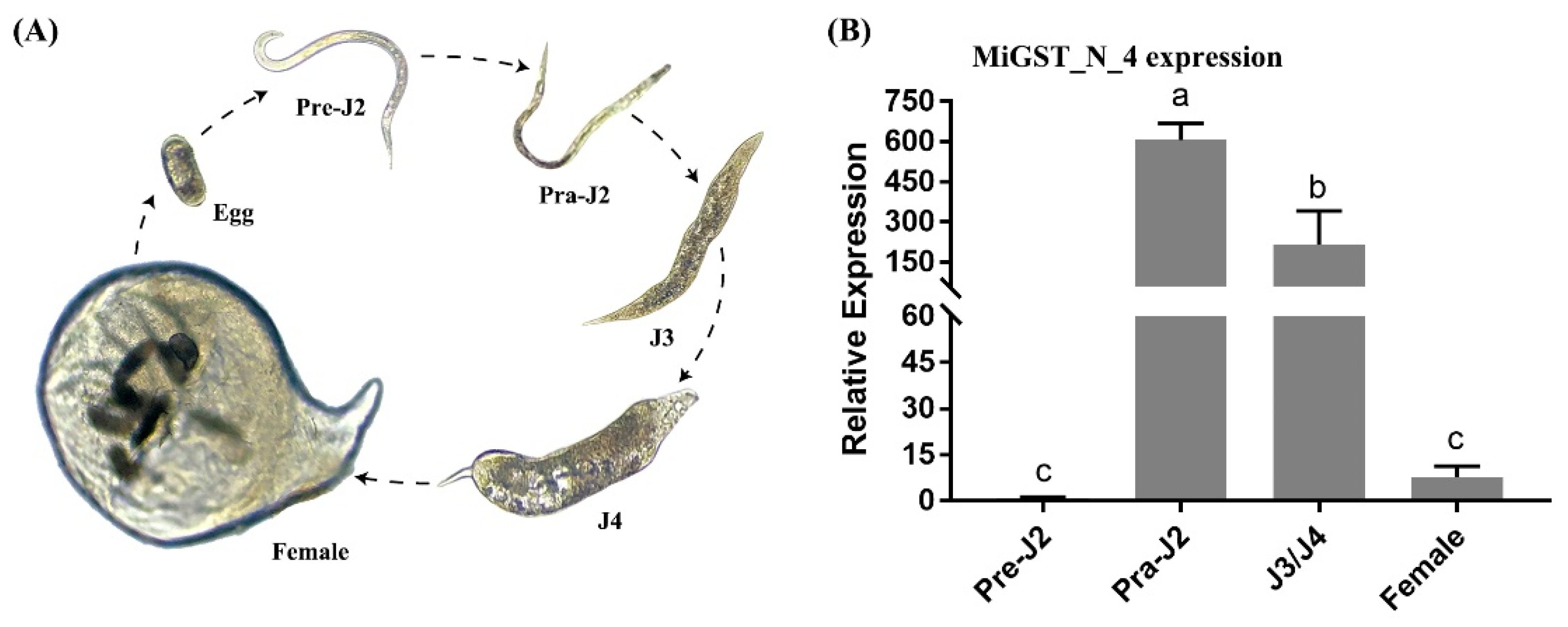
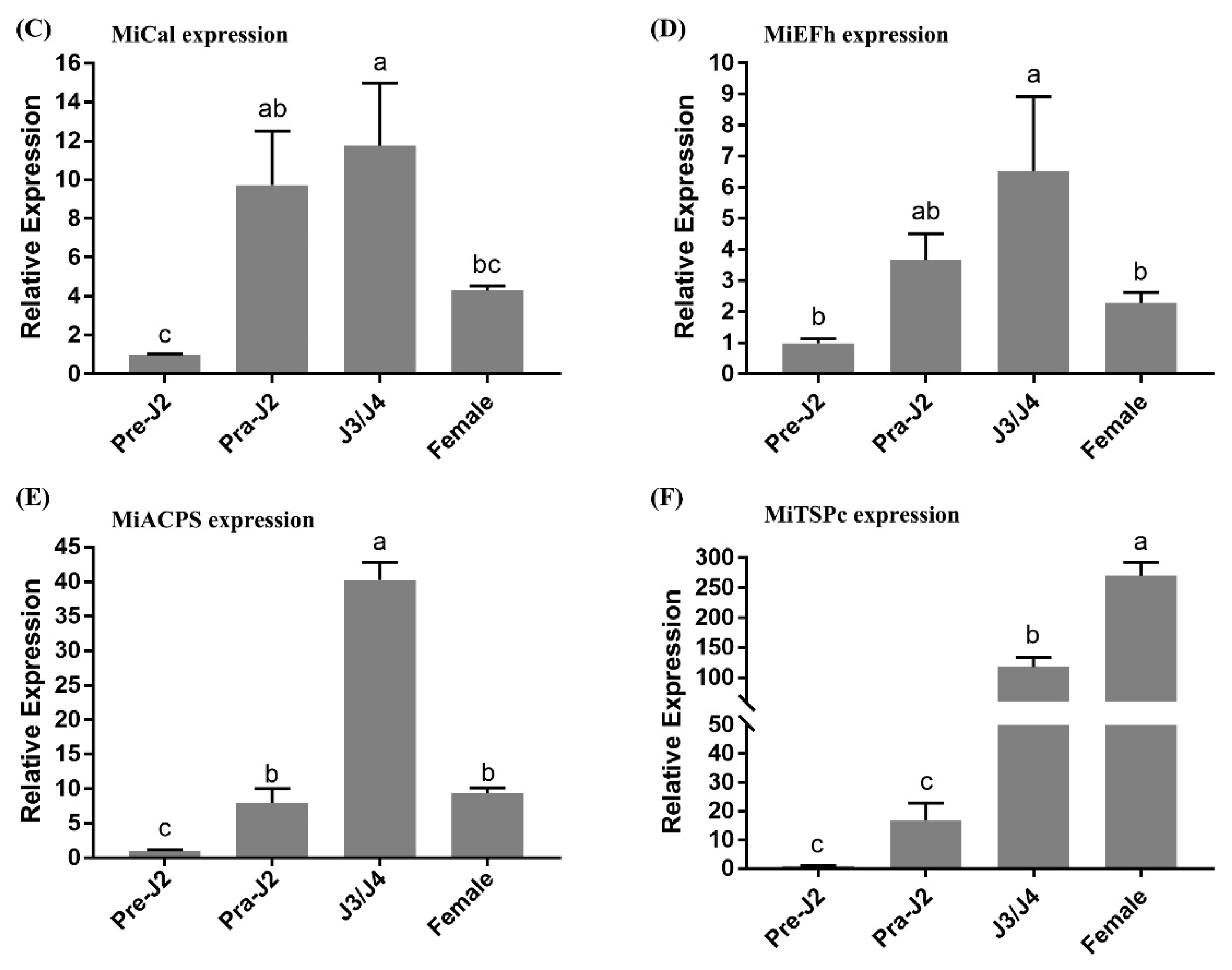
2.4. Expression Pattern Analysis of Five Effectors in M. incognita
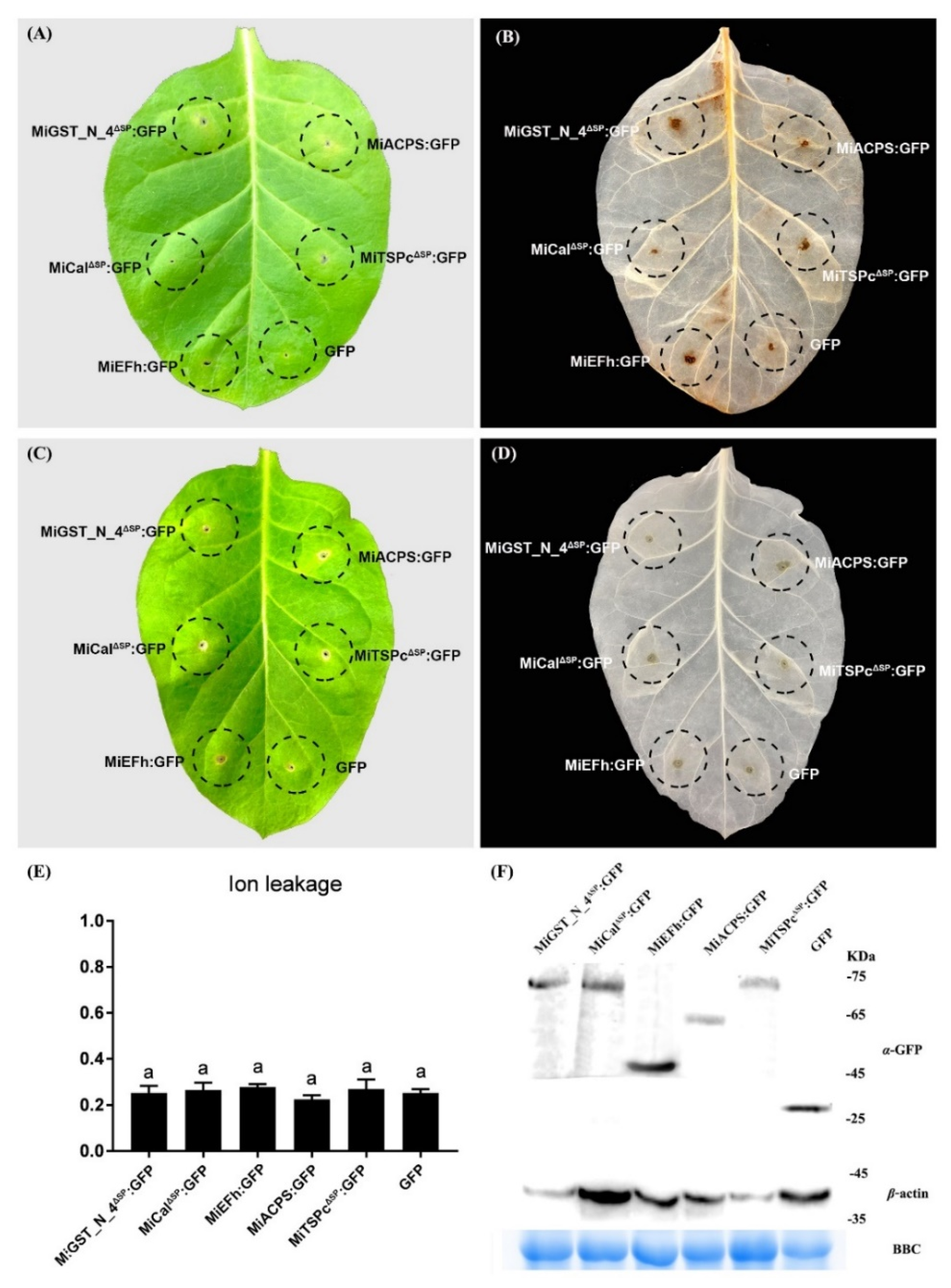
2.5. Effectors Can’t Induce Defense-Related Cell Death in the RKN-Susceptible Tobacco Variety W38
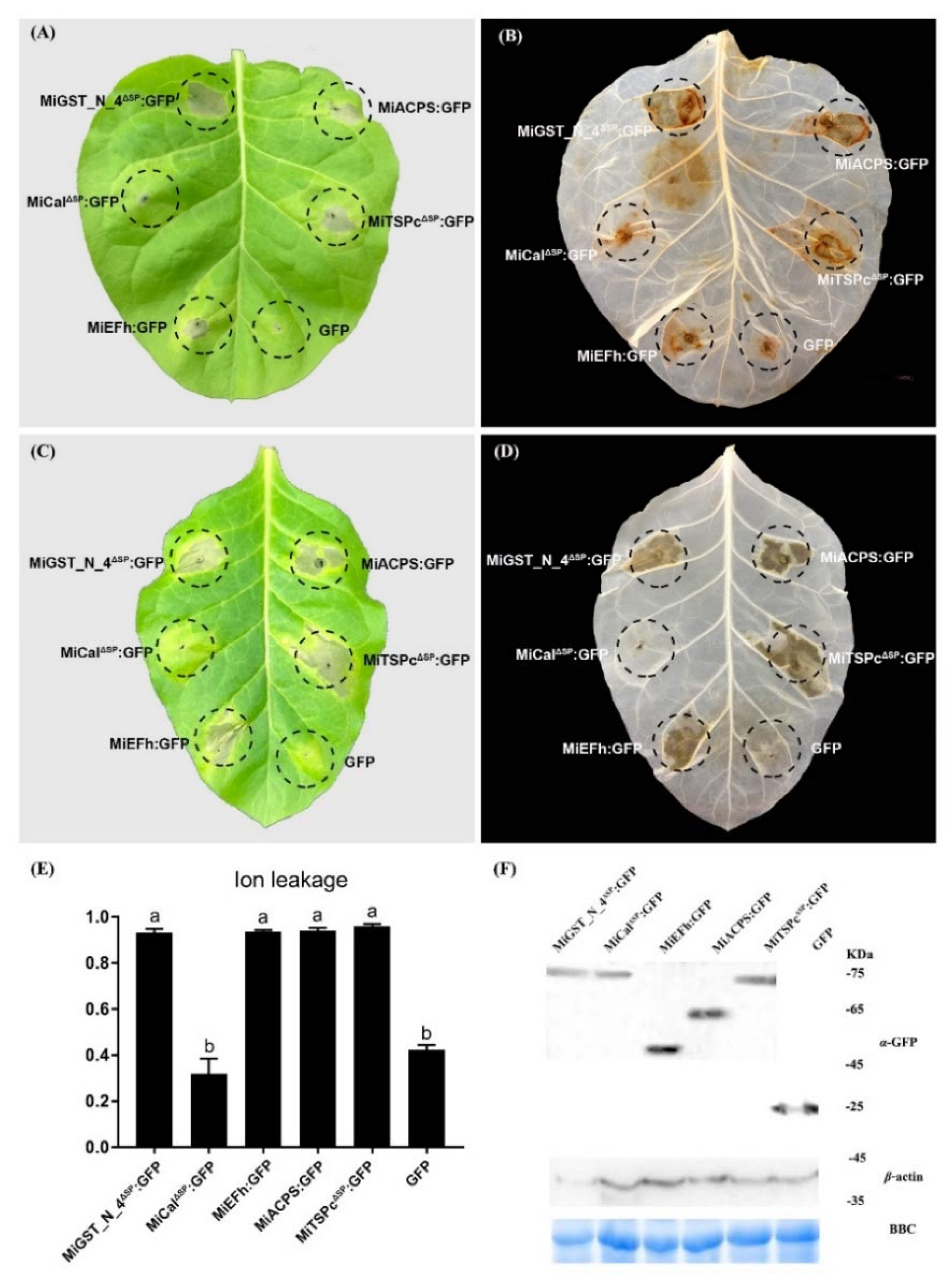
2.6. Effectors Trigger Defense-Related Cell Death in the RKN-Resistance Tobacco Variety K326
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Nematodes
4.2. PsoRPM3 Protein Immunoprecipitation (IP) Experiments and LC-MS Analysis
4.3. Gene Amplification and Sequence Analysis
4.4. In Situ Hybridization of Effectors in Pre-J2
4.5. Developmental Expression Analysis
4.6. Subcellular Localization
4.7. Effector Virulence Testing
4.8. H2O2 Detection
4.9. Electrolyte Leakage Detection
4.10. Protein Extraction and Western Blot Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Nicol, J.M.; Turner, S.J.; Coyne, D.L.; Nijs, L.D.; Hockland, S.; Maafi, Z.T. Current Nematode Threats to World Agriculture. In Genomics and Molecular Genetics of Plant-Nematode Interactions; Jones, J., Gheysen, G., Fenoll, C., Eds.; Springer: Dordrecht, The Netherlands, 2011; pp. 21–43. [Google Scholar]
- Moens, M.; Perry, R.N.; Starr, J.L. Meloidogyne species—A diverse group of novel and important plant parasites. In Root-Knot Nematodes; CABI: Wallingford, UK, 2009; pp. 1–17. [Google Scholar]
- Jones, J.T.; Haegeman, A.; Danchin, E.G.J.; Gaur, H.S.; Helder, J.; Jones, M.G.K.; Kikuchi, T.; Manzanilla-López, R.; Palomares-Rius, J.E.; Wesemael, W.M.L.; et al. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 2013, 14, 946–961. [Google Scholar] [CrossRef] [PubMed]
- Forghani, F.; Hajihassani, A. Recent Advances in the Development of Environmentally Benign Treatments to Control Root-Knot Nematodes. Front. Plant Sci. 2020, 11, 1125. [Google Scholar] [CrossRef] [PubMed]
- Khan, M. Nematode infestation, a potential threat to Indian forests. Indian Phytopathol. 2020, 73, 397–414. [Google Scholar] [CrossRef]
- Abad, P.; Favery, B.; Rosso, M.-N.; Castagnone-Sereno, P. Root-knot nematode parasitism and host response: Molecular basis of a sophisticated interaction. Mol. Plant Pathol. 2003, 4, 217–224. [Google Scholar] [CrossRef]
- Haegeman, A.; Jones, J.T.; Danchin, E.G.J. Horizontal Gene Transfer in Nematodes: A Catalyst for Plant Parasitism? Mol. Plant-Microbe Interact. 2011, 24, 879–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Qiao, F.; Ma, K.; Wang, J.; Hu, J. Roles of CCS52A gene and endoreduplication during the formation of giant cells in Xinjiang wild myrobalan. J. China Agric. Univ. 2018, 23, 43–52. [Google Scholar]
- Chen, X.; Xiao, K.; Zhu, X.; Chen, W.; Yang, Y.; Hu, J. Development of Giant Cells and Roles of CCS52B Gene Work in Meloidogyne incognita Resistant Prunus sogdiana. Acta Hortic. Sin. 2015, 42, 843–852. [Google Scholar]
- Jones, J.T.; Kumar, A.; Pylypenko, L.A.; Thirugnanasambandam, A.; Castelli, L.; Chapman, S.; Cock, P.J.A.; Grenier, E.; Lilley, C.J.; Phillips, M.S.; et al. Identification and functional characterization of effectors in expressed sequence tags from various life cycle stages of the potato cyst nematode Globodera pallida. Mol. Plant Pathol. 2009, 10, 815–828. [Google Scholar] [CrossRef]
- Bellafiore, S.; Shen, Z.; Rosso, M.-N.; Abad, P.; Shih, P.; Briggs, S. Direct Identification of the Meloidogyne incognita Secretome Reveals Proteins with Host Cell Reprogramming Potential. PLoS Pathog. 2008, 4, e1000192. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Koutsovoulos, G.; Kaur, G.; Blaxter, M. Toward 959 nematode genomes. Worm 2012, 1, 42–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petitot, A.-S.; Alexis, D.; Mawusse, A.; Corinne, D.; Julie, G.; Morgane, A.; Fernandez, D. Dual RNA-seq reveals Meloidogyne graminicola transcriptome and candidate effectors during the interaction with rice plants. Mol. Plant Pathol. 2015, 17, 860–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gahoi, S.; Gautam, B. Genome-wide analysis of Excretory/Secretory proteins in root-knot nematode, Meloidogyne incognita provides potential targets for parasite control. Comput. Biol. Chem. 2017, 67, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Kumara Tn, S.; Papolu, P.; Dutta, T.; Kamaraju, D.; Chaudhary, S.; Rao, U. RNAi-induced silencing of an effector confers transcriptional oscillation in another group of effectors in the root-knot nematode, Meloidogyne incognita. Nematology 2016, 18, 857–870. [Google Scholar]
- Siddique, S.; Grundler, F.M.W. Parasitic nematodes manipulate plant development to establish feeding sites. Curr. Opin. Microbiol. 2018, 46, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Kyndt, T.; Vieira, P.; Gheysen, G.; de Almeida-Engler, J. Nematode feeding sites: Unique organs in plant roots. Planta 2013, 238, 807–818. [Google Scholar] [CrossRef]
- Gillet, F.-X.; Bournaud, C.; Antonino, J.; Grossi-de-Sá, M. Plant-parasitic nematodes: Towards understanding molecular players in stress responses. Ann. Bot. 2017, 119, 775–789. [Google Scholar] [CrossRef] [Green Version]
- Castagnone-Sereno, P.; Deleury, E.; Danchin, E.G.J.; Perfus-Barbeoch, L.; Abad, P. Data-mining of the Meloidogyne incognita degradome and comparative analysis of proteases in nematodes. Genomics 2011, 97, 29–36. [Google Scholar] [CrossRef] [Green Version]
- Favery, B.; Quentin, M.; Jaubert-Possamai, S.; Abad, P. Gall-forming root-knot nematodes hijack key plant cellular functions to induce multinucleate and hypertrophied feeding cells. J. Insect Physiol. 2015, 84, 60–69. [Google Scholar] [CrossRef]
- Ali, M.A.; Azeem, F.; Li, H.; Bohlmann, H. Smart Parasitic Nematodes Use Multifaceted Strategies to Parasitize Plants. Front. Plant Sci. 2017, 8, 1699. [Google Scholar] [CrossRef] [Green Version]
- Eves-van den Akker, S.; Laetsch, D.R.; Thorpe, P.; Lilley, C.J.; Danchin, E.G.J.; Da Rocha, M.; Rancurel, C.; Holroyd, N.E.; Cotton, J.A.; Szitenberg, A.; et al. The genome of the yellow potato cyst nematode, Globodera rostochiensis, reveals insights into the basis of parasitism and virulence. Genome Biol. 2016, 17, 124. [Google Scholar] [CrossRef] [Green Version]
- Mejias, J.; Truong, N.M.; Abad, P.; Favery, B.; Quentin, M. Plant Proteins and Processes Targeted by Parasitic Nematode Effectors. Front. Plant Sci. 2019, 10, 970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Rocha, M.; Bournaud, C.; Dazenière, J.; Thorpe, P.; Bailly-Bechet, M.; Pellegrin, C.; Péré, A.; Grynberg, P.; Perfus-Barbeoch, L.; Eves-van den Akker, S.; et al. Genome Expression Dynamics Reveal the Parasitism Regulatory Landscape of the Root-Knot Nematode Meloidogyne incognita and a Promoter Motif Associated with Effector Genes. Genes 2021, 12, 771. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Mao, Z.; Zhang, X.; Zhang, X.; Wang, Y.; Ling, J.; Lin, R.; Li, D.; Kang, X.; Sun, W.; et al. A Meloidogyne incognita effector MiISE5 suppresses programmed cell death to promote parasitism in host plant. Sci. Rep. 2018, 8, 7256. [Google Scholar] [CrossRef] [Green Version]
- Xie, J.; Li, S.; Mo, C.; Wang, G.; Xiao, X.; Xiao, Y. A Novel Meloidogyne incognita Effector Misp12 Suppresses Plant Defense Response at Latter Stages of Nematode Parasitism. Front. Plant Sci. 2016, 7, 964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, J.; Liu, P.; Liu, Q.; Chen, C.; Guo, Q.; Yin, J.; Yang, G.; Jian, H. Msp40 effector of root-knot nematode manipulates plant immunity to facilitate parasitism. Sci. Rep. 2016, 6, 19443. [Google Scholar] [CrossRef] [Green Version]
- Dubreuil, G.; Magliano, M.; Deleury, E.; Abad, P.; Rosso, M.N. Transcriptome analysis of root-knot nematode functions induced in the early stages of parasitism. New Phytol. 2007, 176, 426–436. [Google Scholar] [CrossRef]
- Mejias, J.; Bazin, J.; Truong, N.M.; Chen, Y.; Marteu, N.; Bouteiller, N.; Sawa, S.; Crespi, M.; Vaucheret, H.; Abad, P.; et al. Root-knot nematode effector MiEFF18 interacts with the plant core spliceosomal protein SmD1 required for giant cell formation. New Phytol. 2020, 229, 3408–3423. [Google Scholar] [CrossRef]
- Truong, N.M.; Chen, Y.; Mejias, J.; Soulé, S.; Mulet, K.; Jaouannet, M.; Jaubert-Possamai, S.; Sawa, S.; Abad, P.; Favery, B.; et al. The Meloidogyne incognita Nuclear Effector MiEFF1 Interacts With Arabidopsis Cytosolic Glyceraldehyde-3-Phosphate Dehydrogenases to Promote Parasitism. Front. Plant Sci. 2021, 12, 633. [Google Scholar] [CrossRef]
- Zhao, J.; Li, L.; Liu, Q.; Liu, P.; Li, S.; Yang, D.; Chen, Y.; Pagnotta, S.; Favery, B.; Abad, P.; et al. A MIF-like effector suppresses plant immunity and facilitates nematode parasitism by interacting with plant annexins. J. Exp. Bot. 2019, 70, 5943–5958. [Google Scholar] [CrossRef]
- Shi, Q.; Mao, Z.; Zhang, X.; Ling, J.; Lin, R.; Zhang, X.; Liu, R.; Wang, Y.; Yang, Y.; Cheng, X.; et al. The Novel Secreted Meloidogyne incognita Effector MiISE6 Targets the Host Nucleus and Facilitates Parasitism in Arabidopsis. Front. Plant Sci. 2018, 9, 252. [Google Scholar] [CrossRef] [Green Version]
- Godinho Mendes, R.A.; Basso, M.F.; Paes de Melo, B.; Ribeiro, T.P.; Lima, R.N.; Fernandes de Araújo, J.; Grossi-de-Sa, M.; da Silva Mattos, V.; Togawa, R.C.; Saliba Albuquerque, É.V.; et al. The Mi-EFF1/Minc17998 effector interacts with the soybean GmHub6 protein to promote host plant parasitism by Meloidogyne incognita. Physiol. Mol. Plant Pathol. 2021, 114, 101630. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomma, B.; Nürnberger, T.; Joosten, M.H.A.J. Of PAMPs and effectors: The blurred PTI-ETI dichotomy. Plant Cell 2010, 23, 4–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castel, B.; Ngou, P.M.; Cevik, V.; Redkar, A.; Kim, D.S.; Yang, Y.; Ding, P.; Jones, J.D.G. Diverse NLR immune receptors activate defence via the RPW8-NLR NRG1. New Phytol. 2019, 222, 966–980. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Dangol, S.; Chen, Y.; Choi, J.; Cho, Y.-S.; Lee, J.-E.; Choi, M.-O.; Jwa, N.-S. Magnaporthe oryzae Effector AVR-Pii Helps to Establish Compatibility by Inhibition of the Rice NADP-Malic Enzyme Resulting in Disruption of Oxidative Burst and Host Innate Immunity. Mol. Cells 2016, 39, 426. [Google Scholar] [PubMed] [Green Version]
- Qi, T.; Guo, J.; Liu, P.; He, F.; Wan, C.; Islam, M.A.; Tyler, B.; Kang, Z.; Guo, J. Stripe Rust Effector PstGSRE1 Disrupts Nuclear Localization of ROS-Promoting Transcription Factor TaLOL2 to Defeat ROS-Induced Defense in Wheat. Mol. Plant 2019, 12, 1624–1638. [Google Scholar] [CrossRef]
- Liu, Y.; He, C. Regulation of plant reactive oxygen species (ROS) in stress responses: Learning from AtRBOHD. Plant Cell Rep. 2016, 35, 995–1007. [Google Scholar] [CrossRef]
- Torres, M.A. ROS in biotic interactions. Physiol. Plant. 2009, 138, 414–429. [Google Scholar] [CrossRef]
- Li, Q.; Ai, G.; Shen, D.; Zou, F.; Wang, J.; Bai, T.; Chen, Y.; Li, S.; Zhang, M.; Jing, M.; et al. A Phytophthora capsici Effector Targets ACD11 Binding Partners that Regulate ROS-Mediated Defense Response in Arabidopsis. Mol. Plant 2019, 12, 565–581. [Google Scholar] [CrossRef] [Green Version]
- Jwa, N.-S.; Hwang, B.K. Convergent Evolution of Pathogen Effectors toward Reactive Oxygen Species Signaling Networks in Plants. Front. Plant Sci. 2017, 8, 1687. [Google Scholar] [CrossRef] [Green Version]
- Ali, M.; Cheng, Z.; Ahmad, H.; Hayat, S. Reactive oxygen species (ROS) as defenses against a broad range of plant fungal infections and case study on ROS employed by crops against Verticillium dahliae wilts. J. Plant Interact. 2018, 13, 353–363. [Google Scholar] [CrossRef] [Green Version]
- Rice, S.L.; Leadbeater, B.S.C.; Stone, A.R. Changes in cell structure in roots of resistant potatoes parasitized by potato cyst-nematodes. I. Potatoes with resistance gene H1 derived from Solanum tuberosum ssp. andigenea. Physiol. Plant Pathol. 1985, 27, 219–234. [Google Scholar] [CrossRef]
- Cai, D.; Kleine, M.; Kifle, S.; Harloff, H.-J.; Sandal Niels, N.; Marcker Kjeld, A.; Klein-Lankhorst René, M.; Salentijn Elma, M.J.; Lange, W.; Stiekema Willem, J.; et al. Positional Cloning of a Gene for Nematode Resistance in Sugar Beet. Science 1997, 275, 832–834. [Google Scholar] [CrossRef] [PubMed]
- De Ilarduya, O.M.; Moore, A.E.; Kaloshian, I. The tomato Rme1 locus is required for Mi-1-mediated resistance to root-knot nematodes and the potato aphid. Plant J. 2001, 27, 417–425. [Google Scholar] [CrossRef] [Green Version]
- Claverie, M.; Dirlewanger, E.; Bosselut, N.; Ghelder, C.; Voisin, R.; Kleinhentz, M.; Lafargue, B.; Abad, P.; Rosso, M.-N.; Chalhoub, B.; et al. The Ma Gene for Complete-Spectrum Resistance to Meloidogyne Species in Prunus Is a TNL with a Huge Repeated C-Terminal Post-LRR Region. Plant Physiol. 2011, 156, 779–792. [Google Scholar] [CrossRef] [Green Version]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.-M.; He, S.Y.; Xin, X.-F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef]
- Xiao, K.; Zhu, H.; Zhu, X.; Liu, Z.; Wang, Y.; Pu, W.; Guan, P.; Hu, J. Overexpression of PsoRPM3, an NBS-LRR gene isolated from myrobalan plum, confers resistance to Meloidogyne incognita in tobacco. Plant Mol. Biol. 2021, 107, 129–146. [Google Scholar] [CrossRef]
- Mazo Molina, C.; Mainiero, S.; Haefner, B.; Bednarek, R.; Zhang, J.; Feder, A.; Shi, K.; Strickler, S.; Martin, G. Ptr1 evolved convergently with RPS2 and Mr5 to mediate recognition of AvrRpt2 in diverse solanaceous species. Plant J. 2020, 103, 1433–1445. [Google Scholar] [CrossRef]
- Martin, R.; Qi, T.; Zhang, H.; Liu, F.; King, M.; Toth, C.; Nogales, E.; Staskawicz, B. Structure of the activated Roq1 resistosome directly recognizing the pathogen effector XopQ. Science 2020, 370, eabd9993. [Google Scholar] [CrossRef]
- Toruño, T.Y.; Stergiopoulos, I.; Coaker, G. Plant-Pathogen Effectors: Cellular Probes Interfering with Plant Defenses in Spatial and Temporal Manners. Annu. Rev. Phytopathol. 2016, 54, 419–441. [Google Scholar] [CrossRef] [Green Version]
- Fuller, V.L.; Lilley, C.J.; Urwin, P.E. Nematode resistance. New Phytol. 2008, 180, 27–44. [Google Scholar] [CrossRef] [PubMed]
- Mitchum, M.G.; Hussey, R.S.; Baum, T.J.; Wang, X.; Elling, A.A.; Wubben, M.; Davis, E.L. Nematode effector proteins: An emerging paradigm of parasitism. New Phytol. 2013, 199, 879–894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vieira, P.; Gleason, C. Plant-parasitic nematode effectors—Insights into their diversity and new tools for their identification. Curr. Opin. Plant Biol. 2019, 50, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Grynberg, P.; Togawa, R.; Freitas, L.; Antonino, J.; Rancurel, C.; Costa, M.; Grossi-de-Sá, M.; Miller, R.; Brasileiro, A.; Guimarães, P.; et al. Comparative Genomics Reveals Novel Target Genes towards Specific Control of Plant-Parasitic Nematodes. Genes 2020, 11, 1347. [Google Scholar] [CrossRef]
- Abad, P.; Gouzy, J.; Aury, J.-M.; Castagnone-Sereno, P.; Danchin, E.G.J.; Deleury, E.; Perfus-Barbeoch, L.; Anthouard, V.; Artiguenave, F.; Blok, V.C.; et al. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 2008, 26, 909–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baum, T.; Hussey, R.; Davis, E. Root-Knot and Cyst Nematode Parasitism Genes: The Molecular Basis of Plant Parasitism. Genet. Eng. 2007, 28, 17–43. [Google Scholar]
- Davis, E.L.; Hussey, R.S.; Baum, T.J.; Bakker, J.; Schots, A.; Rosso, M.-N.; Abad, P. Nematode Parasitism Genes. Annu. Rev. Phytopathol. 2000, 38, 365–396. [Google Scholar] [CrossRef]
- Davis, E.; Hussey, R.; Mitchum, M.; Baum, T. Parasitism proteins in nematode-plant interactions. Curr. Opin. Plant Biol. 2008, 11, 360–366. [Google Scholar] [CrossRef]
- Vieira, P.; Danchin, E.; Neveu, C.; Crozat, C.; Jaubert-Possamai, S.; Hussey, R.; Engler, G.; Abad, P.; Engler, J.; Castagnone-Sereno, P.; et al. The plant apoplasm is an important recipient compartment for nematode secreted proteins. J. Exp. Bot. 2011, 62, 1241–1253. [Google Scholar] [CrossRef] [Green Version]
- Rehman, S.; Gupta, V.K.; Goyal, A.K. Identification and functional analysis of secreted effectors from phytoparasitic nematodes. BMC Microbiol. 2016, 16, 48. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Mao, Z.; Sun, Q.; Liu, Q.; Jian, H.; Xie, B. MiMIF-2 Effector of Meloidogyne incognita Exhibited Enzyme Activities and Potential Roles in Plant Salicylic Acid Synthesis. Int. J. Mol. Sci. 2020, 21, 3507. [Google Scholar] [CrossRef]
- Jaouannet, M.; Magliano, M.; Arguel, M.-J.; Gourgues, M.; Evangelisti, E.; Abad, P.; Rosso, M.-N. The Root-Knot Nematode Calreticulin Mi-CRT Is a Key Effector in Plant Defense Suppression. Mol. Plant-Microbe Interact. 2012, 26, 97–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deslandes, L.; Rivas, S. The plant cell nucleus: A true arena for the fight between plants and pathogens. Plant Signal. Behav. 2011, 6, 42–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motion, G.; Amaro, T.; Kulagina, N.; Huitema, E. Nuclear processes associated with plant immunity and pathogen susceptibility. Brief. Funct. Genom. 2015, 14, 243–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Davies, L.J.; Elling, A.A. A Meloidogyne incognita effector is imported into the nucleus and exhibits transcriptional activation activity in planta. Mol. Plant Pathol. 2015, 16, 48–60. [Google Scholar] [CrossRef]
- Huang, G.; Dong, R.; Allen, R.; Davis, E.L.; Baum, T.J.; Hussey, R.S. A Root-Knot Nematode Secretory Peptide Functions as a Ligand for a Plant Transcription Factor. Mol. Plant-Microbe Interact. 2006, 19, 463–470. [Google Scholar] [CrossRef] [Green Version]
- Adamo, N.; Johnson, C.S.; Reed, T.D.; Eisenback, J.D. Reproduction of Meloidogyne arenaria race 2 on Flue-cured tobacco possessing resistance genes Rk1 and/or Rk2. J. Nematol. 2021, 53, e2021-42. [Google Scholar] [CrossRef]
- Huang, G.; Dong, R.; Allen, R.E.X.; Davis, E.L.; Baum, T.J.; Hussey, R.S. Two chorismate mutase genes from the root-knot nematode Meloidogyne incognita. Mol. Plant Pathol. 2005, 6, 23–30. [Google Scholar] [CrossRef]
- Xiao, K. Functional Verification and Mechanism Analysis of PsoRPM3 from Prunus sogdiana in Reponse to Meloidogyne Incognita. Ph.D. Thesis, China Agricultural University, Beijing, China, 2020. [Google Scholar]
- Dareus, R.; Porto, A.C.M.; Bogale, M.; DiGennaro, P.; Chase, C.A.; Rios, E.F. Resistance to Meloidogyne enterolobii and Meloidogyne incognita in Cultivated and Wild Cowpea. HortScience 2021, 56, 460–468. [Google Scholar] [CrossRef]
- Jaouannet, M.; Nguyen, C.-N.; Quentin, M.; Jaubert-Possamai, S.; Rosso, M.-N.; Favery, B. In situ Hybridization (ISH) in Preparasitic and Parasitic Stages of the Plant-parasitic Nematode Meloidogyne spp. Bio-protocol 2018, 8, e2766. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chen, C.; Liu, S.; Liu, Q.; Niu, J.; Liu, P.; Zhao, J.; Jian, H. An ANNEXIN-Like Protein from the Cereal Cyst Nematode Heterodera avenae Suppresses Plant Defense. PLoS ONE 2015, 10, e0122256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, B.Q.; Jung, S. Modulation of chloroplast components and defense responses during programmed cell death in tobacco infected with Pseudomonas syringae. Biochem. Biophys. Res. Commun. 2020, 528, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Labudda, M.; Rozanska, E.; Prabucka, B.; Muszynska, E.; Marecka, D.; Kozak, M.; Dababat, A.A.; Sobczak, M. Activity profiling of barley vacuolar processing enzymes provides new insights into the plant and cyst nematode interaction. Mol. Plant Pathol. 2020, 21, 38–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.-F.; Balint-Kurti, P. Maize Homologs of CCoAOMT and HCT, Two Key Enzymes in Lignin Biosynthesis, Form Complexes with the NLR Rp1 Protein to Modulate the Defense Response. Plant Physiol. 2016, 171, 2166–2177. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pu, W.; Xiao, K.; Luo, S.; Zhu, H.; Yuan, Z.; Gao, C.; Hu, J. Characterization of Five Meloidogyne incognita Effectors Associated with PsoRPM3. Int. J. Mol. Sci. 2022, 23, 1498. https://doi.org/10.3390/ijms23031498
Pu W, Xiao K, Luo S, Zhu H, Yuan Z, Gao C, Hu J. Characterization of Five Meloidogyne incognita Effectors Associated with PsoRPM3. International Journal of Molecular Sciences. 2022; 23(3):1498. https://doi.org/10.3390/ijms23031498
Chicago/Turabian StylePu, Wenjiang, Kun Xiao, Sifang Luo, Haifeng Zhu, Zizhen Yuan, Chaoyuan Gao, and Jianfang Hu. 2022. "Characterization of Five Meloidogyne incognita Effectors Associated with PsoRPM3" International Journal of Molecular Sciences 23, no. 3: 1498. https://doi.org/10.3390/ijms23031498
APA StylePu, W., Xiao, K., Luo, S., Zhu, H., Yuan, Z., Gao, C., & Hu, J. (2022). Characterization of Five Meloidogyne incognita Effectors Associated with PsoRPM3. International Journal of Molecular Sciences, 23(3), 1498. https://doi.org/10.3390/ijms23031498





