Abstract
In recent years, small fishes such as zebrafish and medaka have been widely recognized as model animals. They have high homology in genetics and tissue structure with humans and unique features that mammalian model animals do not have, such as transparency of embryos and larvae, a small body size and ease of experiments, including genetic manipulation. Zebrafish and medaka have been used extensively in the field of neurology, especially to unveil the mechanisms of neurodegenerative diseases such as Parkinson’s and Alzheimer’s disease, and recently, these fishes have also been utilized to understand neurodevelopmental disorders such as autism spectrum disorder. The turquoise killifish has emerged as a new and unique model animal, especially for ageing research due to its unique life cycle, and this fish also seems to be useful for age-related neurological diseases. These small fishes are excellent animal models for the analysis of human neurological disorders and are expected to play increasing roles in this field. Here, we introduce various applications of these model fishes to improve our understanding of human neurological disorders.
1. Introduction
When we think of model animals used in medical research, the typical animal is a mouse. Throughout the history of science, many researchers have standardized and advanced experimental procedures using mice, including genetic techniques, biochemical analysis and behavioral analysis. There is no doubt that mice are at the forefront of model animals. In addition to mice, rats, flies and nematodes have a relatively long history as model animals. Among model animals, small fishes such as zebrafish (Danio rerio), medaka (Oryzias latipes) and turquoise killifish (Nothobranchius furzeri) are relative newcomers but are increasingly present (Table 1) [1,2,3,4,5,6,7].

Table 1.
Characteristics of zebrafish, medaka and turquoise killifish.
Because fish and mammals, such as humans, diverged so recently in the course of evolution, their anatomical and genetic homologies are remarkably well preserved. In the laboratory, small fishes are excellent model animals with many advantages that mammalian models do not have, such as simple maintenance from embryo to adult, excellent tissue visibility with transparent embryos and larvae, easy handling in laboratory experiments, including molecular biology, biochemistry, histology and so on. This review outlines the characteristics of these small fishes, their relevance to humans, the benefits of using small fishes as model animals for neurological disorders and how small fishes are currently being used in research for neurodegenerative diseases and neurodevelopmental disorders.
2. Central Nervous System in Small Fishes
Flies and nematodes, which are used as small model animals, are invertebrates and may be disadvantageous when considered as human disease models. On the other hand, small fishes are vertebrates such as mice and humans, and many of their organ structures are similar to those of humans. The basic structure and function of the central nervous system is conserved from small fishes to humans. In nematodes, for example, the ganglia of the head are sometimes referred to as the “brain”, but they have no direct phylogenetic relationship to the vertebrate brain. Small fishes, on the other hand, have what can accurately be called a brain. Although there are some differences between the brains of small fishes and humans, they are anatomically and functionally similar as a whole. This section describes the anatomical characteristics of the zebrafish brain. An excellent phylogenetic tree of the evolutionary connections among the model animals mentioned above is given in a previous report [6].
The structural and anatomical formation of the central nervous system in zebrafish begins early in development. After the formation of the neural tube at 17 hpf (hours post-fertilization), brain morphogenesis begins. The boundary between the midbrain and hindbrain begins to form, and regions such as the cerebellum and thalamus are developed [8]. Human dopaminergic neurons in the substantia nigra are located in the midbrain, but there are no dopaminergic clusters in the anatomically classified midbrain of zebrafish. There are several clusters of dopaminergic neurons in the neighboring diencephalon, and their formation has begun at 18 hpf [9]. Furthermore, it has been shown that some of the dopaminergic neurons in this diencephalon extend and project long axons to the striatum, and these neurons could be equivalent to human dopaminergic neurons in the substantia nigra [10]. Dopaminergic projections to the forebrain in zebrafish are thought to be homologous to the reward system in mammals [11]. Since the overall composition of dopaminergic neurons is conserved among teleosts, the distribution of dopaminergic neurons in turquoise killifish is similar to zebrafish and medaka. Please refer to the latest report on the analysis of catecholaminergic neurons in the central nervous system of turquoise killifish [12].
The cerebellum has climbing fibers and parallel fibers, and the cell groups present are similar to those of humans, including Purkinje neurons and granule cells [13,14]. Although zebrafish do not have structures corresponding to the deep cerebellar nuclei, cells called eurydendroid cells receive Purkinje cell projections and send efferent projections to various brain regions. Therefore, eurydendroid cells are thought to be functionally homologous to the deep cerebellar nuclei of mammals [15]. In humans, the cerebellum is divided into the vestibulocerebellum, spinocerebellum, and pontocerebellum in terms of phylogeny and functional localization, while the cerebellum of small fishes is mostly regarded as the vestibulocerebellum. We created a functional map of the zebrafish cerebellum and showed that the zebrafish cerebellum contains at least the vestibulocerebellum and spinocerebellum, the details of which can be found in a reference [16]. It is very interesting to determine if there are also cerebellum regions controlling higher brain functions within the telencephalon in small fishes.
Zebrafish also have a telencephalon that corresponds to the human cerebrum, with regions corresponding to the hippocampus and amygdala, which are involved in memory learning and emotional behavior, respectively [17]. In humans, during development, the ventral and dorsal sides of the neural tube form a median constriction, whereas in zebrafish, the dorsal side of the neural tube forms an outward folding [18]. However, unlike humans, zebrafish do not have cortical layer structures. It is important to note that this does not mean that the fish brain does not have structures and functions equivalent to the mammalian cerebral cortex.
The blood–brain barrier (BBB) is also present in zebrafish. Angiogenesis in the hindbrain begins at approximately 20 hpf, and pericytes and glia are found around the vessels by 60 hpf, but the BBB is incomplete until approximately 5–8 dpf (days post-fertilization), allowing various drugs to penetrate the central nervous system [19,20]. The drug can be administered orally or intracorporeally or it can be dissolved in the maintenance water of the larvae and infiltrated into the body tissue by water immersion. This feature is very useful for high-throughput screening using zebrafish larvae. For example, embryos or larvae are individually deposited in each well of 96- and 384-well plates, and various compounds can be dissolved in the water. Thereafter, each fish can be evaluated by gene expression patterns, developmental changes or behavioral analysis. This is less time-consuming and less expensive than the same screening procedure using mammalian models such as mice [21].
One of the major differences between zebrafish and humans is the ability to regenerate the central nervous system, including neurons. When zebrafish are artificially injured in the spinal cord, functional recovery and motor neuron repopulation are observed within 6 to 8 weeks [22,23]. In addition, tissue regeneration of the central nervous system has been observed after artificial damage, even in the telencephalon [24]. The increased regeneration ability of fish, even in the central nervous system, needs to be recognized when using fish as models of human neurological disorders.
Many lines of evidence have been shown using zebrafish. Because zebrafish, medaka and turquoise killifish are closely related teleosts, their major structures in the central nervous system are comparable. A review of these three model fishes including the description of their central nervous system is available [25].
3. Ease of Laboratory Management and Experimentation with Zebrafish, Medaka and Turquoise Killifish
3.1. Visibility and Light Transmission in Small Fishes
One of the important characteristics of small fishes is their high tissue transparency during embryogenesis and the larval stages. In zebrafish and medaka, the developmental process can be observed outside the body of the parent fish, and the embryos and larvae are transparent, making it easy to observe the developmental process and internal structure. In addition, several mutant lines of zebrafish and medaka are available that are more transparent than the wild type, allowing the internal structure of the fish to see, even in the adult stage [26]. This characteristic allows direct observation of the development and tissue and cell activity, which is compatible with live imaging, and allows us to capture developmental and structural changes in the nervous system in greater detail in vivo. When fluorescent proteins are expressed specifically on the cell of interest, it is possible to observe the development and morphological changes of the neurons of interest over time. Chemical or genetically encoded Ca sensors expressed in neurons allow us to observe the activity of neurons with high temporal and spatial resolution [27]. For example, we showed that cerebellar neural activity during behavior can be observed at the cellular level [16]. A very exciting study has also been reported that expresses Ca sensors in neurons of the whole brain and utilizes the sensor signals as in electroencephalography [28]. Using optogenetics techniques, it is also theoretically possible to activate or suppress the neuronal activity of target neurons at any site [16,29]. A unique method to analyze the pathogenesis of diseases by altering the subcellular localization of target proteins has also been reported [30]. In short, small fishes are unique vertebrates that provide, in a live state, macroscopic observation of the nervous system, microscopic observation of precise neural activity and adaptation of light transmission for optogenetics and other applications.
3.2. Ease of Gene Editing with Small Fishes
It is estimated that approximately 71% of human genes have at least one zebrafish gene orthologue [31]. In addition, approximately 84% of the genes known to be associated with human diseases have orthologues in the zebrafish genome [32]. Small fishes have the advantage of simple procedures for gene editing. Efficient genome engineering can be achieved by zinc-finger nucleases (ZFNs) or transcription activator-like effectors (TALENs) in various model organisms, including zebrafish and medaka [33,34,35,36], and the emergence of the CRISPR-Cas9 technique has made it much easier to knockout or edit specific genes in recent years [37,38,39]. In small fishes, oviposition and fertilization occur outside the parents’ body, and gene editing factors are introduced into the fertilized eggs by microinjection, which can then hatch in a petri dish [40]. For example, by injecting GFP mRNA into the embryo, it is possible to observe the results the day after the injection without waiting for days or months [41]. Improved and advanced techniques for gene editing have been reported thus far, and the highly mutagenic CRISPR-Cas9 method enables even injection of F0 embryos mimicking null mutants, making the induction of mutations quick and efficient [42].
Morpholino antisense oligonucleotide-based knockdown of target genes has often been conducted in small fishes, but there can be phenotypic discrepancies between morpholino and knockout phenotypes [43]. This may be due to the presence of morpholino-induced off-target effects or the presence of nonsense-mediated mRNA decay and associated genetic compensation in knockout individuals [44]. In the analysis of gene function in zebrafish, it is strongly recommended to generate mutants and analyze their phenotypes instead of using antisense morpholino oligonucleotides.
The CRISPR-Cas9 system also enables knock-in of the target gene or specific gene variants by means of homology-directed repair (HDR) or other mechanisms [45,46,47,48,49,50]. Stable introduction of exogenous DNA in the zebrafish genome was already achieved in 1988 [51]. Coinjection of the I-SceI meganuclease enzyme together with the transgenic vector carrying the restriction sites [52] or coinjection of tol2 or sleeping beauty transposase with the transgenic vector carrying the transgenic cassette flanked by cis-regulatory repeats [53,54] facilitates germline transmission of the transgene very effectively. These techniques of genome engineering are applicable to zebrafish, medaka and turquoise killifish, but we think it is difficult to conduct microinjection into the eggs of turquoise killifish. This is because of the relatively hard chorion of turquoise killifish compared to zebrafish and medaka, and please kindly see the excellent publications about genome engineering of turquoise killifish [55,56].
If the same thing were to be done with the mouse, gene editing of the embryo would be a complex operation that would require the collection of fertilized eggs from the mated female mouse body, microinjection of the genome-engineering solution into the eggs under clean operations and implantation of the injected eggs into the fallopian tubes of a pseudopregnant mouse, which would be technically much more difficult and expensive. Zebrafish can produce more than 100 eggs per oviposition, making it easy to pick up individuals with the desired gene editing. Zebrafish will mature into adults in 3 months, making it possible to breed the next generation. In the case of medaka and turquoise killifish, the number of eggs per oviposition seems smaller than that of zebrafish, but they lay eggs every day if the condition of the adult fish and aquarium is ideal. Medaka and turquoise killifish can show sexual maturation in approximately 2–3 months and 1 month, respectively. Compared to mice, the cost of subsequent management can also be greatly reduced.
As described above, zebrafish and medaka are the mainstream models for genetic manipulation, while the turquoise killifish provides an excellent model for the studies of aging and age-related disorders (Figure 1). The aging of turquoise killifish is described in the next chapter. When designing a genetic study using small fishes, it is necessary to be conscious of several genetic features. Zebrafish are diploid with 25 chromosomes (25 × 2n) and are close to humans, but their sex chromosomes have not been identified. The mechanism of sex determination is still not fully uncovered, but it has been found that zebrafish sex can be affected by environmental factors [57,58,59]. In contrast, medaka or turquoise killifish sex is determined by XX/XY sex chromosomes [60,61,62]. Vertebrates, including humans, experienced a whole-genome duplication in which the genome doubled twice in our ancestors approximately 500 million years ago. Furthermore, teleosts, including zebrafish, medaka and turquoise killifish, experienced another round of whole-genome duplication [63,64,65]. Therefore, in some genes, there is no 1:1 correspondence between humans and zebrafish, and there can be two orthologous genes. The fact that there are two orthologues means that even if one of the orthologues is knocked out, the other paralogue may be able to compensate for its function. It has also been reported that there are differences in the structure, function and expression patterns between the paralogues of these genes, which might suggest that each gene has its own significance [66]. In some cases, double knockout of those two orthologous genes might be necessary to establish a knockout line with the anticipated phenotypes seen in human diseases or disorders. It is also necessary to check whether the ortholog of the gene of interest exists or not (e.g., the ortholog of SNCA is present in medaka but not in zebrafish).
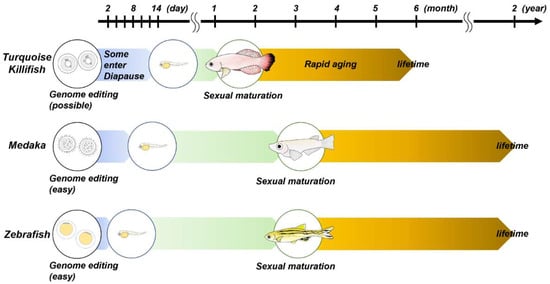
Figure 1.
This schema shows the life cycles of zebrafish, medaka and turquoise killifish. For further details, please refer to the main text.
For zebrafish, ZFIN (Zebrafish Information Network, https://zfin.org/; accessed on 30 November 2021) has a database of zebrafish genes, created transgenic lines and mutant lines. Many lines, including ENU mutagenesis products or transgenic reporter lines, are available for purchase and are managed by ZIRC (Zebrafish International Resource Center) or EZRC (European Zebrafish Resource Center). For medaka, please visit the NBRP medaka website (https://shigen.nig.ac.jp/medaka/top/top.jsp; accessed on 30 November 2021) to look for various mutant and transgenic lines. Additionally, NFIN (The Nothobranchius furzeri Information Network, https://www.nothobranchius.info/; accessed on 30 November 2021) provides an information on laboratory procedures and a gene database of turquoise killifish.
4. Ease of Laboratory Management and Experimentation with Zebrafish, Medaka and Turquoise Killifish
Neurodegeneration is a phenomenon characterized by the progressive loss of neurons, and Alzheimer’s disease, Parkinson’s disease and amyotrophic lateral sclerosis are the major neurodegenerative diseases among humans. They are progressive diseases with a relatively slow chronic clinical course, and the brains show abnormal protein deposition. Each neurodegenerative disease has a relatively selective neuronal loss; for example, dopaminergic neurons and the autonomic nervous system are vulnerable in Parkinson’s disease, and the motor neuron system is selectively lost in amyotrophic lateral sclerosis [67]. Recently, there has been much research on neurodegenerative diseases using the advantageous features of small fishes. To generate and analyze disease models, drug treatment with neurotoxic chemicals, direct microinjection of abnormal proteins, knockout of disease-related genes and knock-in of mutated genes can be used. For Alzheimer’s disease studies, tau transgenic zebrafish [68,69] and zebrafish with direct microinjection of Aβ have been reported [70,71]. For amyotrophic lateral sclerosis studies, TDP-43-, SOD1- or C9orf72-related zebrafish models have been used to analyze the pathogenesis and screen for new drugs [30,72,73,74,75,76]. Here, we discuss Parkinson’s disease; for more information on other neurodegenerative diseases, see other reviews [77,78].
4.1. Zebrafish and Medaka Models of Parkinson’s Disease
Parkinson’s disease is a common disease in the elderly and it is characterized by progressive motor impairment, loss of dopaminergic neurons in the substantia nigra and alpha-synuclein-positive inclusion bodies called Lewy bodies. We and others have used a variety of small fishes to study the pathogenesis of Parkinson’s disease. Toxins such as 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), 6-hydroxydopamine (6-OHDA) and rotenone are known to be toxic to dopaminergic neurons in various animal models. MPTP is a neurotoxin that induces Parkinson’s disease-like symptoms in various animals, including humans. It is metabolized to 1-methyl-4-phenylpyridinium (MPP+) in glial cells and is subsequently incorporated by dopaminergic neurons via dopamine transporters and inhibits the activity of the mitochondrial respiratory chain. Due to this metabolic pathway, dopaminergic neurons are selectively damaged [79,80]. MPTP also induces Parkinson’s disease-like symptoms in medaka. Keeping medaka larvae in MPTP-containing water rapidly induces a reduction in spontaneous swimming movement and a loss of dopaminergic neurons in the diencephalon [81]. MPTP exposure has also been carried out using zebrafish larvae and adults. These studies have demonstrated the effect of MPTP on locomotion and dopaminergic neurons [82,83,84,85,86,87]. 6-OHDA can be taken up by the dopamine transporter, resulting in oxidative stress and selective damage to dopaminergic neurons. 6-OHDA, when administered directly into the cerebrospinal fluid, also induces a loss of dopamine neurons and a decrease in spontaneous swimming movements in medaka [88]. The injection of 6-OHDA into the ventral diencephalon of adult zebrafish or by keeping zebrafish larvae in 6-OHDA-containing water can cause a reduction in dopaminergic neurons and impaired swimming movements [89,90,91,92]. Keeping zebrafish larvae in rotenone-containing water also causes a reduction in dopamine and impaired swimming movements [93,94]. Other chemicals, including proteasome inhibitors [88], ammonium chloride or tunicamycin [95], can induce Parkinson’s disease-like phenotypes in small fishes. These toxin-induced models can be useful for Parkinson’s disease research and drug screening.
Next, we will introduce genetic models of Parkinson’s disease using zebrafish and medaka. It is surprisingly difficult to simulate human Parkinson’s disease by genetic manipulation in mice, which is one of the most representative model animals. For example, even in triple knockout mice with Parkin, PINK1 and DJ-1, the gene products responsible for autosomal recessive familial Parkinson’s disease, there is no dopaminergic neuron loss [96]. Similar to mice, Parkin or Pink1 single knockout medaka does not show dopaminergic cell loss [97,98]. We analyzed Parkin and Pink1 double knockout medaka and found that a loss of dopaminergic neurons occurred, which was not observed in single knockout fish [98]. In the case of zebrafish, single depletion of Pink1 is sufficient to induce the loss of dopaminergic neurons [99,100,101]. DJ-1 knockout zebrafish and medaka models have also been created, which need further pathological evaluations but are promising for the production of new fish models of Parkinson’s disease [102,103,104,105].
ATP13A2 is another gene product that is responsible for autosomal recessive early-onset parkinsonism that is characterized by levodopa responsiveness, supranuclear gaze palsy, pyramidal signs and dementia [106]. We created Atp13a2 mutant medaka, which shows a mutation similar to that seen in human patients, and this fish shows loss of dopaminergic neurons with a decrease in cathepsin D activity and fingerprint-like inclusion body formation [107]. Atp13a2 knockout zebrafish also show similar phenotypes [108]. GBA, which is also the causative gene of Gaucher disease, is one of the high-risk genes for idiopathic Parkinson’s disease, and Gba knockout medaka and zebrafish exhibit not only dopaminergic neurodegeneration but also alpha-synuclein accumulation [109,110,111]. LRRK2 mutations are a relatively common cause of autosomal dominant familial Parkinson’s disease and they are also related to idiopathic Parkinson’s disease [112,113]. There is a difficulty in understanding such autosomal dominant disease because it might not be clear whether a loss of the normal function of the gene is a major cause of the phenotype or whether a gain of toxic function can explain the disease. There have been several zebrafish models of Lrrk2, but we will await further evaluations and consistent findings [114,115,116,117].
In this way, various models of Parkinson’s disease can be generated by treating zebrafish or medaka with chemicals or by conducting genetic modification of the zebrafish or medaka genome. These models are very useful to analyze the function of molecules related to Parkinson’s disease in vivo and to understand the pathophysiology of human Parkinson’s disease. Furthermore, it has already been widely used in the field of drug discovery. High-throughput screening using the characteristics of zebrafish has picked up many compounds with the potential to improve the pathology of Parkinson’s disease. Please refer to the recent review on drug discovery for major neurodegenerative diseases including Parkinson’s disease [77]. Of course, one limitation of using small fishes as model animals is that there can be a discrepancy between fishes and humans. As is the case with most human diseases, the onset of disease is complicated by a number of factors, such as aging, environmental factors and multifactorial genetic effects. Therefore, we should carefully examine various models including cell lines, small fishes or mammalians and it is also important to investigate human samples. By going back and forth between models such as fish and human samples, we can understand the human pathology more reliably.
4.2. Idiopathic Parkinson’s Disease Phenotypes Seen in Turquoise Killifish
Next, we focused on turquoise killifish to understand idiopathic Parkinson’s disease, which is not hereditary and accounts for 90–95% of all Parkinson’s disease cases. Turquoise killifish is a small fish species that lives in ponds, swamps and puddles in Mozambique and other countries [7]. Its habitat has a long dry season and a short rainy season, and during the dry season, the water in which the turquoise killifish lives dries up and the adult fish cannot survive. However, it has been able to survive as a species by adopting a life history in which it spawns drought-resistant eggs in the soil, which hatch during the next or future rainy season. In such a life cycle, a positive selection pressure for anti-ageing does not work [118]. Most likely, turquoise killifish have a short lifespan and exhibit an ageing phenotype in a very short period of time. Specifically, the lifespan of turquoise killifish is approximately four to six months, and around the age of three months, they present various signs of ageing, including organ atrophy, spine curvature and increased levels of senescence-associated beta-galactosidase [119,120,121]. Although Parkinson’s disease is strongly associated with ageing in humans, most experimental animals may not present a sufficient disease phenotype during ageing. We have found that turquoise killifish exhibit degeneration of dopaminergic and noradrenergic neurons and the progression of alpha-synuclein pathology with ageing [122]. These pathological phenotypes are similar to those observed in human Parkinson’s disease. Genetic depletion of alpha-synuclein by CRISPR-Cas9 system ameliorates neurodegeneration, suggesting that alpha-synuclein is not a bystander in the pathogenesis of Parkinson’s disease but is a causative protein of neurodegeneration. The turquoise killifish has the potential to reveal the mechanisms of Parkinson’s disease, especially the majority of cases of idiopathic Parkinson’s disease. This unique fish will also be useful for other age-related disorders in the brain and other organs.
5. Human Neurodevelopmental Disorders in Small Fishes
Human neurodevelopmental disorders are diagnosed based on the relative relationship between a person’s behavior and society, such as developmental characteristics and difficulties in social life, not based on genetic diagnosis or biomarkers such as MRI scans [123]. One limitation of using small fishes to study neurodevelopmental disorders is that it is not probable that small fishes will meet the diagnostic criteria for those human neurodevelopmental disorders. Although it is difficult to apply the complex higher-order functions of humans to zebrafish, there have been reports in recent years that zebrafish can be used as a model animal for neurodevelopmental disorders by applying behavioral analysis that imitates human social responses. Furthermore, as has already been mentioned, the utility of small fishes in the laboratory as a model animal for neurodevelopmental disorders has led to many interesting findings in terms of consistent observations from the cellular and molecular scale to tissue, developmental and behavioral analysis.
Autism spectrum disorder (ASD) is one of the most common neurodevelopmental disorders. Although the pathogenesis of ASD has not been established, findings from comprehensive genetic analysis of patients with ASD have been accumulated, and a database of risk genes for the onset of ASD has been created. SFARI (https://gene.sfari.org/; accessed on 30 November 2021), a database operated by the Simon Foundation in the United States, is available for reference. There are currently 1023 registered genes classified by risk intensity. Moreover, genetic factors have been recognized in the pathogenesis of attention-deficit/hyperactivity disorder (ADHD), and in recent years, findings from meta-analyses of genome-wide association analyses have accumulated [124,125,126]. The following is a summary of research reports using zebrafish mutant models of genes thought to be associated with these neurodevelopmental disorders (Table 2).

Table 2.
Studies using behavioral analysis in zebrafish with gene mutations associated with neurodevelopmental disorders.
DYRK1A is a serine/threonine kinase that is essential for brain development and function, and overactivation of this protein is observed in Down syndrome [133]. In addition, DYRK1A belongs to score 1 in the SFARI database and is considered to be a highly relevant risk gene for ASD. Kim et al. generated and analyzed Dyrk1aa knockout zebrafish, an orthologue of DYRK1A. They showed that adult knockout fish showed microcephaly, behavioral analysis showed that anxiety behavior was reduced by the novel tank test, and social interaction was impaired by the shoaling test and social preference test. They concluded that this was an autistic-like behavioral change in fish [127]. In the same way, zebrafish orthologue knockout lines were generated for SHANK3 and NRXN2, which belong to score 1 of ASD risk genes in the SFARI database. SHANK3 is widely expressed in the brain and is mainly involved in the formation of postsynaptic scaffolds and neurotransmission [134]. Liu et al. generated Shank3b knockout zebrafish that showed impaired social interactions by behavioral analysis, and reported reduced expression of Homer1, a SHANK-binding protein, in the adult fish brain [128]. NRXN2 is a transmembrane protein that resides in the presynaptic terminal and is involved in synapse construction and neurotransmitter release mechanisms [135]. NRXN2α knockout mice have been used as a model for autism and have been shown to exhibit increased anxiety-like behavior in assays such as the light/dark box test and the elevated plus maze test [136]. Koh et al. generated Nrxn2a knockout zebrafish and found increased anxiety-like behavior in the novel tank test, suggesting that autism-like behavioral changes also occur in zebrafish [129].
PER1 is known as a clock gene, and genome-wide association analysis of ADHD patients suggests that this gene is a risk gene for ADHD [124]. Huang et al. created Per1b knockout zebrafish and showed that juveniles were hyperactive, had increased attack frequency in the mirror-image attack test and were rescued by microinjection of per1b mRNA. They also showed that the dopamine content was decreased in Per1b knockout zebrafish brains and that the overactive phenotype could be rescued by selegiline (monoamine oxidase B inhibitor) or methylphenidate (dopamine transporter inhibitor, human ADHD treatment). They also analyzed PER1 knockout mice. Similar to the zebrafish model, PER1 knockout mice showed hyperactivity and decreased dopamine content in brain samples, suggesting the possibility that PER1 abnormalities may be involved in dopaminergic neural abnormalities in ADHD [131]. This report is quite impressive because it is suggestive of a highly conserved phenotype among vertebrate species, including behavioral characteristics.
To summarize how the behavioral characteristics of zebrafish express the symptoms of human neurodevelopmental disorders, “reactivity to anxiety” corresponds to sensory hypersensitivity/sensory deprivation in autism spectrum disorders, “lack of crowding” as a difficulty in social communication and interpersonal interactions and “hyperactivity and aggression” as phenotypes for hyperactivity/impulsivity symptoms in ADHD, can be evaluated in each assay. Even if the anatomical and physiological differences are not clear in a disease model, if some phenotype can be obtained through behavioral analysis, it might be used as a milestone to assess whether some intervention can provide rescue, such as pharmacological high-throughput screening [130,131,132]. What should be carefully considered is its interpretation in behavioral analysis. While the behavioral analysis of mice has a long history and has been standardized by many researchers, the behavioral analysis of zebrafish is still in its development phase. For example, the novel tank test tracks the behavior of zebrafish after they have been transferred to a new tank and aggregates and statistically processes how much time they spent at which water depth and how far they travelled. In this assay, zebrafish first spend time hiding at the bottom of the tank and then they gradually expand their range of activities to the surface. If it is observed that the zebrafish spend less time at the bottom of the tank and immediately start to move closer to the surface, it may have different meanings depending on whether it is explained as “not feeling anxious easily” or “hyperactivity and impulsiveness”. See the references for a list of zebrafish behaviors [137], a summary of behavioral analysis and its limitations and a contrast with behavioral analysis in mice [138,139,140]. Behavioral analysis looks at the habits of fish, but it is necessary to critically consider them when applying them to humans. It would be more convincing if trends in phenotypes could be observed in multiple assays, rather than making assumptions based on the result of a single behavioral analysis. In addition to behavioral analysis, other types of methods that can evaluate stress responses are also being considered; for example, by evaluating the level of cortisol, which is one of the stress hormones [141,142,143]. There are still many unknown aspects of using small fish as a model for human higher brain functions and human neurodevelopmental disorders, and we hope that increasing research will be accumulated.
In addition, zebrafish are also used in the field of psychiatry to analyze schizophrenia and depression. It is very interesting to see the phenotype of zebrafish as a model animal for psychiatric symptoms [144,145]. Even though the fields are different, zebrafish are used in similar ways as described in this review. For more information, refer to other excellent publications [139,146,147].
6. Conclusions
In this review, we have discussed the features of zebrafish, medaka and turquoise killifish in the laboratory and the actual analysis of neurodegenerative diseases and neurodevelopmental disorders using these small fishes. In the analysis of human neurological disorders, small fishes are very good model animals and will be further developed in the future. At this point, we need to have a sense of humility towards mammalian model animals. Even if various experimental results are shown in small fishes, if the same thing can be shown in mice, the impact can be greater in mice. To demonstrate the meaning and value of using small fishes, research designs are expected to take advantage of the characteristics of small fishes and their benefits in the laboratory, as described in this review. In addition, we should not forget that we are looking at the human nervous system through small fishes. It may not be clear what the changes in the RNA and proteins in small fishes in the context of human diseases and disorders mean, if we only pay attention to small fishes. The same thing is applicable to the meaning of the changes in morphology and physiological functions at the organ level and the meaning of changes in behaviors obtained through behavioral analysis. The meaning of the results obtained from small fishes will become clear when the results are bridged to mammalian model animals such as mice and then to human analysis. If such a relationship can be established between small fishes and other samples, these fishes can become increasingly powerful and useful tools for solving human neurological disorders.
Author Contributions
H.M. and K.K. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from AMED (Grant Numbers JP19gm6110028 and JP19dm0107154 (H.M.)), the Takeda Science Foundation (H.M.), JSPS KAKENHI (Grant Numbers JP 14516799 (H.M.), JP 16690735 (H.M.) and JP 17925674 (H.M.)) and JST [Moonshot R&D] [Grant Number JPMJMS2024] (H.M.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data and tools described in this manuscript are available upon request.
Acknowledgments
We acknowledge Shinano Kobayashi and Noriko Matsui for participating in helpful discussions and providing continuous support. We acknowledge Ai Ito for her work on the illustrations.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Delomas, T.A.; Dabrowski, K. Larval rearing of zebrafish at suboptimal temperatures. J. Therm. Biol. 2018, 74, 170–173. [Google Scholar] [CrossRef]
- Shima, A.; Mitani, H. Medaka as a research organism: Past, present and future. Mech. Dev. 2004, 121, 599–604. [Google Scholar] [CrossRef]
- Kirchmaier, S.; Naruse, K.; Wittbrodt, J.; Loosli, F. The genomic and genetic toolbox of the teleost medaka (Oryzias latipes). Genetics 2015, 199, 905–918. [Google Scholar] [CrossRef]
- Reichard, M.; Polacik, M.; Sedlácek, O. Distribution, colour polymorphism and habitat use of the African killifish Nothobranchius furzeri, the vertebrate with the shortest life span. J. Fish. Biol. 2009, 74, 198–212. [Google Scholar] [CrossRef]
- Dodzian, J.; Kean, S.; Seidel, J.; Valenzano, D.R. A Protocol for Laboratory Housing of Turquoise Killifish (Nothobranchius furzeri). J. Vis. Exp. 2018, 134, 57073. [Google Scholar] [CrossRef]
- Valenzano, D.R.; Benayoun, B.A.; Singh, P.P.; Zhang, E.; Etter, P.D.; Hu, C.K.; Clément-Ziza, M.; Willemsen, D.; Cui, R.; Harel, I.; et al. The African Turquoise Killifish Genome Provides Insights into Evolution and Genetic Architecture of Lifespan. Cell 2015, 163, 1539–1554. [Google Scholar] [CrossRef]
- Poeschla, M.; Valenzano, D.R. The turquoise killifish: A genetically tractable model for the study of aging. J. Exp. Biol. 2020, 223 (Suppl. 1), jeb209296. [Google Scholar] [CrossRef]
- Lowery, L.A.; Sive, H. Initial formation of zebrafish brain ventricles occurs independently of circulation and requires the nagie oko and snakehead/atp1a1a.1 gene products. Development 2005, 132, 2057–2067. [Google Scholar] [CrossRef]
- Holzschuh, J.; Ryu, S.; Aberger, F.; Driever, W. Dopamine transporter expression distinguishes dopaminergic neurons from other catecholaminergic neurons in the developing zebrafish embryo. Mech. Dev. 2001, 101, 237–243. [Google Scholar] [CrossRef]
- Tay, T.L.; Ronneberger, O.; Ryu, S.; Nitschke, R.; Driever, W. Comprehensive catecholaminergic projectome analysis reveals single-neuron integration of zebrafish ascending and descending dopaminergic systems. Nat. Commun. 2011, 2, 171. [Google Scholar] [CrossRef]
- Rink, E.; Wullimann, M.F. Connections of the ventral telencephalon and tyrosine hydroxylase distribution in the zebrafish brain (Danio rerio) lead to identification of an ascending dopaminergic system in a teleost. Brain Res. Bull. 2002, 57, 385–387. [Google Scholar] [CrossRef]
- Borgonovo, J.; Ahumada-Galleguillos, P.; Oñate-Ponce, A.; Allende-Castro, C.; Henny, P.; Concha, M.L. Organization of the Catecholaminergic System in the Short-Lived Fish Nothobranchius furzeri. Front. Neuroanat. 2021, 15, 728720. [Google Scholar] [CrossRef]
- Bae, Y.K.; Kani, S.; Shimizu, T.; Tanabe, K.; Nojima, H.; Kimura, Y.; Higashijima, S.; Hibi, M. Anatomy of zebrafish cerebellum and screen for mutations affecting its development. Dev. Biol. 2009, 330, 406–426. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, M.; Hibi, M. Development and evolution of cerebellar neural circuits. Dev. Growth Differ. 2012, 54, 373–389. [Google Scholar] [CrossRef] [PubMed]
- Pouwels, E. On the development of the cerebellum of the trout, Salmo gairdneri. IV. Development of the pattern of connectivity. Anat. Embryol. 1978, 153, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Matsui, H.; Namikawa, K.; Babaryka, A.; Köster, R.W. Functional regionalization of the teleost cerebellum analyzed in vivo. Proc. Natl. Acad. Sci. USA 2014, 111, 11846–11851. [Google Scholar] [CrossRef]
- von Trotha, J.W.; Vernier, P.; Bally-Cuif, L. Emotions and motivated behavior converge on an amygdala-like structure in the zebrafish. Eur. J. Neurosci. 2014, 40, 3302–3315. [Google Scholar] [CrossRef]
- Mueller, T.; Dong, Z.; Berberoglu, M.A.; Guo, S. The dorsal pallium in zebrafish, Danio rerio (Cyprinidae, Teleostei). Brain Res. 2011, 1381, 95–105. [Google Scholar] [CrossRef]
- Quiñonez-Silvero, C.; Hübner, K.; Herzog, W. Development of the brain vasculature and the blood-brain barrier in zebrafish. Dev. Biol. 2020, 457, 181–190. [Google Scholar] [CrossRef]
- Fleming, A.; Diekmann, H.; Goldsmith, P. Functional characterisation of the maturation of the blood-brain barrier in larval zebrafish. PLoS ONE 2013, 8, e77548. [Google Scholar] [CrossRef]
- d’Amora, M.; Giordani, S. The Utility of Zebrafish as a Model for Screening Developmental Neurotoxicity. Front. Neurosci. 2018, 12, 976. [Google Scholar] [CrossRef] [PubMed]
- Reimer, M.M.; Sörensen, I.; Kuscha, V.; Frank, R.E.; Liu, C.; Becker, C.G.; Becker, T. Motor neuron regeneration in adult zebrafish. J. Neurosci. 2008, 28, 8510–8516. [Google Scholar] [CrossRef]
- Klatt Shaw, D.; Saraswathy, V.M.; Zhou, L.; McAdow, A.R.; Burris, B.; Butka, E.; Morris, S.A.; Dietmann, S.; Mokalled, M.H. Localized EMT reprograms glial progenitors to promote spinal cord repair. Dev. Cell. 2021, 56, 613–626.e7. [Google Scholar] [CrossRef]
- Kizil, C.; Kaslin, J.; Kroehne, V.; Brand, M. Adult neurogenesis and brain regeneration in zebrafish. Dev. Neurobiol. 2012, 72, 429–461. [Google Scholar] [CrossRef]
- D’Angelo, L.; Lossi, L.; Merighi, A.; de Girolamo, P. Anatomical features for the adequate choice of experimental animal models in biomedicine: I. Fishes. Ann. Anat. 2016, 205, 75–84. [Google Scholar] [CrossRef]
- White, R.M.; Sessa, A.; Burke, C.; Bowman, T.; LeBlanc, J.; Ceol, C.; Bourque, C.; Dovey, M.; Goessling, W.; Burns, C.E.; et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008, 2, 183–189. [Google Scholar] [CrossRef]
- Chen, T.W.; Wardill, T.J.; Sun, Y.; Pulver, S.R.; Renninger, S.L.; Baohan, A.; Schreiter, E.R.; Kerr, R.A.; Orger, M.B.; Jayaraman, V.; et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 2013, 499, 295–300. [Google Scholar] [CrossRef]
- Leung, L.C.; Wang, G.X.; Madelaine, R.; Skariah, G.; Kawakami, K.; Deisseroth, K.; Urban, A.E.; Mourrain, P. Neural signatures of sleep in zebrafish. Nature 2019, 571, 198–204. [Google Scholar] [CrossRef]
- Del Bene, F.; Wyart, C. Optogenetics: A new enlightenment age for zebrafish neurobiology. Dev. Neurobiol. 2012, 72, 404–414. [Google Scholar] [CrossRef]
- Asakawa, K.; Handa, H.; Kawakami, K. Optogenetic modulation of TDP-43 oligomerization accelerates ALS-related pathologies in the spinal motor neurons. Nat. Commun. 2020, 11, 1004. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef]
- Kosuta, C.; Daniel, K.; Johnstone, D.L.; Mongeon, K.; Ban, K.; LeBlanc, S.; MacLeod, S.; Et-Tahiry, K.; Ekker, M.; MacKenzie, A.; et al. High-throughput DNA Extraction and Genotyping of 3dpf Zebrafish Larvae by Fin Clipping. J. Vis. Exp. 2018, 136, 58024. [Google Scholar] [CrossRef]
- Sander, J.D.; Cade, L.; Khayter, C.; Reyon, D.; Peterson, R.T.; Joung, J.K.; Yeh, J.R. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol. 2011, 29, 697–698. [Google Scholar] [CrossRef]
- Huang, P.; Xiao, A.; Zhou, M.; Zhu, Z.; Lin, S.; Zhang, B. Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 2011, 29, 699–700. [Google Scholar] [CrossRef]
- Meng, X.; Noyes, M.B.; Zhu, L.J.; Lawson, N.D.; Wolfe, S.A. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 2008, 26, 695–701. [Google Scholar] [CrossRef] [PubMed]
- Doyon, Y.; McCammon, J.M.; Miller, J.C.; Faraji, F.; Ngo, C.; Katibah, G.E.; Amora, R.; Hocking, T.D.; Zhang, L.; Rebar, E.J. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 2008, 26, 702–708. [Google Scholar] [CrossRef]
- Varshney, G.K.; Pei, W.; LaFave, M.C.; Idol, J.; Xu, L.; Gallardo, V.; Carrington, B.; Bishop, K.; Jones, M.; Li, M.; et al. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 2015, 25, 1030–1042. [Google Scholar] [CrossRef] [PubMed]
- Ablain, J.; Durand, E.M.; Yang, S.; Zhou, Y.; Zon, L.I. A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish. Dev. Cell. 2015, 32, 756–764. [Google Scholar] [CrossRef] [PubMed]
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Tsai, S.Q.; Sander, J.D.; Peterson, R.T.; Yeh, J.R.; Joung, J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 227–229. [Google Scholar] [CrossRef] [PubMed]
- Rosen, J.N.; Sweeney, M.F.; Mably, J.D. Microinjection of Zebrafish Embryos to Analyze Gene Function. J. Vis. Exp. 2009, 25, 1115. [Google Scholar] [CrossRef] [PubMed]
- Finckbeiner, S.; Ko, P.J.; Carrington, B.; Sood, R.; Gross, K.; Dolnick, B.; Sufrin, J.; Liu, P. Transient knockdown and overexpression reveal a developmental role for the zebrafish enosf1b gene. Cell Biosci. 2011, 1, 32. [Google Scholar] [CrossRef] [PubMed]
- Hoshijima, K.; Jurynec, M.J.; Klatt Shaw, D.; Jacobi, A.M.; Behlke, M.A.; Grunwald, D.J. Highly Efficient CRISPR-Cas9-Based Methods for Generating Deletion Mutations and F0 Embryos that Lack Gene Function in Zebrafish. Dev. Cell. 2019, 51, 645–657.e4. [Google Scholar] [CrossRef] [PubMed]
- Kok, F.O.; Shin, M.; Ni, C.W.; Gupta, A.; Grosse, A.S.; van Impel, A.; Kirchmaier, B.C.; Peterson-Maduro, J.; Kourkoulis, G.; Male, I.; et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev. Cell. 2015, 32, 97–108. [Google Scholar] [CrossRef] [PubMed]
- El-Brolosy, M.A.; Kontarakis, Z.; Rossi, A.; Kuenne, C.; Günther, S.; Fukuda, N.; Kikhi, K.; Boezio, G.L.M.; Takacs, C.M.; Lai, S.L.; et al. Genetic compensation triggered by mutant mRNA degradation. Nature 2019, 568, 193–197. [Google Scholar] [CrossRef]
- Irion, U.; Krauss, J.; Nüsslein-Volhard, C. Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development 2014, 141, 4827–4830. [Google Scholar] [CrossRef]
- Prykhozhij, S.V.; Fuller, C.; Steele, S.L.; Veinotte, C.J.; Razaghi, B.; Robitaille, J.M.; McMaster, C.R.; Shlien, A.; Malkin, D.; Berman, J.N. Optimized knock-in of point mutations in zebrafish using CRISPR/Cas9. Nucleic Acids Res. 2018, 46, 9252. [Google Scholar] [CrossRef]
- Tessadori, F.; Roessler, H.I.; Savelberg, S.M.C.; Chocron, S.; Kamel, S.M.; Duran, K.J.; van Haelst, M.M.; van Haaften, G.; Bakkers, J. Effective CRISPR/Cas9-based nucleotide editing in zebrafish to model human genetic cardiovascular disorders. Dis. Model. Mech. 2018, 11, dmm035469. [Google Scholar] [CrossRef]
- de Vrieze, E.; de Bruijn, S.E.; Reurink, J.; Broekman, S.; van de Riet, V.; Aben, M.; Kremer, H.; van Wijk, E. Efficient Generation of Knock-In Zebrafish Models for Inherited Disorders Using CRISPR-Cas9 Ribonucleoprotein Complexes. Int. J. Mol. Sci. 2021, 22, 9429. [Google Scholar] [CrossRef]
- Auer, T.O.; Del Bene, F. CRISPR/Cas9 and TALEN-mediated knock-in approaches in zebrafish. Methods 2014, 69, 142–150. [Google Scholar] [CrossRef]
- Kimura, Y.; Hisano, Y.; Kawahara, A.; Higashijima, S. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci. Rep. 2014, 4, 6545. [Google Scholar] [CrossRef]
- Stuart, G.W.; McMurray, J.V.; Westerfield, M. Replication, integration and stable germ-line transmission of foreign sequences injected into early zebrafish embryos. Development 1988, 103, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Thermes, V.; Grabher, C.; Ristoratore, F.; Bourrat, F.; Choulika, A.; Wittbrodt, J.; Joly, J.S. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech. Dev. 2002, 118, 91–98. [Google Scholar] [CrossRef]
- Kawakami, K.; Shima, A.; Kawakami, N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc. Natl. Acad. Sci. USA 2000, 97, 11403–11408. [Google Scholar] [CrossRef] [PubMed]
- Grabher, C.; Henrich, T.; Sasado, T.; Arenz, A.; Wittbrodt, J.; Furutani-Seiki, M. Transposon-mediated enhancer trapping in medaka. Gene 2003, 322, 57–66. [Google Scholar] [CrossRef]
- Harel, I.; Valenzano, D.R.; Brunet, A. Efficient genome engineering approaches for the short-lived African turquoise killifish. Nat. Protoc. 2016, 11, 2010–2028. [Google Scholar] [CrossRef]
- Hartmann, N.; Englert, C. A microinjection protocol for the generation of transgenic killifish (Species: Nothobranchius furzeri). Dev. Dyn. 2012, 241, 1133–1141. [Google Scholar] [CrossRef]
- Baroiller, J.F.; D’Cotta, H. Environment and sex determination in farmed fish. Comp. Biochem. Physiol. C. Toxicol. Pharmacol. 2001, 130, 399–409. [Google Scholar] [CrossRef]
- Orn, S.; Holbech, H.; Madsen, T.H.; Norrgren, L.; Petersen, G.I. Gonad development and vitellogenin production in zebrafish (Danio rerio) exposed to ethinylestradiol and methyltestosterone. Aquat. Toxicol. 2003, 65, 397–411. [Google Scholar] [CrossRef]
- Abozaid, H.; Wessels, S.; Hörstgen-Schwark, G. Elevated temperature applied during gonadal transformation leads to male bias in zebrafish (Danio rerio). Sex Dev. 2012, 6, 201–209. [Google Scholar] [CrossRef]
- Schartl, M. A comparative view on sex determination in medaka. Mech. Dev. 2004, 121, 639–645. [Google Scholar] [CrossRef]
- Valenzano, D.R.; Kirschner, J.; Kamber, R.A.; Zhang, E.; Weber, D.; Cellerino, A.; Englert, C.; Platzer, M.; Reichwald, K.; Brunet, A. Mapping loci associated with tail color and sex determination in the short-lived fish Nothobranchius furzeri. Genetics 2009, 183, 1385–1395. [Google Scholar] [CrossRef] [PubMed]
- Reichwald, K.; Petzold, A.; Koch, P.; Downie, B.R.; Hartmann, N.; Pietsch, S.; Baumgart, M.; Chalopin, D.; Felder, M.; Bens, M.; et al. Insights into Sex Chromosome Evolution and Aging from the Genome of a Short-Lived Fish. Cell 2015, 163, 1527–1538. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.H.; Dewar, K.; Wagner, G.P.; Takahashi, K.; Ruddle, F.; Ledje, C.; Bartsch, P.; Scemama, J.L.; Stellwag, E.; Fried, C.; et al. Bichir HoxA cluster sequence reveals surprising trends in ray-finned fish genomic evolution. Genome Res. 2004, 14, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Hoegg, S.; Brinkmann, H.; Taylor, J.S.; Meyer, A. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J. Mol. Evol. 2004, 59, 190–203. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.M.; Brunet, F.; Petit, J.L.; Stange-Thomann, N.; Mauceli, E.; Bouneau, L.; Fischer, C.; Ozouf-Costaz, C.; Bernot, A.; et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 2004, 431, 946–957. [Google Scholar] [CrossRef]
- Lechermeier, C.G.; Zimmer, F.; Lüffe, T.M.; Lesch, K.P.; Romanos, M.; Lillesaar, C.; Drepper, C. Transcript Analysis of Zebrafish GLUT3 Genes, slc2a3a and slc2a3b, Define Overlapping as Well as Distinct Expression Domains in the Zebrafish (Danio rerio) Central Nervous System. Front. Mol. Neurosci. 2019, 12, 199. [Google Scholar] [CrossRef]
- Dugger, B.N.; Dickson, D.W. Pathology of Neurodegenerative Diseases. Cold Spring Harb. Perspect. Biol. 2017, 9, a028035. [Google Scholar] [CrossRef]
- Bai, Q.; Garver, J.A.; Hukriede, N.A.; Burton, E.A. Generation of a transgenic zebrafish model of Tauopathy using a novel promoter element derived from the zebrafish eno2 gene. Nucleic Acids Res. 2007, 35, 6501–6516. [Google Scholar] [CrossRef]
- Paquet, D.; Bhat, R.; Sydow, A.; Mandelkow, E.M.; Berg, S.; Hellberg, S.; Fälting, J.; Distel, M.; Köster, R.W.; Schmid, B.; et al. A zebrafish model of tauopathy allows in vivo imaging of neuronal cell death and drug evaluation. J. Clin. Investig. 2009, 119, 1382–1395. [Google Scholar] [CrossRef]
- Bhattarai, P.; Thomas, A.K.; Cosacak, M.I.; Papadimitriou, C.; Mashkaryan, V.; Zhang, Y.; Kizil, C. Modeling Amyloid-beta42 Toxicity and Neurodegeneration in Adult Zebrafish Brain. J. Vis. Exp. 2017, 128, 56014. [Google Scholar] [CrossRef]
- Bhattarai, P.; Thomas, A.K.; Zhang, Y.; Kizil, C. The effects of aging on Amyloid-beta42-induced neurodegeneration and regeneration in adult zebrafish brain. Neurogenesis 2017, 4, e1322666. [Google Scholar] [CrossRef] [PubMed]
- Lissouba, A.; Liao, M.; Kabashi, E.; Drapeau, P. Transcriptomic Analysis of Zebrafish TDP-43 Transgenic Lines. Front. Mol. Neurosci. 2018, 11, 463. [Google Scholar] [CrossRef]
- Ramesh, T.; Lyon, A.N.; Pineda, R.H.; Wang, C.; Janssen, P.M.; Canan, B.D.; Burghes, A.H.; Beattie, C.E. A genetic model of amyotrophic lateral sclerosis in zebrafish displays phenotypic hallmarks of motoneuron disease. Dis. Model. Mech. 2010, 3, 652–662. [Google Scholar] [CrossRef] [PubMed]
- Shaw, M.P.; Higginbottom, A.; McGown, A.; Castelli, L.M.; James, E.; Hautbergue, G.M.; Shaw, P.J.; Ramesh, T.M. Stable transgenic C9orf72 zebrafish model key aspects of the ALS/FTD phenotype and reveal novel pathological features. Acta Neuropathol. Commun. 2018, 6, 125. [Google Scholar] [CrossRef] [PubMed]
- Ohki, Y.; Wenninger-Weinzierl, A.; Hruscha, A.; Asakawa, K.; Kawakami, K.; Haass, C.; Edbauer, D.; Schmid, B. Glycine-alanine dipeptide repeat protein contributes to toxicity in a zebrafish model of C9orf72 associated neurodegeneration. Mol. Neurodegener. 2017, 12, 6. [Google Scholar] [CrossRef][Green Version]
- Kabashi, E.; Bercier, V.; Lissouba, A.; Liao, M.; Brustein, E.; Rouleau, G.A.; Drapeau, P. FUS and TARDBP but not SOD1 interact in genetic models of amyotrophic lateral sclerosis. PloS Genet. 2011, 7, e1002214. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, J.B.; He, K.J.; Wang, F.; Liu, C.F. Advances of Zebrafish in Neurodegenerative Disease: From Models to Drug Discovery. Front. Pharmacol. 2021, 12, 713963. [Google Scholar] [CrossRef]
- Wang, J.; Cao, H. Zebrafish and Medaka: Important Animal Models for Human Neurodegenerative Diseases. Int. J. Mol. Sci. 2021, 22, 10766. [Google Scholar] [CrossRef]
- Davis, G.C.; Williams, A.C.; Markey, S.P.; Ebert, M.H.; Caine, E.D.; Reichert, C.M.; Kopin, I.J. Chronic Parkinsonism secondary to intravenous injection of meperidine analogues. Psychiatry Res. 1979, 1, 249–254. [Google Scholar] [CrossRef]
- Gerlach, M.; Riederer, P.; Przuntek, H.; Youdim, M.B. MPTP mechanisms of neurotoxicity and their implications for Parkinson’s disease. Eur. J. Pharmacol. 1991, 208, 273–286. [Google Scholar] [CrossRef]
- Matsui, H.; Taniguchi, Y.; Inoue, H.; Uemura, K.; Takeda, S.; Takahashi, R. A chemical neurotoxin, MPTP induces Parkinson’s disease like phenotype, movement disorders and persistent loss of dopamine neurons in medaka fish. Neurosci. Res. 2009, 65, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Lam, C.S.; Korzh, V.; Strahle, U. Zebrafish embryos are susceptible to the dopaminergic neurotoxin MPTP. Eur. J. Neurosci. 2005, 21, 1758–1762. [Google Scholar] [CrossRef] [PubMed]
- Sallinen, V.; Torkko, V.; Sundvik, M.; Reenilä, I.; Khrustalyov, D.; Kaslin, J.; Panula, P. MPTP and MPP+ target specific aminergic cell populations in larval zebrafish. J. Neurochem. 2009, 108, 719–731. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Casado, M.E.; Lima, E.; García, J.A.; Doerrier, C.; Aranda, P.; Sayed, R.K.; Guerra-Librero, A.; Escames, G.; López, L.C.; Acuña-Castroviejo, D. Melatonin rescues zebrafish embryos from the parkinsonian phenotype restoring the parkin/PINK1/DJ-1/MUL1 network. J. Pineal. Res. 2016, 61, 96–107. [Google Scholar] [CrossRef]
- Kalyn, M.; Hua, K.; Mohd Noor, S.; Wong, C.E.D.; Ekker, M. Comprehensive Analysis of Neurotoxin-Induced Ablation of Dopaminergic Neurons in Zebrafish Larvae. Biomedicines 2019, 8, 1. [Google Scholar] [CrossRef]
- Zhao, Y.; Han, Y.; Wang, Z.; Chen, T.; Qian, H.; He, J.; Li, J.; Han, B.; Wang, T. Rosmarinic acid protects against 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced dopaminergic neurotoxicity in zebrafish embryos. Toxicol. Vitr. 2020, 65, 104823. [Google Scholar] [CrossRef]
- Kalyn, M.; Ekker, M. Cerebroventricular Microinjections of MPTP on Adult Zebrafish Induces Dopaminergic Neuronal Death, Mitochondrial Fragmentation, and Sensorimotor Impairments. Front. Neurosci. 2021, 15, 718244. [Google Scholar] [CrossRef]
- Matsui, H.; Ito, H.; Taniguchi, Y.; Inoue, H.; Takeda, S.; Takahashi, R. Proteasome inhibition in medaka brain induces the features of Parkinson’s disease. J. Neurochem. 2010, 115, 178–187. [Google Scholar] [CrossRef]
- Vijayanathan, Y.; Lim, F.T.; Lim, S.M.; Long, C.M.; Tan, M.P.; Majeed, A.B.A.; Ramasamy, K. 6-OHDA-Lesioned Adult Zebrafish as a Useful Parkinson’s Disease Model for Dopaminergic Neuroregeneration. Neurotox. Res. 2017, 32, 496–508. [Google Scholar] [CrossRef]
- Vaz, R.L.; Sousa, S.; Chapela, D.; van der Linde, H.C.; Willemsen, R.; Correia, A.D.; Outeiro, T.F.; Afonso, N.D. Identification of antiparkinsonian drugs in the 6-hydroxydopamine zebrafish model. Pharmacol. Biochem. Behav. 2020, 189, 172828. [Google Scholar] [CrossRef]
- Li, M.; Zhou, F.; Xu, T.; Song, H.; Lu, B. Acteoside protects against 6-OHDA-induced dopaminergic neuron damage via Nrf2-ARE signaling pathway. Food Chem. Toxicol. 2018, 119, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Cronin, A.; Grealy, M. Neuroprotective and Neuro-restorative Effects of Minocycline and Rasagiline in a Zebrafish 6-Hydroxydopamine Model of Parkinson’s Disease. Neuroscience 2017, 367, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, W.; Yang, J.; Wang, F.; Sima, Y.; Zhong, Z.M.; Wang, H.; Hu, L.F.; Liu, C.F. Parkinson’s disease-like motor and non-motor symptoms in rotenone-treated zebrafish. Neurotoxicology 2017, 58, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Byrnes, J.; Ganetzky, R.; Lightfoot, R.; Tzeng, M.; Nakamaru-Ogiso, E.; Seiler, C.; Falk, M.J. Pharmacologic modeling of primary mitochondrial respiratory chain dysfunction in zebrafish. Neurochem. Int. 2018, 117, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Matsui, H.; Ito, H.; Taniguchi, Y.; Takeda, S.; Takahashi, R. Ammonium chloride and tunicamycin are novel toxins for dopaminergic neurons and induce Parkinson’s disease-like phenotypes in medaka fish. J. Neurochem. 2010, 115, 1150–1160. [Google Scholar] [CrossRef] [PubMed]
- Kitada, T.; Tong, Y.; Gautier, C.A.; Shen, J. Absence of nigral degeneration in aged parkin/DJ-1/PINK1 triple knockout mice. J. Neurochem. 2009, 111, 696–702. [Google Scholar] [CrossRef]
- Matsui, H.; Taniguchi, Y.; Inoue, H.; Kobayashi, Y.; Sakaki, Y.; Toyoda, A.; Uemura, K.; Kobayashi, D.; Takeda, S.; Takahashi, R. Loss of PINK1 in medaka fish (Oryzias latipes) causes late-onset decrease in spontaneous movement. Neurosci. Res. 2010, 66, 151–161. [Google Scholar] [CrossRef]
- Matsui, H.; Gavinio, R.; Asano, T.; Uemura, N.; Ito, H.; Taniguchi, Y.; Kobayashi, Y.; Maki, T.; Shen, J.; Takeda, S.; et al. PINK1 and Parkin complementarily protect dopaminergic neurons in vertebrates. Hum. Mol. Genet. 2013, 22, 2423–2434. [Google Scholar] [CrossRef]
- Matsui, H.; Sugie, A. An optimized method for counting dopaminergic neurons in zebrafish. PLoS ONE 2017, 12, e0184363. [Google Scholar] [CrossRef]
- Flinn, L.J.; Keatinge, M.; Bretaud, S.; Mortiboys, H.; Matsui, H.; De Felice, E.; Woodroof, H.I.; Brown, L.; McTighe, A.; Soellner, R.; et al. TigarB causes mitochondrial dysfunction and neuronal loss in PINK1 deficiency. Ann. Neurol. 2013, 74, 837–847. [Google Scholar] [CrossRef]
- Soman, S.; Keatinge, M.; Moein, M.; Da Costa, M.; Mortiboys, H.; Skupin, A.; Sugunan, S.; Bazala, M.; Kuznicki, J.; Bandmann, O. Inhibition of the mitochondrial calcium uniporter rescues dopaminergic neurons in pink1(-/-) zebrafish. Eur. J. Neurosci. 2017, 45, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Edson, A.J.; Hushagen, H.A.; Frøyset, A.K.; Elda, I.; Khan, E.A.; Di Stefano, A.; Fladmark, K.E. Dysregulation in the Brain Protein Profile of Zebrafish Lacking the Parkinson’s Disease-Related Protein DJ-1. Mol. Neurobiol. 2019, 56, 8306–8322. [Google Scholar] [CrossRef] [PubMed]
- Chin, H.Y.; Lardelli, M.; Collins-Praino, L.; Barthelson, K. Loss of park7 activity has differential effects on expression of iron responsive element (IRE) gene sets in the brain transcriptome in a zebrafish model of Parkinson’s disease. Mol. Brain. 2021, 14, 83. [Google Scholar] [CrossRef] [PubMed]
- Ansai, S.; Kinoshita, M. Targeted mutagenesis using CRISPR/Cas system in medaka. Biol. Open 2014, 3, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Ansai, S.; Sakuma, T.; Yamamoto, T.; Ariga, H.; Uemura, N.; Takahashi, R.; Kinoshita, M. Efficient targeted mutagenesis in medaka using custom-designed transcription activator-like effector nucleases. Genetics 2013, 193, 739–749. [Google Scholar] [CrossRef]
- Najim al-Din, A.S.; Wriekat, A.; Mubaidin, A.; Dasouki, M.; Hiari, M. Pallido-pyramidal degeneration, supranuclear upgaze paresis and dementia: Kufor-Rakeb syndrome. Acta Neurol. Scand. 1994, 89, 347–352. [Google Scholar] [CrossRef]
- Matsui, H.; Sato, F.; Sato, S.; Koike, M.; Taruno, Y.; Saiki, S.; Funayama, M.; Ito, H.; Taniguchi, Y.; Uemura, N.; et al. ATP13A2 deficiency induces a decrease in cathepsin D activity, fingerprint-like inclusion body formation, and selective degeneration of dopaminergic neurons. FEBS Lett. 2013, 587, 1316–1325. [Google Scholar] [CrossRef]
- Nyuzuki, H.; Ito, S.; Nagasaki, K.; Nitta, Y.; Matsui, N.; Saitoh, A.; Matsui, H. Degeneration of dopaminergic neurons and impaired intracellular trafficking in Atp13a2 deficient zebrafish. IBRO Rep. 2020, 9, 1–8. [Google Scholar] [CrossRef]
- Uemura, N.; Koike, M.; Ansai, S.; Kinoshita, M.; Ishikawa-Fujiwara, T.; Matsui, H.; Naruse, K.; Sakamoto, N.; Uchiyama, Y.; Todo, T.; et al. Viable neuronopathic Gaucher disease model in Medaka (Oryzias latipes) displays axonal accumulation of alpha-synuclein. PloS Genet. 2015, 11, e1005065. [Google Scholar] [CrossRef]
- Keatinge, M.; Bui, H.; Menke, A.; Chen, Y.C.; Sokol, A.M.; Bai, Q.; Ellett, F.; Da Costa, M.; Burke, D.; Gegg, M.; et al. Glucocerebrosidase 1 deficient Danio rerio mirror key pathological aspects of human Gaucher disease and provide evidence of early microglial activation preceding alpha-synuclein-independent neuronal cell death. Hum. Mol. Genet. 2015, 24, 6640–6652. [Google Scholar] [CrossRef]
- Matsui, H.; Ito, J.; Matsui, N.; Uechi, T.; Onodera, O.; Kakita, A. Cytosolic dsDNA of mitochondrial origin induces cytotoxicity and neurodegeneration in cellular and zebrafish models of Parkinson’s disease. Nat. Commun. 2021, 12, 3101. [Google Scholar] [CrossRef] [PubMed]
- Zimprich, A.; Biskup, S.; Leitner, P.; Lichtner, P.; Farrer, M.; Lincoln, S.; Kachergus, J.; Hulihan, M.; Uitti, R.J.; Calne, D.B.; et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 2004, 44, 601–607. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Tang, K.F.; Li, Y.; Xiong, Y.Y.; Shen, L.; Wei, Z.Y.; Zhou, K.J.; Niu, J.M.; Han, X.; Yang, L.; et al. Quantitative assessment of the effect of LRRK2 exonic variants on the risk of Parkinson’s disease: A meta-analysis. Parkinsonism Relat. Disord. 2012, 18, 722–730. [Google Scholar] [CrossRef] [PubMed]
- Sheng, D.; See, K.; Hu, X.; Yu, D.; Wang, Y.; Liu, Q.; Li, F.; Lu, M.; Zhao, J.; Liu, J. Disruption of LRRK2 in Zebrafish leads to hyperactivity and weakened antibacterial response. Biochem. Biophys. Res. Commun. 2018, 497, 1104–1109. [Google Scholar] [CrossRef] [PubMed]
- Suzzi, S.; Ahrendt, R.; Hans, S.; Semenova, S.A.; Chekuru, A.; Wirsching, P.; Kroehne, V.; Bilican, S.; Sayed, S.; Winkler, S.; et al. Deletion of lrrk2 causes early developmental abnormalities and age-dependent increase of monoamine catabolism in the zebrafish brain. PloS Genet. 2021, 17, e1009794. [Google Scholar] [CrossRef]
- Ren, G.; Xin, S.; Li, S.; Zhong, H.; Lin, S. Disruption of LRRK2 does not cause specific loss of dopaminergic neurons in zebrafish. PLoS ONE 2011, 6, e20630. [Google Scholar] [CrossRef]
- Sheng, D.; Qu, D.; Kwok, K.H.; Ng, S.S.; Lim, A.Y.; Aw, S.S.; Lee, C.W.; Sung, W.K.; Tan, E.K.; Lufkin, T.; et al. Deletion of the WD40 domain of LRRK2 in Zebrafish causes Parkinsonism-like loss of neurons and locomotive defect. PloS Genet. 2010, 6, e1000914. [Google Scholar] [CrossRef]
- Cui, R.; Medeiros, T.; Willemsen, D.; Iasi, L.N.M.; Collier, G.E.; Graef, M.; Reichard, M.; Valenzano, D.R. Relaxed Selection Limits Lifespan by Increasing Mutation Load. Cell 2019, 178, 385–399.e20. [Google Scholar] [CrossRef]
- Genade, T.; Benedetti, M.; Terzibasi, E.; Roncaglia, P.; Valenzano, D.R.; Cattaneo, A.; Cellerino, A. Annual fishes of the genus Nothobranchius as a model system for aging research. Aging Cell 2005, 4, 223–233. [Google Scholar] [CrossRef]
- Valenzano, D.R.; Terzibasi, E.; Cattaneo, A.; Domenici, L.; Cellerino, A. Temperature affects longevity and age-related locomotor and cognitive decay in the short-lived fish Nothobranchius furzeri. Aging Cell 2006, 5, 275–278. [Google Scholar] [CrossRef]
- Harel, I.; Benayoun, B.A.; Machado, B.; Singh, P.P.; Hu, C.K.; Pech, M.F.; Valenzano, D.R.; Zhang, E.; Sharp, S.C.; Artandi, S.E.; et al. A platform for rapid exploration of aging and diseases in a naturally short-lived vertebrate. Cell 2015, 160, 1013–1026. [Google Scholar] [CrossRef] [PubMed]
- Matsui, H.; Kenmochi, N.; Namikawa, K. Age- and α-Synuclein-Dependent Degeneration of Dopamine and Noradrenaline Neurons in the Annual Killifish Nothobranchius furzeri. Cell Rep. 2019, 26, 1727–1733.e6. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.; Elsabbagh, M.; Baird, G.; Veenstra-Vanderweele, J. Autism spectrum disorder. Lancet 2018, 392, 508–520. [Google Scholar] [CrossRef]
- Lasky-Su, J.; Neale, B.M.; Franke, B.; Anney, R.J.; Zhou, K.; Maller, J.B.; Vasquez, A.A.; Chen, W.; Asherson, P.; Buitelaar, J.; et al. Genome-wide association scan of quantitative traits for attention deficit hyperactivity disorder identifies novel associations and confirms candidate gene associations. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2008, 147B, 1345–1354. [Google Scholar] [CrossRef]
- Hawi, Z.; Cummins, T.D.; Tong, J.; Johnson, B.; Lau, R.; Samarrai, W.; Bellgrove, M.A. The molecular genetic architecture of attention deficit hyperactivity disorder. Mol. Psychiatry 2015, 20, 289–297. [Google Scholar] [CrossRef]
- Demontis, D.; Walters, R.K.; Martin, J.; Mattheisen, M.; Als, T.D.; Agerbo, E.; Baldursson, G.; Belliveau, R.; Bybjerg-Grauholm, J.; Bækvad-Hansen, M.; et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 2019, 51, 63–75. [Google Scholar] [CrossRef]
- Kim, O.H.; Cho, H.J.; Han, E.; Hong, T.I.; Ariyasiri, K.; Choi, J.H.; Hwang, K.S.; Jeong, Y.M.; Yang, S.Y.; Yu, K.; et al. Zebrafish knockout of Down syndrome gene, DYRK1A, shows social impairments relevant to autism. Mol. Autism. 2017, 8, 50. [Google Scholar] [CrossRef]
- Liu, C.X.; Li, C.Y.; Hu, C.C.; Wang, Y.; Lin, J.; Jiang, Y.H.; Li, Q.; Xu, X. CRISPR/Cas9-induced shank3b mutant zebrafish display autism-like behaviors. Mol. Autism. 2018, 9, 23. [Google Scholar] [CrossRef]
- Koh, A.; Tao, S.; Jing Goh, Y.; Chaganty, V.; See, K.; Purushothaman, K.; Orbán, L.; Mathuru, A.S.; Wohland, T.; Winkler, C. A Neurexin2aa deficiency results in axon pathfinding defects and increased anxiety in zebrafish. Hum. Mol. Genet. 2021, 29, 3765–3780. [Google Scholar] [CrossRef]
- Hoffman, E.J.; Turner, K.J.; Fernandez, J.M.; Cifuentes, D.; Ghosh, M.; Ijaz, S.; Jain, R.A.; Kubo, F.; Bill, B.R.; Baier, H.; et al. Estrogens Suppress a Behavioral Phenotype in Zebrafish Mutants of the Autism Risk Gene, CNTNAP2. Neuron 2016, 89, 725–733. [Google Scholar] [CrossRef]
- Huang, J.; Zhong, Z.; Wang, M.; Chen, X.; Tan, Y.; Zhang, S.; He, W.; He, X.; Huang, G.; Lu, H.; et al. Circadian Modulation of Dopamine Levels and Dopaminergic Neuron Development Contributes to Attention Deficiency and Hyperactive Behavior. J. Neurosci. 2015, 35, 2572–2587. [Google Scholar] [CrossRef] [PubMed]
- Dark, C.; Williams, C.; Bellgrove, M.A.; Hawi, Z.; Bryson-Richardson, R.J. Functional validation of CHMP7 as an ADHD risk gene. Transl. Psychiatry 2020, 10, 385. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, Y.; Nonaka, Y.; Goto, T.; Ohnishi, E.; Hiramatsu, T.; Kii, I.; Yoshida, M.; Ikura, T.; Onogi, H.; Shibuya, H.; et al. Development of a novel selective inhibitor of the Down syndrome-related kinase DYRK1A. Nat. Commun. 2010, 1, 86. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, P.; Feng, G. SHANK proteins: Roles at the synapse and in autism spectrum disorder. Nat. Rev. Neurosci. 2017, 18, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Südhof, T.C. Neuroligins and neurexins link synaptic function to cognitive disease. Nature 2008, 455, 903–911. [Google Scholar] [CrossRef] [PubMed]
- Born, G.; Grayton, H.M.; Langhorst, H.; Dudanova, I.; Rohlmann, A.; Woodward, B.W.; Collier, D.A.; Fernandes, C.; Missler, M. Genetic targeting of NRXN2 in mice unveils role in excitatory cortical synapse function and social behaviors. Front. Synaptic Neurosci. 2015, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Kalueff, A.V.; Gebhardt, M.; Stewart, A.M.; Cachat, J.M.; Brimmer, M.; Chawla, J.S.; Craddock, C.; Kyzar, E.J.; Roth, A.; Landsman, S.; et al. Towards a comprehensive catalog of zebrafish behavior 1.0 and beyond. Zebrafish 2013, 10, 70–86. [Google Scholar] [CrossRef]
- Vaz, R.; Hofmeister, W.; Lindstrand, A. Zebrafish Models of Neurodevelopmental Disorders: Limitations and Benefits of Current Tools and Techniques. Int. J. Mol. Sci. 2019, 20, 1296. [Google Scholar] [CrossRef]
- Lachowicz, J.; Niedziałek, K.; Rostkowska, E.; Szopa, A.; Świąder, K.; Szponar, J.; Serefko, A. Zebrafish as an Animal Model for Testing Agents with Antidepressant Potential. Life 2021, 11, 792. [Google Scholar] [CrossRef]
- de Abreu, M.S.; Giacomini, A.C.V.V.; Genario, R.; Fontana, B.D.; Parker, M.O.; Marcon, L.; Scolari, N.; Bueno, B.; Demin, K.A.; Galstyan, D.; et al. Zebrafish models of impulsivity and impulse control disorders. Eur. J. Neurosci. 2020, 52, 4233–4248. [Google Scholar] [CrossRef]
- Alsop, D.; Vijayan, M.M. Development of the corticosteroid stress axis and receptor expression in zebrafish. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008, 294, R711–R719. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, J.M.; Feist, G.W.; Varga, Z.M.; Westerfield, M.; Kent, M.L.; Schreck, C.B. Whole-body cortisol response of zebrafish to acute net handling stress. Aquaculture 2009, 297, 157–162. [Google Scholar] [CrossRef] [PubMed]
- De Abreu, M.S.; Demin, K.A.; Giacomini, A.C.V.V.; Amstislavskaya, T.G.; Strekalova, T.; Maslov, G.O.; Kositsin, Y.; Petersen, E.V.; Kalueff, A.V. Understanding how stress responses and stress-related behaviors have evolved in zebrafish and mammals. Neurobiol. Stress 2021, 15, 100405. [Google Scholar] [CrossRef] [PubMed]
- Langova, V.; Vales, K.; Horka, P.; Horacek, J. The Role of Zebrafish and Laboratory Rodents in Schizophrenia Research. Front. Psychiatry 2020, 11, 703. [Google Scholar] [CrossRef] [PubMed]
- de Abreu, M.S.; Friend, A.J.; Demin, K.A.; Amstislavskaya, T.G.; Bao, W.; Kalueff, A.V. Zebrafish models: Do we have valid paradigms for depression? J. Pharmacol. Toxicol. Methods 2018, 94 Pt 2, 16–22. [Google Scholar] [CrossRef]
- Thyme, S.B.; Pieper, L.M.; Li, E.H.; Pandey, S.; Wang, Y.; Morris, N.S.; Sha, C.; Choi, J.W.; Herrera, K.J.; Soucy, E.R.; et al. Phenotypic Landscape of Schizophrenia-Associated Genes Defines Candidates and Their Shared Functions. Cell 2019, 177, 478–491.e20. [Google Scholar] [CrossRef]
- Demin, K.A.; Meshalkina, D.A.; Volgin, A.D.; Yakovlev, O.V.; de Abreu, M.S.; Alekseeva, P.A.; Friend, A.J.; Lakstygal, A.M.; Zabegalov, K.; Amstislavskaya, T.G.; et al. Developing zebrafish experimental animal models relevant to schizophrenia. Neurosci. Biobehav. Rev. 2019, 105, 126–133. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).


