Aptamers—Diagnostic and Therapeutic Solution in SARS-CoV-2
Abstract
1. Introduction
1.1. Characteristics of Coronavirus SARS-CoV-2
1.1.1. Transmission
1.1.2. Symptoms
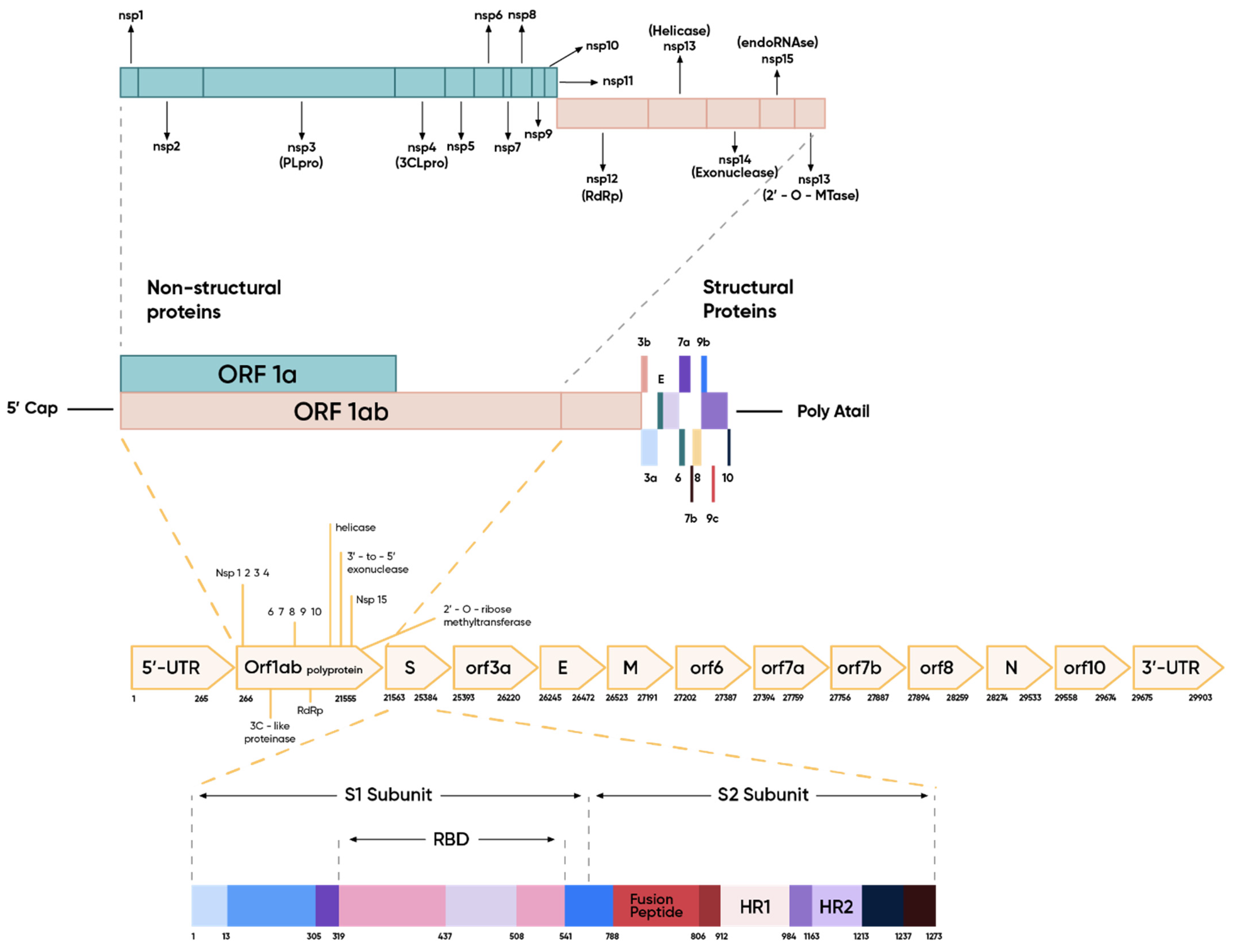
1.1.3. Structure of Genome and Virion
The Spike Protein and Initiation of the Infection Cycle
Other Structural Proteins
1.2. Aptamers
1.2.1. Characteristics
1.2.2. Advantages over Monoclonal Antibodies
2. Diagnostics Tools in SARS-CoV-2
2.1. Conventional Diagnostics Methods for the SARS-CoV-2 Infection
2.1.1. Polymerase Chain Reaction
2.1.2. Serological Tests
2.2. Aptamers as Diagnostics Tool in SARS-CoV-2
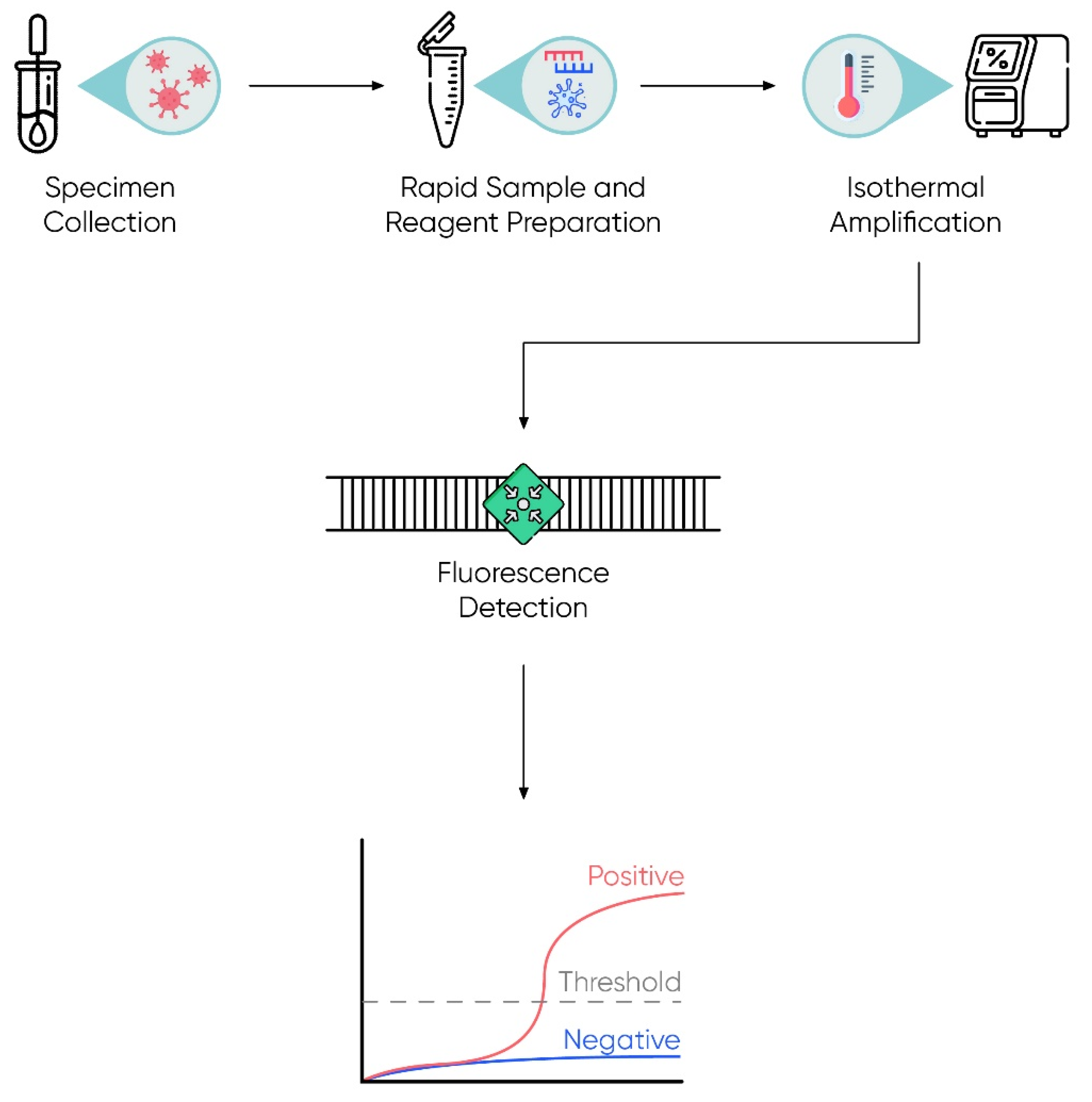
2.2.1. Isothermal Detection
2.2.2. Repurposing of SARS-CoV Aptamers
2.2.3. Aptamers Dedicated to SARS-CoV-2
Aptamers and Aptasensors
3. Aptamers in SARS-CoV-2 Infection Therapy
3.1. Repurposing of SARS-CoV and Other Aptamers
3.2. SARS-CoV-2 Aptamers
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Acquah, C.; Jeevanandam, J.; Tan, K.X.; Danquah, M.K. Engineered Aptamers for Enhanced COVID-19 Theranostics. Cell. Mol. Bioeng. 2021, 14, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.-W.; Yuan, S.; Kok, K.-H.; To, K.K.-W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.-Y.; Poon, R.W.-S.; et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef]
- World Health Organization. Coronavirus Disease (COVID-19) Pandemic. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 21 April 2020).
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef]
- World Health Organization. Virtual Press Conference on COVID-19, 11 March 2020. Available online: https://www.who.int/docs/default-source/%0Acoronaviruse/transcripts/who-audioemergencies-coronavirus-press-conference-fulland-fnal-11mar2020.pdf?sfvrsn=cb432bb3_2 (accessed on 11 March 2020).
- Our World in Data. COVID-19 Data Explorer. Available online: https://ourworldindata.org/explorers/coronavirus-data-explorer (accessed on 2 January 2022).
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The Proximal Origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef]
- Gussow, A.B.; Auslander, N.; Faure, G.; Wolf, Y.I.; Zhang, F.; Koonin, E.V. Genomic Determinants of Pathogenicity in SARS-CoV-2 and Other Human Coronaviruses. Proc. Natl. Acad. Sci. USA 2020, 117, 15193–15199. [Google Scholar] [CrossRef]
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.W.; Müller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.A.; Zaki, A.; Fouchier, R.A.M.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef]
- Lai, C.-C.; Liu, Y.H.; Wang, C.-Y.; Wang, Y.-H.; Hsueh, S.-C.; Yen, M.-Y.; Ko, W.-C.; Hsueh, P.-R. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): Facts and myths. J. Microbiol. Immunol. Infect. 2020, 53, 404–412. [Google Scholar] [CrossRef]
- Bai, Y.; Yao, L.; Wei, T.; Tian, F.; Jin, D.Y.; Chen, L.; Wang, M. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA 2020, 323, 1406–1407. [Google Scholar] [CrossRef]
- Liu, Y.-C.; Liao, C.-H.; Chang, C.-F.; Chou, C.-C.; Lin, Y.-R. A Locally Transmitted Case of SARS-CoV-2 Infection in Taiwan. N. Engl. J. Med. 2020, 382, 1070–1072. [Google Scholar] [CrossRef]
- Rothe, C.; Schunk, M.; Sothmann, P.; Bretzel, G.; Froeschl, G.; Wallrauch, C.; Zimmer, T.; Thiel, V.; Janke, C.; Guggemos, W.; et al. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N. Engl. J. Med. 2020, 382, 970–971. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Thilakavathy, K.; Kumar, S.S.; He, G.; Liu, S.V. Potential Factors Influencing Repeated SARS Outbreaks in China. Int. J. Environ. Res. Public Health 2020, 17, 1633. [Google Scholar] [CrossRef] [PubMed]
- Kolifarhood, G.; Aghaali, M.; Saadati, H.M.; Taherpour, N.; Rahimi, S.; Izadi, N.; Nazari, S.S.H. Epidemiological and Clinical Aspects of COVID-19: A Narrative Review. Arch. Acad. Emerg. Med. 2020, 8, e41. [Google Scholar] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Gupta, A.; Madhavan, M.V.; Sehgal, K.; Nair, N.; Mahajan, S.; Sehrawat, T.S.; Bikdeli, B.; Ahluwalia, N.; Ausiello, J.C.; Wan, E.Y.; et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020, 26, 1017–1032. [Google Scholar] [CrossRef]
- Colafrancesco, S.; Scrivo, R.; Barbati, C.; Conti, F.; Priori, R. Targeting the Immune System for Pulmonary Inflammation and Cardiovascular Complications in COVID-19 Patients. Front. Immunol. 2020, 11, 1439. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904.e9. [Google Scholar] [CrossRef]
- Di Micco, P.; Camporese, G.; Russo, V.; Cardillo, G.; Imbalzano, E.; Tufano, A.; Bernardi, E.; Fontanella, A. Clinical Differences between COVID-19 and a COVID-Like Syndrome. J. Clin. Med. 2021, 10, 2519. [Google Scholar] [CrossRef]
- Tan, W.; Xhao, Z.; Ma, X.; Wang, W.; Niu, P.; Xu, W.; Gao, G.F.; Wu, G. A novel corona virus genome identified in a cluster of pneumonia cases—Wuhan, China 2019–2020. China CDC Wkly. 2020, 2, 61–62. [Google Scholar] [CrossRef]
- John Hopkins University. Racial Data Transparency. Available online: https://coronavirus.jhu.edu/data/racial-data-transparency (accessed on 21 July 2021).
- Lai, C.C.; Shih, T.P.; Ko, W.C.; Tang, H.J.; Hsueh, P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents 2020, 55, 105924. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef]
- Chauhan, S. Comprehensive review of coronavirus disease 2019 (COVID-19). Biomed. J. 2020, 43, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Anjorin, A.A. The coronavirus disease 2019 (COVID-19) pandemic: A review and an update on cases in Africa. Asian Pac. J. Trop. Med. 2020, 13, 199. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef]
- Chan, J.F.-W.; Kok, K.-H.; Zhu, Z.; Chu, H.; To, K.K.-W.; Yuan, S.; Yuen, K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef]
- De Haan, C.A.M.; Kuo, L.; Masters, P.S.; Vennema, H.; Rottier, P.J.M. Coronavirus Particle Assembly: Primary Structure Requirements of the Membrane Protein. J. Virol. 1998, 72, 6838–6850. [Google Scholar] [CrossRef]
- Pandey, P.; Rane, J.S.; Chatterjee, A.; Kumar, A.; Khan, R.; Prakash, A.; Ray, S. Targeting SARS-CoV-2 Spike Protein of COVID-19 with Naturally Occurring Phytochemicals: An in silico Study for Drug Development. J. Biomol. Struct. Dyn. 2020, 39, 6306–6316. [Google Scholar] [CrossRef]
- Bar-On, Y.M.; Flamholz, A.; Phillips, R.; Milo, R. SARS-CoV-2 (COVID-19) by the numbers. Elife 2020, 9, e57309. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef]
- Rajarshi, K.; Chatterjee, A.; Ray, S. Combating COVID-19 with mesenchymal stem cell therapy. Biotechnol. Rep. 2020, 26, e00467. [Google Scholar] [CrossRef] [PubMed]
- Rajarshi, K.; Chatterjee, A.; Ray, S. BCG vaccination strategy implemented to reduce the impact of COVID-19: Hype or hope? Med. Drug Discov. 2020, 7, 100049. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Huang, Y.; Lau, S.K.P.; Yuen, K.-Y. Coronavirus Genomics and Bioinformatics Analysis. Viruses 2010, 2, 1803–1820. [Google Scholar] [CrossRef] [PubMed]
- Khailany, R.A.; Safdar, M.; Ozaslan, M. Genomic Characterization of a Novel SARS-CoV-2. Gene Rep. 2020, 19, 100682. [Google Scholar] [CrossRef]
- Astuti, I.; Ysrafil. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An Overview of Viral Structure and Host Response. Diabetes Metab. Syndr. 2020, 14, 407–412. [Google Scholar] [CrossRef]
- Ahmed, S.F.; Quadeer, A.A.; McKay, M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses 2020, 12, 254. [Google Scholar] [CrossRef]
- Beck, B.R.; Shin, B.; Choi, Y.; Park, S.; Kang, K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug target interaction deep learning model. Comput. Struct. Biotechnol. J. 2020, 18, 784–790. [Google Scholar] [CrossRef]
- Van Boheemen, S.; de Graaf, M.; Lauber, C.; Bestebroer, T.M.; Raj, V.S.; Zaki, A.M.; Osterhaus, A.D.M.E.; Haagmans, B.L.; Gorbalenya, A.E.; Snijder, E.J.; et al. Genomic Characterization of a Newly Discovered Coronavirus Associated with Acute Respiratory Distress Syndrome in Humans. mBio 2012, 3, e00473. [Google Scholar] [CrossRef]
- Yang, D.; Leibowitz, J.L. The structure and functions of coronavirus genomic 3′ and 5′ ends. Virus Res. 2015, 206, 120–133. [Google Scholar] [CrossRef]
- Albini, A.; di Guardo, G.; Noonan, D.M.; Lombardo, M. The SARS-CoV-2 Receptor, ACE-2, Is Expressed on Many Different Cell Types: Implications for ACE-Inhibitor and Angiotensin II Receptor Blocker-Based Cardiovascular Therapies. Int. Emerg. Med. 2020, 15, 759. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Petitjean, S.J.L.; Koehler, M.; Zhang, Q.; Dumitru, A.C.; Chen, W.; Derclaye, S.; Vincent, S.P.; Soumillion, P.; Alsteens, D. Molecular Interaction and Inhibition of SARS-CoV-2 Binding to the ACE2 Receptor. Nat. Commun. 2020, 11, 4541. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.K.; Dorosky, D.; Sharma, P.; Abbasi, S.A.; Dye, J.M.; Kranz, D.M.; Herbert, A.S.; Procko, E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science 2020, 369, 1261–1265. [Google Scholar] [CrossRef] [PubMed]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Yan, R.H.; Zhang, Y.Y.; Li, Y.; Xia, L.; Guo, Y.Y.; Zhou, Q. A novel screening strategy of anti-SARS-CoV-2 drugs via blocking interaction between Spike RBD and ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Forster, P.; Forster, L.; Renfrew, C.; Forster, M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. USA 2020, 117, 9241–9243. [Google Scholar] [CrossRef]
- Pramanik, A.; Gao, Y.; Patibandla, S.; Mitra, D.; McCandless, M.G.; Fassero, L.A.; Gates, K.; Tandon, R.; Ray, P.C. Aptamer Conjugated Gold Nanostar-Based Distance-Dependent Nanoparticle Surface Energy Transfer Spectroscopy for Ultrasensitive Detection and Inactivation of Corona Virus. J. Phys. Chem. Lett. 2021, 12, 2166–2171. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef] [PubMed]
- Rane, J.S.; Pandey, P.; Chatterjee, A.; Khan, R.; Kumar, A.; Prakash, A.; Ray, S. Targeting virus–host interaction by novel pyrimidine derivative: An in silico approach towards discovery of potential drug against COVID-19. J. Biomol. Struct. Dyn. 2021, 39, 5768–5778. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Song, J.; Wei, X.; Huang, M.; Sun, M.; Zhu, L.; Lin, B.; Shen, H.; Zhu, Z.; Yang, C. Discovery of Aptamers Targeting the Receptor-Binding Domain of the SARS-CoV-2 Spike Glycoprotein. Anal. Chem. 2020, 92, 9895–9900. [Google Scholar] [CrossRef]
- Westerbeck, J.W.; Machamer, C.E. The Infectious Bronchitis Coronavirus Envelope Protein Alters Golgi pH To Protect the Spike Protein and Promote the Release of Infectious Virus. J. Virol. 2019, 93, e00015–e00019. [Google Scholar] [CrossRef]
- Rouka, E.; Hatzoglou, C.; Gourgoulianis, K.; Zarogiannis, S. Antibody epitope and mimotope prediction of the viroporins M2 of the Influenza A virus, E of the Human SARS coronavirus and SH of the Respiratory Syncytial virus. Eur. Respir. J. 2019, 54, 2387. [Google Scholar] [CrossRef]
- McBride, R.; van Zyl, M.; Fielding, B.C. The Coronavirus Nucleocapsid Is a Multifunctional Protein. Viruses 2014, 6, 2991–3018. [Google Scholar] [CrossRef]
- Van Nguyen, T.H.; Lichière, J.; Canard, B.; Papageorgiou, N.; Attoumani, S.; Ferron, F.; Coutard, B. Structure and oligomerization state of the C-terminal region of the Middle East respiratory syndrome coronavirus nucleoprotein. Acta Crystallogr. Sect. D Struct. Biol. 2019, 75, 8–15. [Google Scholar] [CrossRef]
- Surjit, M.; Liu, B.; Kumar, P.; Chow, V.T.; Lal, S.K. The nucleocapsid protein of the SARS coronavirus is capable of self-association through a C-terminal 209 amino acid interaction domain. Biochem. Biophys. Res. Commun. 2004, 317, 1030–1036. [Google Scholar] [CrossRef]
- Lin, S.-Y.; Liu, C.-L.; Chang, Y.-M.; Zhao, J.; Perlman, S.; Hou, M.-H. Structural Basis for the Identification of the N-Terminal Domain of Coronavirus Nucleocapsid Protein as an Antiviral Target. J. Med. Chem. 2014, 57, 2247–2257. [Google Scholar] [CrossRef]
- Cui, L.; Wang, H.; Ji, Y.; Yang, J.; Xu, S.; Huang, X.; Wang, Z.; Qin, L.; Tien, P.; Zhou, X.; et al. The Nucleocapsid protein of coronaviruses acts as a Vidal suppressor of RNA silencing in mammalian cells. J. Virol. 2015, 89, 9029–9043. [Google Scholar] [CrossRef] [PubMed]
- Wandtke, T.; Woźniak, J.; Kopiński, P. Aptamers in Diagnostics and Treatment of Viral Infections. Viruses 2015, 7, 751–780. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Yang, G.; Ghulam, M.; Li, L.; Qu, F. Evolution of multi-functional capillary electrophoresis for high-efficiency selection of aptamers. Biotechnol. Adv. 2019, 37, 107432. [Google Scholar] [CrossRef]
- Abid, S.A.; Muneer, A.A.; Al-Kadmy, I.M.; Sattar, A.A.; Beshbishy, A.M.; Batiha, G.E.-S.; Hetta, H.F. Biosensors as a future diagnostic approach for COVID-19. Life Sci. 2021, 273, 119117. [Google Scholar] [CrossRef]
- Acquah, C.; Danquah, M.K.; Yon, J.L.; Sidhu, A.; Ongkudon, C.M. A review on immobilised aptamers for high throughput biomolecular detection and screening. Anal. Chim. Acta 2015, 888, 10–18. [Google Scholar] [CrossRef]
- Tan, S.Y.; Acquah, C.; Sidhu, A.; Ongkudon, C.M.; Yon, L.S.; Danquah, M.K. SELEX Modifications and Bioanalytical Techniques for Aptamer–Target Binding Characterization. Crit. Rev. Anal. Chem. 2016, 46, 521–537. [Google Scholar] [CrossRef]
- Li, H.-Y.; Jia, W.-N.; Li, X.-Y.; Zhang, L.; Liu, C.; Wu, J. Advances in detection of infectious agents by aptamer-based technologies. Emerg. Microbes Infect. 2020, 9, 1671–1681. [Google Scholar] [CrossRef]
- Wang, Y.; Ye, Z.-Z.; Si, C.-Y.; Ying, Y.-B. Application of Aptamer Based Biosensors for Detection of Pathogenic Microorganisms. Chin. J. Anal. Chem. 2012, 40, 634–642. [Google Scholar] [CrossRef]
- Han, K.; Liang, Z.; Zhou, N. Design Strategies for Aptamer-Based Biosensors. Sensors 2010, 10, 4541–4557. [Google Scholar] [CrossRef]
- Zou, X.; Wu, J.; Gu, J.; Shen, L.; Mao, L. Application of Aptamers in Virus Detection and Antiviral Therapy. Front. Microbiol. 2019, 10, 1462. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Chen, X. Aptamer-based targeted therapy. Adv. Drug Deliv. Rev. 2018, 134, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Blind, M.; Blank, M. Aptamer Selection Technology and Recent Advances. Mol. Ther. Nucleic Acids 2015, 4, e223. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Wang, R.; Hargis, B.; Lu, H.; Li, Y. A SPR Aptasensor for Detection of Avian Influenza Virus H5N1. Sensors 2012, 12, 12506–12518. [Google Scholar] [CrossRef] [PubMed]
- Robertson, D.L.; Joyce, G.F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 1990, 344, 467–468. [Google Scholar] [CrossRef]
- Darmostuk, M.; Rimpelova, S.; Gbelcova, H.; Ruml, T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol. Adv. 2015, 33, 1141–1161. [Google Scholar] [CrossRef]
- Low, S.Y.; Hill, J.E.; Peccia, J. DNA aptamers bind specifically and selectively to (1→3)-β-d-glucans. Biochem. Biophys. Res. Commun. 2009, 378, 701–705. [Google Scholar] [CrossRef][Green Version]
- Tan, S.Y.; Acquah, C.; Tan, S.Y.; Ongkudon, C.M.; Danquah, M.K. Characterisation of charge distribution and stability of aptamer-thrombin binding interaction. Process Biochem. 2017, 60, 42–51. [Google Scholar] [CrossRef]
- Wang, T.; Chen, C.; Larcher, L.M.; Barrero, R.A.; Veedu, R.N. Three decades of nucleic acid aptamer technologies: Lessons learned, progress and opportunities on aptamer development. Biotechnol. Adv. 2019, 37, 28–50. [Google Scholar] [CrossRef]
- Aspermair, P.; Mishyn, V.; Bintinger, J.; Happy, H.; Bagga, K.; Subramanian, P.; Knoll, W.; Boukherroub, R.; Szunerits, S. Reduced graphene oxide–based field effect transistors for the detection of E7 protein of human papillomavirus in saliva. Anal. Bioanal. Chem. 2020, 413, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Valencia-Reséndiz, D.G.; Palomino-Vizcaino, G.; Tapia-Vieyra, J.V.; Benítez-Hess, M.L.; Leija-Montoya, A.G.; Alvarez-Salas, L.M. Inhibition of Human Papillomavirus Type 16 Infection Using an RNA Aptamer. Nucleic Acid Ther. 2018, 28, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Yadavalli, T.; Agelidis, A.; Jaishankar, D.; Mangano, K.; Thakkar, N.; Penmetcha, K.; Shukla, D. Targeting Herpes Simplex Virus-1 gD by a DNA Aptamer Can Be an Effective New Strategy to Curb Viral Infection. Mol. Ther. Nucleic Acids 2017, 9, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.I.; Han, S.R.; Lee, S.-W. Development of RNA aptamer that inhibits methyltransferase activity of dengue virus. Biotechnol. Lett. 2018, 40, 315–324. [Google Scholar] [CrossRef]
- Tanaka, K.; Kasahara, Y.; Miyamoto, Y.; Takumi, O.; Kasai, T.; Onodera, K.; Kuwahara, M.; Oka, M.; Yoneda, Y.; Obika, S. Development of oligonucleotide-based antagonists of Ebola virus protein 24 inhibiting its interaction with karyopherin alpha 1. Org. Biomol. Chem. 2018, 16, 4456–4463. [Google Scholar] [CrossRef]
- Nguyen, P.D.M.; Zheng, J.; Gremminger, T.J.; Qiu, L.; Zhang, D.; Tuske, S.; Lange-Osborn, M.; Griffin, P.R.; Arnold, E.; Chen, S.-J.; et al. Binding interface and impact on protease cleavage for an RNA aptamer to HIV-1 reverse transcriptase. Nucleic Acids Res. 2020, 48, 2709–2722. [Google Scholar] [CrossRef]
- Virgilio, A.; Amato, T.; Petraccone, L.; Esposito, F.; Grandi, N.; Tramontano, E.; Romero, R.; Haider, S.; Gomez-Monterrey, I.; Novellino, E.; et al. Improvement of the activity of the anti-HIV-1 integrase aptamer T30175 by introducing a modified thymidine into the loops. Sci. Rep. 2018, 8, 7447. [Google Scholar] [CrossRef]
- Dearborn, A.D.; Eren, E.; Watts, N.R.; Palmer, I.W.; Kaufman, J.D.; Steven, A.C.; Wingfield, P.T. Structure of an RNA Aptamer that Can Inhibit HIV-1 by Blocking Rev-Cognate RNA (RRE) Binding and Rev-Rev Association. Structure 2018, 26, 1187–1195.e4. [Google Scholar] [CrossRef]
- Zheng, H.; Lang, Y.; Yu, J.; Han, Z.; Chen, B.; Wang, Y. Affinity binding of aptamers to agarose with DNA tetrahedron for removal of hepatitis B virus surface antigen. Colloids Surf. B Biointerfaces 2019, 178, 80–86. [Google Scholar] [CrossRef]
- Trausch, J.J.; Shank-Retzlaff, M.; Verch, T. Development and Characterization of an HPV Type-16 Specific Modified DNA Aptamer for the Improvement of Potency Assays. Anal. Chem. 2017, 89, 3554–3561. [Google Scholar] [CrossRef]
- Ghanbari, K.; Roushani, M.; Azadbakht, A. Ultra-sensitive aptasensor based on a GQD nanocomposite for detection of hepatitis C virus core antigen. Anal. Biochem. 2017, 534, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Pleshakova, T.O.; Kaysheva, A.L.; Bayzyanova, J.M.; Anashkina, A.S.; Uchaikin, V.F.; Ziborov, V.S.; Konev, V.A.; Archakov, A.I.; Ivanov, Y.D. The detection of hepatitis c virus core antigen using afm chips with immobolized aptamers. J. Virol. Methods 2018, 251, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Pleshakova, T.O.; Kaysheva, A.L.; Shumov, I.D.; Ziborov, V.S.; Bayzyanova, J.M.; Konev, V.A.; Uchaikin, V.F.; Archakov, A.I.; Ivanov, Y.D. Detection of Hepatitis C Virus Core Protein in Serum Using Aptamer-Functionalized AFM Chips. Micromachines 2019, 10, 129. [Google Scholar] [CrossRef] [PubMed]
- Dolai, S.; Tabib-Azar, M. Whole virus detection using aptamers and paper-based sensor potentiometry. Med. Devices Sens. 2020, 3, e10112. [Google Scholar] [CrossRef]
- Lee, K.H.; Zeng, H. Aptamer-Based ELISA Assay for Highly Specific and Sensitive Detection of Zika NS1 Protein. Anal. Chem. 2017, 89, 12743–12748. [Google Scholar] [CrossRef]
- Basso, C.R.; Crulhas, B.P.; Magro, M.; Vianello, F.; Pedrosa, V.A. A new immunoassay of hybrid nanomater conjugated to aptamers for the detection of dengue virus. Talanta 2019, 197, 482–490. [Google Scholar] [CrossRef]
- Kim, B.; Chung, K.W.; Lee, J.H. Non-stop aptasensor capable of rapidly monitoring norovirus in a sample. J. Pharm. Biomed. Anal. 2018, 152, 315–321. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, Z.; Yin, Y.; Jia, F.; Wu, Q.; Tian, P.; Wang, D. Development and evaluation of a novel in situ target-capture approach for aptamer selection of human noroviruses. Talanta 2019, 193, 199–205. [Google Scholar] [CrossRef]
- Chand, R.; Neethirajan, S. Microfluidic platform integrated with graphene-gold nano-composite aptasensor for one-step detection of norovirus. Biosens. Bioelectron. 2017, 98, 47–53. [Google Scholar] [CrossRef]
- Wang, C.-H.; Chang, C.-P.; Lee, G.-B. Integrated microfluidic device using a single universal aptamer to detect multiple types of influenza viruses. Biosens. Bioelectron. 2016, 86, 247–254. [Google Scholar] [CrossRef]
- Tseng, Y.-T.; Wang, C.-H.; Chang, C.-P.; Lee, G.-B. Integrated microfluidic system for rapid detection of influenza H1N1 virus using a sandwich-based aptamer assay. Biosens. Bioelectron. 2016, 82, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Bai, C.; Lu, Z.; Jiang, H.; Yang, Z.; Liu, X.; Ding, H.; Li, H.; Dong, J.; Huang, A.; Fang, T.; et al. Aptamer selection and application in multivalent binding-based electrical impedance detection of inactivated H1N1 virus. Biosens. Bioelectron. 2018, 110, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, J.; Chaudhary, N.; Kim, H.; Jang, J. Subtyping of influenza A H1N1 virus using a label-free electrochemical biosensor based on the DNA aptamer targeting the stem region of HA protein. Anal. Chim. Acta 2019, 1064, 94–103. [Google Scholar] [CrossRef]
- Chen, C.; Zou, Z.; Chen, L.; Ji, X.; He, Z. Functionalized magnetic microparticle-based colorimetric platform for influenza a virus detection. Nanotechnology 2016, 27, 435102. [Google Scholar] [CrossRef] [PubMed]
- Le, T.T.; Chang, P.; Benton, D.J.; McCauley, J.W.; Iqbal, M.; Cass, A.E.G. Dual Recognition Element Lateral Flow Assay toward Multiplex Strain Specific Influenza Virus Detection. Anal. Chem. 2017, 89, 6781–6786. [Google Scholar] [CrossRef]
- Shubham, S.; Hoinka, J.; Banerjee, S.; Swanson, E.; Dillard, J.A.; Lennemann, N.J.; Przytycka, T.M.; Maury, W.; Nilsen-Hamilton, M. A 2′FY-RNA Motif Defines an Aptamer for Ebolavirus Secreted Protein. Sci. Rep. 2018, 8, 12373. [Google Scholar] [CrossRef]
- Caglayan, M.O.; Ustundag, Z. Spectrophotometric ellipsometry based Tat-protein RNA-aptasensor for HIV-1 diagnosis. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 227, 117748. [Google Scholar] [CrossRef]
- Huang, R.; Xi, Z.; Deng, Y.; He, N. Fluorescence based Aptasensors for the determination of hepatitis B virus e antigen. Sci. Rep. 2016, 6, 31103. [Google Scholar] [CrossRef]
- Bibby, K. Metagenomic identification of viral pathogens. Trends Biotechnol. 2013, 31, 275–279. [Google Scholar] [CrossRef]
- Actor, J.K. Introductory Immunology; Basic Concepts for Interdisciplinary Applications, 2nd ed.; Academic Press: London, UK, 2019; p. 194. ISBN 9780128165720. [Google Scholar]
- Pedersen, J.C. Hemagglutination-inhibition assay for influenza virus subtype identification and the detection and quantitation of serum antibodies to influenza virus, in Animal Influenza Virus. Methods Mol. Biol. 2014, 1161, 11–25. [Google Scholar] [CrossRef]
- Goldsmith, C.S.; Miller, S.E. Modern Uses of Electron Microscopy for Detection of Viruses. Clin. Microbiol. Rev. 2009, 22, 552–563. [Google Scholar] [CrossRef] [PubMed]
- Souf, S. Recent advances in diagnostic testing for viral infections. Biosci. Horizons Int. J. Stud. Res. 2016, 9. [Google Scholar] [CrossRef]
- Storch, G.A. Diagnostic Virology. Clin. Infect. Dis. 2000, 31, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Zu, Z.Y.; di Jiang, M.; Xu, P.P.; Chen, W.; Ni, Q.Q.; Lu, G.M.; Zhang, L.J. Coronavirus Disease 2019 (COVID-19): A Perspective from China. Radiology 2020, 296, E15–E25. [Google Scholar] [CrossRef]
- Obande, G.A.; Singh, K.K.B. Current and Future Perspectives on Isothermal Nucleic Acid Amplification Technologies for Diagnosing Infections. Infect. Drug Resist. 2020, 13, 455–483. [Google Scholar] [CrossRef]
- Chinese Society of Radiology. Radiological diagnosis of new coronavirus infected pneumonitis: Expert recommendation from the Chinese Society of Radiology (First edition). Chin. J. Radiol. 2020, 54, E001. [Google Scholar]
- Shen, M.; Zhou, Y.; Ye, J.; Al-Maskri, A.A.A.; Kang, Y.; Zeng, S.; Cai, S. Recent advances and perspectives of nucleic acid detection for coronavirus. J. Pharm. Anal. 2020, 10, 97–101. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A. CRISPR−Cas12-Based Detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef]
- Bwire, G.M.; Majigo, M.V.; Njiro, B.J.; Mawazo, A. Detection profile of SARS-CoV-2 using RT-PCR in different types of clinical specimens: A systematic review and meta-analysis. J. Med. Virol. 2021, 93, 719–725. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Hahn, S.M. Coronavirus (COVID-19) Update: FDA Expedites Review of Diagnostic Tests to Combat COVID-19. 2020. Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-expedites-review-diagnostic-tests-combat-covid-19 (accessed on 27 July 2021).
- Khan, P.; Aufdembrink, L.M.; Engelhart, A.E. Isothermal SARS-CoV-2 Diagnostics: Tools for Enabling Distributed Pandemic Testing as a Means of Supporting Safe Reopenings. ACS Synth. Biol. 2020, 9, 2861–2880. [Google Scholar] [CrossRef] [PubMed]
- Krishna, V.D.; Wu, K.; Perez, A.M.; Wang, J.-P. Giant Magnetoresistance-based Biosensor for Detection of Influenza A Virus. Front. Microbiol. 2016, 7, 400. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Laboratory Testing for Coronavirus Disease 2019 (COVID-19) in Suspected Human Cases: Interim Guidance, 2 March 2020; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Nguyen, T.; Bang, D.D.; Wolff, A. 2019 Novel Coronavirus Disease (COVID-19): Paving the Road for Rapid Detection and Point-of-Care Diagnostics. Micromachines 2020, 11, 306. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Zhu, J.; Zhang, Z.; Han, Y. A Familial Cluster of Infection Associated With the 2019 Novel Coronavirus Indicating Possible Person-to-Person Transmission During the Incubation Period. J. Infect. Dis. 2020, 221, 1757–1761. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wu, Q.; Chen, J.; Ni, X.; Dai, J. A DNA Aptamer Based Method for Detection of SARS-CoV-2 Nucleocapsid Protein. Virol. Sin. 2020, 35, 351–354. [Google Scholar] [CrossRef] [PubMed]
- Kucirka, L.M.; Lauer, S.A.; Laeyendecker, O.; Boon, D.; Lessler, J. Variation in False-Negative Rate of Reverse Transcriptase Polymerase Chain Reaction–Based SARS-CoV-2 Tests by Time Since Exposure. Ann. Intern. Med. 2020, 173, 262–267. [Google Scholar] [CrossRef]
- Service, R.F. Fast, cheap tests could enable safer reopening. Science 2020, 369, 608–609. [Google Scholar] [CrossRef]
- Hindson, J. COVID-19: Faecal–oral transmission? Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 259. [Google Scholar] [CrossRef]
- To, K.K.-W.; Tsang, O.T.-Y.; Yip, C.C.-Y.; Chan, K.-H.; Wu, T.-C.; Chan, J.M.-C.; Leung, W.-S.; Chik, T.S.-H.; Choi, C.Y.-C.; Kandamby, D.H.; et al. Consistent Detection of 2019 Novel Coronavirus in Saliva. Clin. Infect. Dis. 2020, 71, 841–843. [Google Scholar] [CrossRef]
- Zhang, G.; Guo, J.; Wang, X. Immunochromatographic Lateral Flow Strip Tests. Methods Mol. Biol. 2009, 504, 169–183. [Google Scholar] [CrossRef] [PubMed]
- Tomita, M.; Tsumoto, K. Hybridoma technologies for antibody production. Immunotherapy 2011, 3, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.Y.-P.; Lin, R.T.P.; Renia, L.; Ng, L.F.P. Serological Approaches for COVID-19: Epidemiologic Perspective on Surveillance and Control. Front. Immunol. 2020, 11, 879. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.-S.; Chiu, S.-C.; Tseng, T.-C.; Lin, S.-F.; Lin, J.-H.; Hsu, Y.-F.; Wang, M.-C.; Lin, T.-L.; Yang, W.-Z.; Ferng, T.-L.; et al. Serologic and Molecular Biologic Methods for SARS-associated Coronavirus Infection, Taiwan. Emerg. Infect. Dis. 2004, 10, 305–310. [Google Scholar] [CrossRef]
- Haveri, A.; Smura, T.; Kuivanen, S.; Österlund, P.; Hepojoki, J.; Ikonen, N.; Pitkäpaasi, M.; Blomqvist, S.; Rönkkö, E.; Kantele, A.; et al. Serological and molecular findings during SARS-CoV-2 infection: The first case study in Finland, January to February 2020. Eurosurveillance 2020, 25, 2000266. [Google Scholar] [CrossRef]
- Udugama, B.; Kadhiresan, P.; Kozlowski, H.N.; Malekjahani, A.; Osborne, M.; Li, V.Y.C.; Chen, H.; Mubareka, S.; Gubbay, J.B.; Chan, W.C.W. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 2020, 14, 3822–3835. [Google Scholar] [CrossRef]
- Serologic Testing for IgG Antibodies Against SARS-CoV-2-Insights. Available online: https://news.mayocliniclabs.com/2020/06/08/serologictesting-for-igg-antibodies-against-sars-cov-2/ (accessed on 19 July 2021).
- World Health Organization. Advice on the Use of Point-of-Care Immunodiagnostic Tests for COVID-19. Scientific Brief. Available online: https://www.who.int/news-room/commentaries/detail/advice-on-the-use-of-point-of-care-immunodiagnostic-tests-for-covid-19 (accessed on 20 December 2021).
- Weitzel, T.; Legarraga, P.; Iruretagoyena, M.; Pizarro, G.; Vollrath, V.; Araos, R.; Munita, J.M.; Porte, L. Comparative evaluation of four rapid SARS-CoV-2 antigen detection tests using universal transport medium. Travel Med. Infect. Dis. 2021, 39, 101942. [Google Scholar] [CrossRef]
- Mak, G.C.; Cheng, P.K.; Lau, S.S.; Wong, K.K.; Lau, C.S.; Lam, E.T.; Chan, R.C.W.; Tsang, D.N.C. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020, 129, 104500. [Google Scholar] [CrossRef]
- World Health Organization. Laboratory Testing Strategy Recommendations for COVID-19: Interim Guidance. Available online: https://apps.who.int/iris/handle/10665/331509 (accessed on 20 December 2021).
- Che, X.-Y.; Hao, W.; Wang, Y.; Di, B.; Yin, K.; Xu, Y.-C.; Feng, C.-S.; Wan, Z.-Y.; Cheng, V.C.; Yuen, K.-Y. Nucleocapsid Protein as Early Diagnostic Marker for SARS. Emerg. Infect. Dis. 2004, 10, 1947–1949. [Google Scholar] [CrossRef]
- Li, Y.-H.; Li, J.; Liu, X.-E.; Wang, L.; Li, T.; Zhou, Y.-H.; Zhuang, H. Detection of the nucleocapsid protein of severe acute respiratory syndrome coronavirus in serum: Comparison with results of other viral markers. J. Virol. Methods 2005, 130, 45–50. [Google Scholar] [CrossRef]
- Acquah, C.; Chan, Y.W.; Pan, S.; Yon, L.S.; Ongkudon, C.M.; Guo, H.-B.; Danquah, M.K. Characterisation of aptamer-anchored poly(EDMA-co-GMA) monolith for high throughput affinity binding. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Acquah, C.; Danquah, M.K.; Agyei, D.; Moy, C.K.S.; Sidhu, A.; Ongkudon, C.M. Deploying aptameric sensing technology for rapid pandemic monitoring. Crit. Rev. Biotechnol. 2016, 36, 1010–1022. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Fang, X.; Liu, X.; Ou, H.; Zhang, H.; Wang, J.; Li, Q.; Cheng, H.; Zhang, W.; Luo, Z. Discovery of sandwich type COVID-19 nucleocapsid protein DNA aptamers. Chem. Commun. 2020, 56, 10235–10238. [Google Scholar] [CrossRef] [PubMed]
- Lavania, S.; DAS, R.; Dhiman, A.; Myneedu, V.P.; Verma, A.; Singh, N.; Sharma, T.K.; Tyagi, J.S. Aptamer-Based TB Antigen Tests for the Rapid Diagnosis of Pulmonary Tuberculosis: Potential Utility in Screening for Tuberculosis. ACS Infect. Dis. 2018, 4, 1718–1726. [Google Scholar] [CrossRef]
- Ye, H.; Duan, N.; Gu, H.; Wang, H.; Wang, Z. Fluorometric determination of lipopolysaccharides via changes of the graphene oxide-enhanced fluorescence polarization caused by truncated aptamers. Mikrochim. Acta 2019, 186, 173. [Google Scholar] [CrossRef]
- Babamiri, B.; Salimi, A.; Hallaj, R. A molecularly imprinted electrochemiluminescence sensor for ultrasensitive HIV-1 gene detection using EuS nanocrystals as luminophore. Biosens. Bioelectron. 2018, 117, 332–339. [Google Scholar] [CrossRef]
- Weerathunge, P.; Ramanathan, R.; Torok, V.A.; Hodgson, K.; Xu, Y.; Goodacre, R.; Behera, B.K.; Bansal, V. Ultrasensitive Colorimetric Detection of Murine Norovirus Using NanoZyme Aptasensor. Anal. Chem. 2019, 91, 3270–3276. [Google Scholar] [CrossRef]
- Mudiyanselage, A.P.K.K.K.; Yu, Q.; Leon-Duque, M.A.; Zhao, B.; Wu, R.; You, M. Genetically Encoded Catalytic Hairpin Assembly for Sensitive RNA Imaging in Live Cells. J. Am. Chem. Soc. 2018, 140, 8739–8745. [Google Scholar] [CrossRef]
- Li, J.; Fu, H.-E.; Wu, L.-J.; Zheng, A.-X.; Chen, G.-N.; Yang, H.-H. General Colorimetric Detection of Proteins and Small Molecules Based on Cyclic Enzymatic Signal Amplification and Hairpin Aptamer Probe. Anal. Chem. 2012, 84, 5309–5315. [Google Scholar] [CrossRef]
- Ma, C.; Wu, K.; Zhao, H.; Liu, H.; Wang, K.; Xia, K. Fluorometric aptamer-based determination of ochratoxin A based on the use of graphene oxide and RNase H-aided amplification. Microchim. Acta 2018, 185, 347. [Google Scholar] [CrossRef]
- Abdolahzadeh, A.; Dolgosheina, E.V.; Unrau, P.J. RNA detection with high specificity and sensitivity using nested fluorogenic Mango NASBA. RNA 2019, 25, 1806–1813. [Google Scholar] [CrossRef] [PubMed]
- Aufdembrink, L.M.; Khan, P.; Gaut, N.J.; Adamala, K.P.; Engelhart, A.E. Highly specific, multiplexed isothermal pathogen detection with fluorescent aptamer readout. RNA 2020, 26, 1283–1290. [Google Scholar] [CrossRef] [PubMed]
- Science X Staff. Coronavirus Testing Kits to Be Developed using New RNA Imaging Technology. Available online: https://phys.org/news/2020-03-coronavirus-kits-rna-imaging-technology.html (accessed on 28 July 2021).
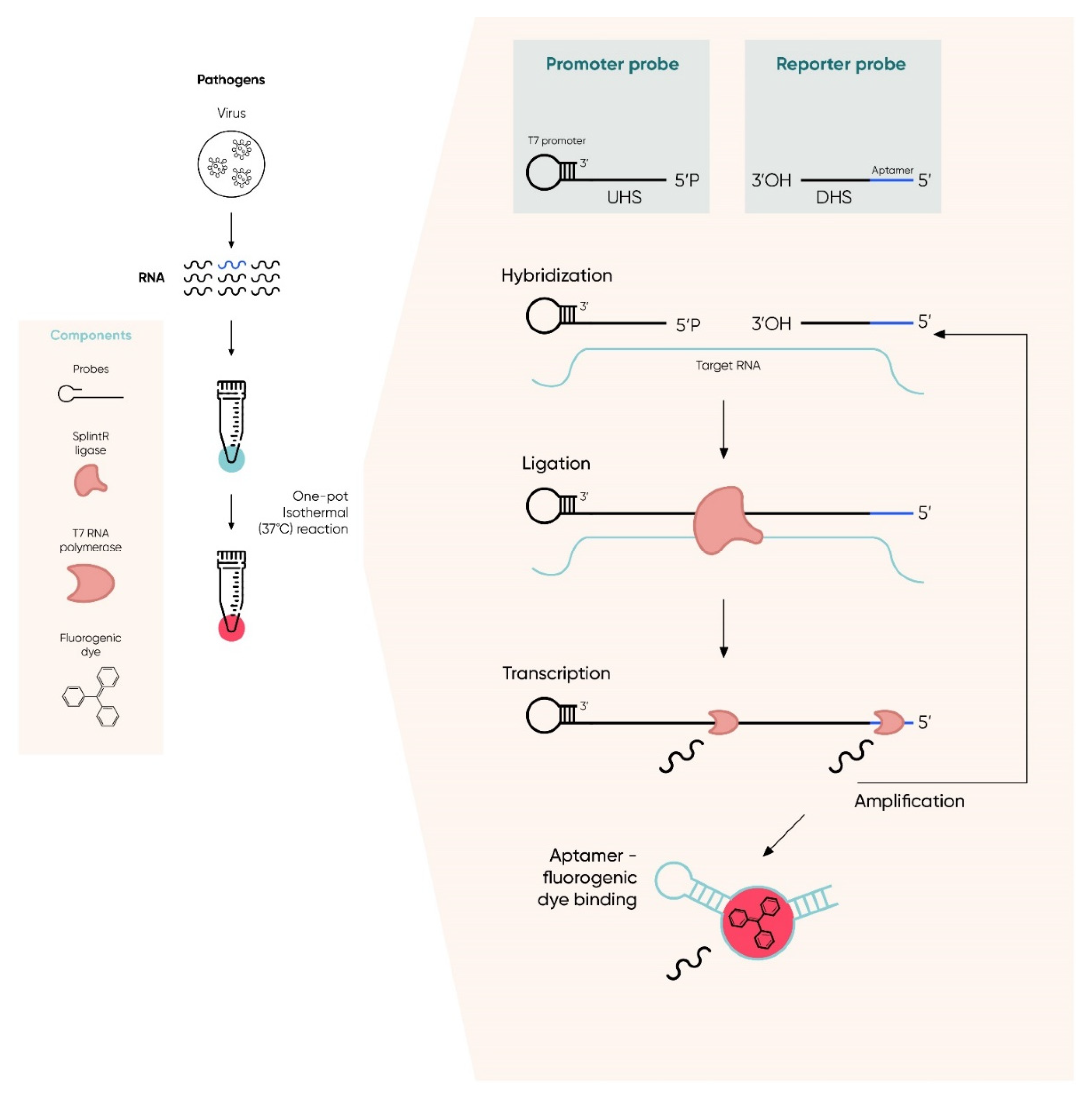
- Woo, C.H.; Jang, S.; Shin, G.; Jung, G.Y.; Lee, J.W. Sensitive fluorescence detection of SARS-CoV-2 RNA in clinical samples via one-pot isothermal ligation and transcription. Nat. Biomed. Eng. 2020, 4, 1168–1179. [Google Scholar] [CrossRef] [PubMed]
- Ahn, D.-G.; Jeon, I.-J.; Kim, J.D.; Song, M.-S.; Han, S.-R.; Lee, S.-W.; Jung, H.; Oh, J.-W. RNA aptamer-based sensitive detection of SARS coronavirus nucleocapsid protein. Analyst 2009, 134, 1896–1901. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.-J.; Woo, H.-M.; Kim, K.-S.; Oh, J.-W.; Jeong, Y.-J. Novel system for detecting SARS coronavirus nucleocapsid protein using an ssDNA aptamer. J. Biosci. Bioeng. 2011, 112, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.; Feng, J.; Zhang, J.; Hu, K.; Wang, D.; Zha, L.; Hu, X.; Li, R. Aptamer/antibody sandwich method for digital detection of SARS-CoV2 nucleocapsid protein. Talanta 2021, 236, 122847. [Google Scholar] [CrossRef]
- Jia, H.; Zhang, A.; Yang, Y.; Cui, Y.; Xu, J.; Jiang, H.; Tao, S.; Zhang, D.; Zeng, H.; Hou, Z.; et al. A graphene oxide coated tapered microfiber acting as a super-sensor for rapid detection of SARS-CoV-2. Lab A Chip 2021, 21, 2398–2406. [Google Scholar] [CrossRef]
- Perumal, V.; Hashim, U. Advances in biosensors: Principle, architecture and applications. J. Appl. Biomed. 2014, 12, 1–15. [Google Scholar] [CrossRef]
- Vidic, J.; Manzano, M.; Chang, C.-M.; Jaffrezic-Renault, N. Advanced biosensors for detection of pathogens related to livestock and poultry. Vet. Res. 2017, 48, 11. [Google Scholar] [CrossRef]
- Scott, K.; Yu, E.H. Microbial Electrochemical and Fuel Cells: Fundamentals and Applications; Woodhead Publishing: Cambridge, UK, 2019; p. 410. ISBN 9781782423966. [Google Scholar]
- Caygill, R.L.; Blair, G.E.; Millner, P.A. A review on viral biosensors to detect human pathogens. Anal. Chim. Acta 2010, 681, 8–15. [Google Scholar] [CrossRef]
- Arranz, A.; Ripoll, J. Advances in optical imaging for pharmacological studies. Front. Pharmacol. 2015, 6, 189. [Google Scholar] [CrossRef] [PubMed]
- Orooji, Y.; Sohrabi, H.; Hemmat, N.; Oroojalian, F.; Baradaran, B.; Mokhtarzadeh, A.; Mohaghegh, M.; Karimi-Maleh, H. An Overview on SARS-CoV-2 (COVID-19) and Other Human Coronaviruses and Their Detection Capability via Amplification Assay, Chemical Sensing, Biosensing, Immunosensing, and Clinical Assays. Nano-Micro Lett. 2021, 13, 18. [Google Scholar] [CrossRef] [PubMed]
- Pashchenko, O.; Shelby, T.; Banerjee, T.; Santra, S. A Comparison of Optical, Electrochemical, Magnetic, and Colorimetric Point-of-Care Biosensors for Infectious Disease Diagnosis. ACS Infect. Dis. 2018, 4, 1162–1178. [Google Scholar] [CrossRef] [PubMed]
- Zakashansky, J.A.; Imamura, A.H.; Salgado, D.F.; Romero Mercieca, H.C.; Aguas, R.F.L.; Lao, A.M.; Pariser, J.; Arroyo-Currás, N.; Khine, M. Detection of the SARS-CoV-2 spike protein in saliva with Shrinky-Dink© electrodes. Anal. Methods 2021, 13, 874–883. [Google Scholar] [CrossRef] [PubMed]
- Torres-Chavolla, E.; Alocilja, E.C. Aptasensors for detection of microbial and viral pathogens. Biosens. Bioelectron. 2009, 24, 3175–3182. [Google Scholar] [CrossRef]
- Yoo, H.; Jo, H.; Oh, S.S. Detection and beyond: Challenges and advances in aptamer-based biosensors. Mater. Adv. 2020, 1, 2663–2687. [Google Scholar] [CrossRef]
- Chen, H.; Park, S.-G.; Choi, N.; Kwon, H.-J.; Kang, T.; Lee, M.-K.; Choo, J. Sensitive Detection of SARS-CoV-2 Using a SERS-Based Aptasensor. ACS Sens. 2021, 6, 2378–2385. [Google Scholar] [CrossRef]
- Stanborough, T.; Given, F.M.; Koch, B.; Sheen, C.R.; Stowers-Hull, A.B.; Waterland, M.R.; Crittenden, D.L. Optical Detection of CoV-SARS-2 Viral Proteins to Sub-Picomolar Concentrations. ACS Omega 2021, 6, 6404–6413. [Google Scholar] [CrossRef]
- Li, M.; Wu, J.; Ma, M.; Feng, Z.; Mi, Z.; Rong, P.; Liu, D. Alkyne- and Nitrile-Anchored Gold Nanoparticles for Multiplex SERS Imaging of Biomarkers in Cancer Cells and Tissues. Nanotheranostics 2019, 3, 113–119. [Google Scholar] [CrossRef]
- Nishi, K.; Isobe, S.-I.; Zhu, Y.; Kiyama, R. Fluorescence-Based Bioassays for the Detection and Evaluation of Food Materials. Sensors 2015, 15, 25831–25867. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Qian, J.; Cai, F.; He, S. Raman reporter-coated gold nanorods and their applications in multimodal optical imaging of cancer cells. Anal. Bioanal. Chem. 2011, 400, 2793–2800. [Google Scholar] [CrossRef] [PubMed]
- Zavyalova, E.; Ambartsumyan, O.; Zhdanov, G.; Gribanyov, D.; Gushchin, V.; Tkachuk, A.; Rudakova, E.; Nikiforova, M.; Kuznetsova, N.; Popova, L.; et al. SERS-Based Aptasensor for Rapid Quantitative Detection of SARS-CoV-2. Nanomaterials 2021, 11, 1394. [Google Scholar] [CrossRef] [PubMed]
- Kacherovsky, N.; Yang, L.F.; Dang, H.V.; Cheng, E.L.; Cardle, I.I.; Walls, A.C.; McCallum, M.; Sellers, D.L.; DiMaio, F.; Salipante, S.J.; et al. Discovery and Characterization of Spike N-Terminal Domain-Binding Aptamers for Rapid SARS-CoV-2 Detection. Angew. Chem. Int. Ed. 2021, 60, 21211–21215. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Li, Z.; Mohammed, I.; Zhao, L.; Wei, W.; Xiao, H.; Guo, W.; Zhao, Y.; Qu, F.; Huang, Y. Identification of SARS-CoV-2-against aptamer with high neutralization activity by blocking the RBD domain of spike protein 1. Signal Transduct. Target. Ther. 2021, 6, 227. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Anand, A.; Jain, N.; Goswami, S.; Anantharaj, A.; Patil, S.; Singh, R.; Kumar, A.; Shrivastava, T.; Bhatnagar, S.; et al. A novel G-quadruplex aptamer-based spike trimeric antigen test for the detection of SARS-CoV-2. Mol. Ther. Nucleic Acids 2021, 26, 321–332. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Z.; Gu, J.; Stacey, H.D.; Ang, J.C.; Capretta, A.; Filipe, C.D.M.; Mossman, K.L.; Balion, C.; Salena, B.J.; et al. Diverse high-affinity DNA aptamers for wild-type and B.1.1.7 SARS-CoV-2 spike proteins from a pre-structured DNA library. Nucleic Acids Res. 2021, 49, 7267–7279. [Google Scholar] [CrossRef]
- Zhang, Z.; Pandey, R.; Li, J.; Gu, J.; White, D.; Stacey, H.D.; Ang, J.C.; Steinberg, C.-J.; Capretta, A.; Filipe, C.D.M.; et al. High-Affinity Dimeric Aptamers Enable the Rapid Electrochemical Detection of Wild-Type and B.1.1.7 SARS-CoV-2 in Unprocessed Saliva. Angew. Chem. Int. Ed. 2021, 60, 24266–24274. [Google Scholar] [CrossRef]
- Humeniuk, R.; Mathias, A.; Cao, H.; Osinusi, A.; Shen, G.; Chng, E.; Ling, J.; Vu, A.; German, P. Safety, Tolerability, and Pharmacokinetics of Remdesivir, An Antiviral for Treatment of COVID-19, in Healthy Subjects. Clin. Transl. Sci. 2020, 13, 896–906. [Google Scholar] [CrossRef]
- Weisshoff, H.; Krylova, O.; Nikolenko, H.; Düngen, H.-D.; Dallmann, A.; Becker, S.; Göttel, P.; Müller, J.; Haberland, A. Aptamer BC 007-Efficient binder of spreading-crucial SARS-CoV-2 proteins. Heliyon 2020, 6, e05421. [Google Scholar] [CrossRef]
- Sridharan, K.; Gogtay, N.J. Therapeutic nucleic acids: Current clinical status. Br. J. Clin. Pharmacol. 2016, 82, 659–672. [Google Scholar] [CrossRef]
- Asha, K.; Kumar, P.; Sanicas, M.; Meseko, C.A.; Khanna, M.; Kumar, B. Advancements in Nucleic Acid Based Therapeutics against Respiratory Viral Infections. J. Clin. Med. 2018, 8, 6. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-J.; Huang, H.-W.; Liu, C.-Y.; Hong, C.-F.; Chan, Y.-L. Inhibition of SARS-CoV replication by siRNA. Antivir. Res. 2005, 65, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Ismail, S.I.; Alshaer, W. Therapeutic aptamers in discovery, preclinical and clinical stages. Adv. Drug Deliv. Rev. 2018, 134, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, P.; Kurniawan, H.; Byrne, M.E.; Wower, J. Therapeutic RNA aptamers in clinical trials. Eur. J. Pharm. Sci. 2013, 48, 259–271. [Google Scholar] [CrossRef]
- Radom, F.; Jurek, P.M.; Mazurek, M.P.; Otlewski, J.; Jeleń, F. Aptamers: Molecules of great potential. Biotechnol. Adv. 2013, 31, 1260–1274. [Google Scholar] [CrossRef]
- Parashar, N.C.; Poddar, J.; Chakrabarti, S.; Parashar, G. Repurposing of SARS-CoV nucleocapsid protein specific nuclease resistant RNA aptamer for therapeutics against SARS-CoV-2. Infect Genet. Evol. 2020, 85, 104497. [Google Scholar] [CrossRef]
- Shum, K.T.; Tanner, J.A. Differential Inhibitory Activities and Stabilisation of DNA Aptamers against the SARS Coronavirus Helicase. ChemBioChem 2008, 9, 3037–3045. [Google Scholar] [CrossRef]
- Jang, K.J.; Lee, N.-R.; Yeo, W.-S.; Jeong, Y.-J.; Kim, D.-E. Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/Helicase. Biochem. Biophys. Res. Commun. 2008, 366, 738–744. [Google Scholar] [CrossRef]
- Adedeji, A.O.; Marchand, B.; Velthuis, A.J.W.T.; Snijder, E.J.; Weiss, S.; Eoff, R.L.; Singh, K.; Sarafianos, S.G. Mechanism of Nucleic Acid Unwinding by SARS-CoV Helicase. PLoS ONE 2012, 7, e36521. [Google Scholar] [CrossRef]
- Haberland, A.; Krylova, O.; Nikolenko, H.; Göttel, P.; Dallmann, A.; Müller, J.; Weisshoff, H. Aptamer BC 007’s Affinity to Specific and Less-Specific Anti-SARS-CoV-2 Neutralizing Antibodies. Viruses 2021, 13, 932. [Google Scholar] [CrossRef]
- Haberland, A.; Holtzhauer, M.; Schlichtiger, A.; Bartel, S.; Schimke, I.; Müller, J.; Dandel, M.; Luppa, P.B.; Wallukat, G. Aptamer BC 007 - a broad spectrum neutralizer of pathogenic autoantibodies against G-protein-coupled receptors. Eur. J. Pharmacol. 2016, 789, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Haberland, A.; Becker, N.P.; Wenzel, K.; Wallukat, G.; Goettel, P.; Schulze-Rothe, S.; Schimke, I.; Golor, G.; Grossmann, M.; et al. The DNA-based drug BC 007 neutralizes agonistically acting autoantibodies directed against G protein–coupled receptors-Successful mode of action demonstrated in clinical phase 1 trial. J. Am. Coll. Cardiol. 2018, 71, A645. [Google Scholar] [CrossRef]
- Wallukat, G.; Schimke, I. Agonistic autoantibodies directed against G-protein-coupled receptors and their relationship to cardiovascular diseases. Semin. Immunopathol. 2014, 36, 351–363. [Google Scholar] [CrossRef] [PubMed]
- Becker, N.-P.; Haberland, A.; Wenzel, K.; Göttel, P.; Wallukat, G.; Davideit, H.; Schulze-Rothe, S.; Hönicke, A.-S.; Schimke, I.; Bartel, S.; et al. A Three-Part, Randomised Study to Investigate the Safety, Tolerability, Pharmacokinetics and Mode of Action of BC 007, Neutraliser of Pathogenic Autoantibodies Against G-Protein Coupled Receptors in Healthy, Young and Elderly Subjects. Clin. Drug Investig. 2020, 40, 433–447. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, Y.; Wu, J.; Qi, J.; Zeng, Z.; Wan, Q.; Chen, Z.; Manandhar, P.; Cavener, V.S.; Boyle, N.R.; et al. Neutralizing Aptamers Block S/RBD-ACE2 Interactions and Prevent Host Cell Infection. Angew. Chem. Int. Ed. 2021, 60, 10273–10278. [Google Scholar] [CrossRef]
- Schmitz, A.; Weber, A.; Bayin, M.; Breuers, S.; Fieberg, V.; Famulok, M.; Mayer, G. A SARS-CoV-2 Spike Binding DNA Aptamer that Inhibits Pseudovirus Infection by an RBD-Independent Mechanism. Angew. Chem. Int. Ed. 2021, 60, 10279–10285. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Liu, S.; Wei, X.; Wan, S.; Huang, M.; Song, T.; Lu, Y.; Weng, X.; Lin, Z.; Chen, H.; et al. Aptamer Blocking Strategy Inhibits SARS-CoV-2 Virus Infection. Angew. Chem. Int. Ed. 2021, 60, 10266–10272. [Google Scholar] [CrossRef]
- Ferreira-Bravo, I.A.; DeStefano, J.J. Xeno-Nucleic Acid (XNA) 2′-Fluoro-Arabino Nucleic Acid (FANA) Aptamers to the Receptor-Binding Domain of SARS-CoV-2 S Protein Block ACE2 Binding. Viruses 2021, 13, 1983. [Google Scholar] [CrossRef]
- Ambartsumyan, O.; Gribanyov, D.; Kukushkin, V.; Kopylov, A.; Zavyalova, E. SERS-Based Biosensors for Virus Determination with Oligonucleotides as Recognition Elements. Int. J. Mol. Sci. 2020, 21, 3373. [Google Scholar] [CrossRef]
- Villa, A.; Brunialti, E.; Dellavedova, J.; Meda, C.; Rebecchi, M.; Conti, M.; Donnici, L.; De Francesco, R.; Reggiani, A.; Lionetti, V.; et al. DNA aptamers masking angiotensin converting enzyme 2 as an innovative way to treat SARS-CoV-2 pandemic. Pharmacol. Res. 2021, 175, 105982. [Google Scholar] [CrossRef]


| Virus | Aptamer Name | Nucleic Acid Type | Target | Ref. |
|---|---|---|---|---|
| THERAPY OF VIRAL DISEASES | ||||
| HPV | Sc5-c3 | RNA | HPV-16 VLPs | [85,86] |
| HSV | DApt | DNA | gD | [87] |
| Dengue | n/d | RNA | MTase | [88] |
| Ebola | VPKS-2; VPKS-5 | DNA | VP24 | [89] |
| HIV | 148.1–38 m | RNA | RT | [90] |
| T30175 | DNA | Integrase | [91] | |
| RBA-14 | RNA | Rev | [92] | |
| HBV | H01 | DNA | HbSAg | [93] |
| DIAGNOSTICS OF VIRAL DISEASES | ||||
| HPV | HPV-07 | DNA | HPV-16 VLPs | [94] |
| HCV | C4 | DNA | Core | [95] |
| A12, A14, A15 | DNA | Core | [96,97] | |
| Zika | n/d | DNA | Capsid | [98] |
| 2, 10 | DNA | NS1 | [99] | |
| Dengue | n/d | DNA | DENV | [100] |
| Norovirus | n/d | DNA | VLPs | [101] |
| APTL-1 | DNA | Capsid | [102] | |
| n/d | DNA | Capsid | [103] | |
| Influenza | n/d | DNA | H1N1 virus | [104,105] |
| A20 | DNA | H1N1 virus | [106] | |
| V46 | DNA | H1N1 HA | [107] | |
| A22 | DNA | H3N2 HA | [108] | |
| P30-10-16 | RNA | H3N2 HA | [109] | |
| Ebola | 39SGP1A | RNA | GP | [110] |
| HIV | AntiTat5 | RNA | Tat | [111] |
| HBV | Aptamer 2-19 | DNA | HbEAg | [112] |
| Aptasensor by [Ref.] | Advantages | General Advantages of Aptasensors | General Disadvantages of Aptasensors |
|---|---|---|---|
| Zahanshansky et al. [176] | Saliva as the diagnostic material | Different diagnostic material can be used depending on the type of aptasensor Short detection time High sensitivity and specificity Some of the aptasensor undergoes miniaturization and can be used in a physician’s office No need to perform nucleic acid aplification | RNA aptamers as recognition elements are sensitive to exonuclease degradation [177] Necessity of RNA aptamer modification in order to improve their stability [177] Quenching of fluorophores conjugated with aptamers in optical aptasensors by biological components included in the tested material [177] Split aptamers as detection molecules can be applied in aptasensors only in a closed system [178] |
| Collection of saliva is less invasive for the patient and safer for the medical staff | |||
| Detection time | |||
| Cost production | |||
| Chen et al. [179] | Sensitivity and selectivity | ||
| Detection time | |||
| Qualitatively and quantitatively evaluation | |||
| Pramanik et al. [55] | Detection time | ||
| Needs only 8 viral particles in 1 mL of diagnostic material | |||
| Stanborough et al. [180] | Detection threshold in sub-picomolar range | ||
| Sensitivity and specificity | |||
| Zhang et al. [153] | Detection threshold in picomolar range | ||
| Detection time | |||
| Diagnostic strip form | |||
| Different types of diagnostic material—sputum, urine, serum | |||
| Can be performed in a physician’s office |
| Virus Variant | Nucleic Acid Type | Target | Aptamer Name |
Binding Affinity (Kd) [nM] | Detection Technique | Limit of Detection | Sens | Spec | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| WILD-TYPE | DNA | S protein (N-terminal domain) | SNAP1 | 39.32 ± 0.12 | Lateral Flow Assay | 250 pM; 2.5 × 107 copies/mL−1 | n/d | n/d | [185] |
| ELISA | 10 pM; 5 × 105 copies/mL−1 | ||||||||
| SNAP3 | 76.59 ± 0.12 | n/d | n/d | ||||||
| S1 protein (RBD) | nCoV-S1-Apt1 | 0.33 ± 0.02 | AuNPSs colorimetric assay (serum) | 3.13 nM | [186] | ||||
| DNA (G-quadruplex) | S protein | S14 | 21.8 | ALISA (swab) | 2 nM | 91% | 98% | [187] | |
| DNA (hairpin) | S1 protein | MSA1 | 0.023 | Colorimetric sandwich assay (saliva) | 400 fM 2.4 × 108 particles/mL−1 | n/d | n/d | [188] | |
| MSA5 | 0.012 | n/d | |||||||
| WILD-TYPE | DNA (dimers) | S protein | DSA1N5 | 0.12 ± 0.02 | CoV-eChip (saliva) | saliva: 1 × 103 particles/mL−1 spike protein: 1 fM | 81% | 100% | [189] |
| ALPHA | 0.29 ± 0.04 | saliva: 5 × 103 particles/mL−1 spike protein: 2.8 fM | |||||||
| DELTA | 0.48 ± 0.06 | spike protein: 3.6 fM | n/d | n/d |
|
Aptamer Name | Nucleic Acid Type | Target | Aptamer Application Method | Inhibitory Effect | Kd/IC50 | Ref. |
|---|---|---|---|---|---|---|
| SP6 | DNA | S protein (different site than RBD) | Application into ACE2 expressing Vero E6 cell line culture infected with the CoV-2 pseudotyped virus | SP6 concentration-dependent reduction of infection rate | Kd = 13.9 ± 0.6 nM | [208] |
| cb-CoV-2-6C3 | S1 protein (RBD) | Application into 293T cell line culture expressing ACE2 infected with the SARS-CoV-2 pseudovirus incubated previously with cb-CoV2-6C3 | Reduction in viral RNA amount in the cells by 87.01% | Kd = 0.13 nM IC50 = 9.68 nM | [209] | |
| nCoV-S1-Apt1 | Competitive binding assay of ACE2 protein and anti-S1 IgG with nCoV-S1-Apt1 | n/d | Kd = 0.1 nM IC50 = 80.12 nM | [186] | ||
| FANA-R8-9 | FANA (2′-fluoro-arabinonucleid acid) | S protein (RBD and the larger domain) | ACE2 ELISA assay | The same aptamer effectiveness as the neutralizing RBD-specific antibodies | Kd = 14.4 ± 4.6 nM IC50 = 1.30 ± 0.2 µg/mL | [210] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wandtke, T.; Wędrowska, E.; Szczur, M.; Przybylski, G.; Libura, M.; Kopiński, P. Aptamers—Diagnostic and Therapeutic Solution in SARS-CoV-2. Int. J. Mol. Sci. 2022, 23, 1412. https://doi.org/10.3390/ijms23031412
Wandtke T, Wędrowska E, Szczur M, Przybylski G, Libura M, Kopiński P. Aptamers—Diagnostic and Therapeutic Solution in SARS-CoV-2. International Journal of Molecular Sciences. 2022; 23(3):1412. https://doi.org/10.3390/ijms23031412
Chicago/Turabian StyleWandtke, Tomasz, Ewelina Wędrowska, Marcin Szczur, Grzegorz Przybylski, Marek Libura, and Piotr Kopiński. 2022. "Aptamers—Diagnostic and Therapeutic Solution in SARS-CoV-2" International Journal of Molecular Sciences 23, no. 3: 1412. https://doi.org/10.3390/ijms23031412
APA StyleWandtke, T., Wędrowska, E., Szczur, M., Przybylski, G., Libura, M., & Kopiński, P. (2022). Aptamers—Diagnostic and Therapeutic Solution in SARS-CoV-2. International Journal of Molecular Sciences, 23(3), 1412. https://doi.org/10.3390/ijms23031412





