Fosfomycin Resistance Evolutionary Pathways of Stenotrophomonas maltophilia in Different Growing Conditions
Abstract
1. Introduction
2. Results
2.1. Experimental Evolution upon Fosfomycin Challenge Leads to High Levels of Resistance in S. maltophilia Populations under all Tested Conditions
2.2. Selected Mutations in the Presence of Fosfomycin
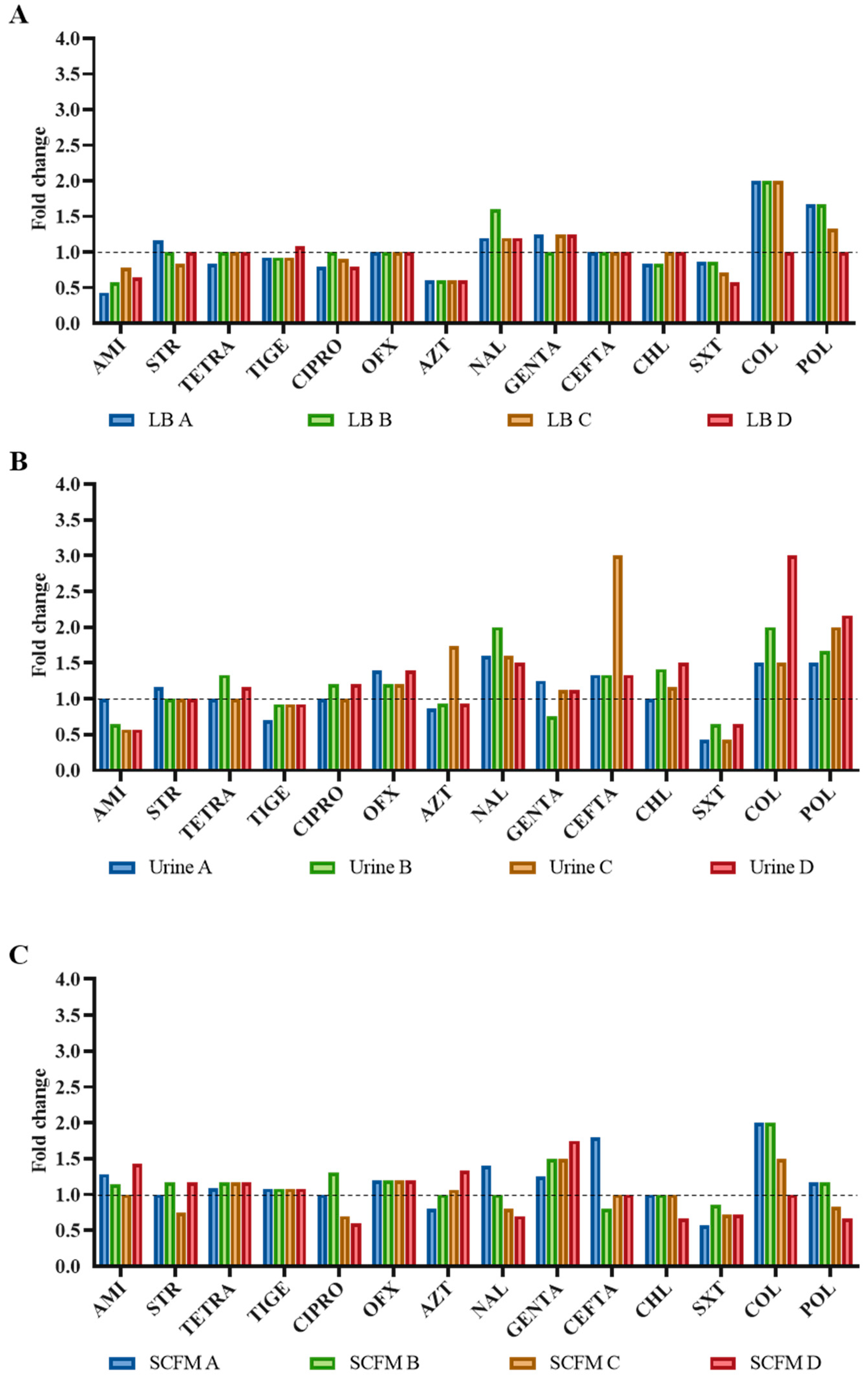
2.3. Antibiotic Susceptibility Profile of the Fosfomycin-Evolved Populations
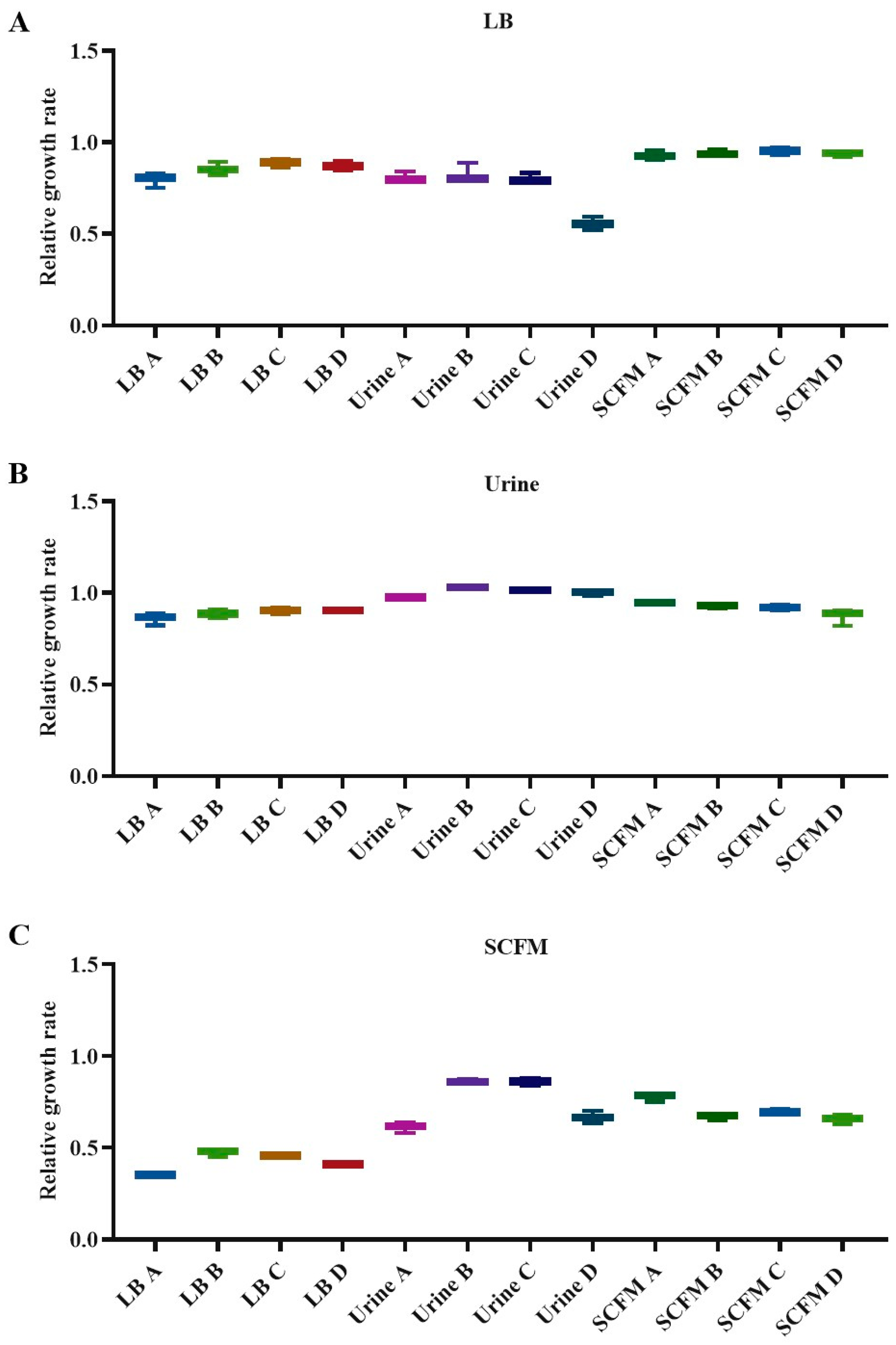
2.4. Fitness Costs Associated with Fosfomycin Resistance
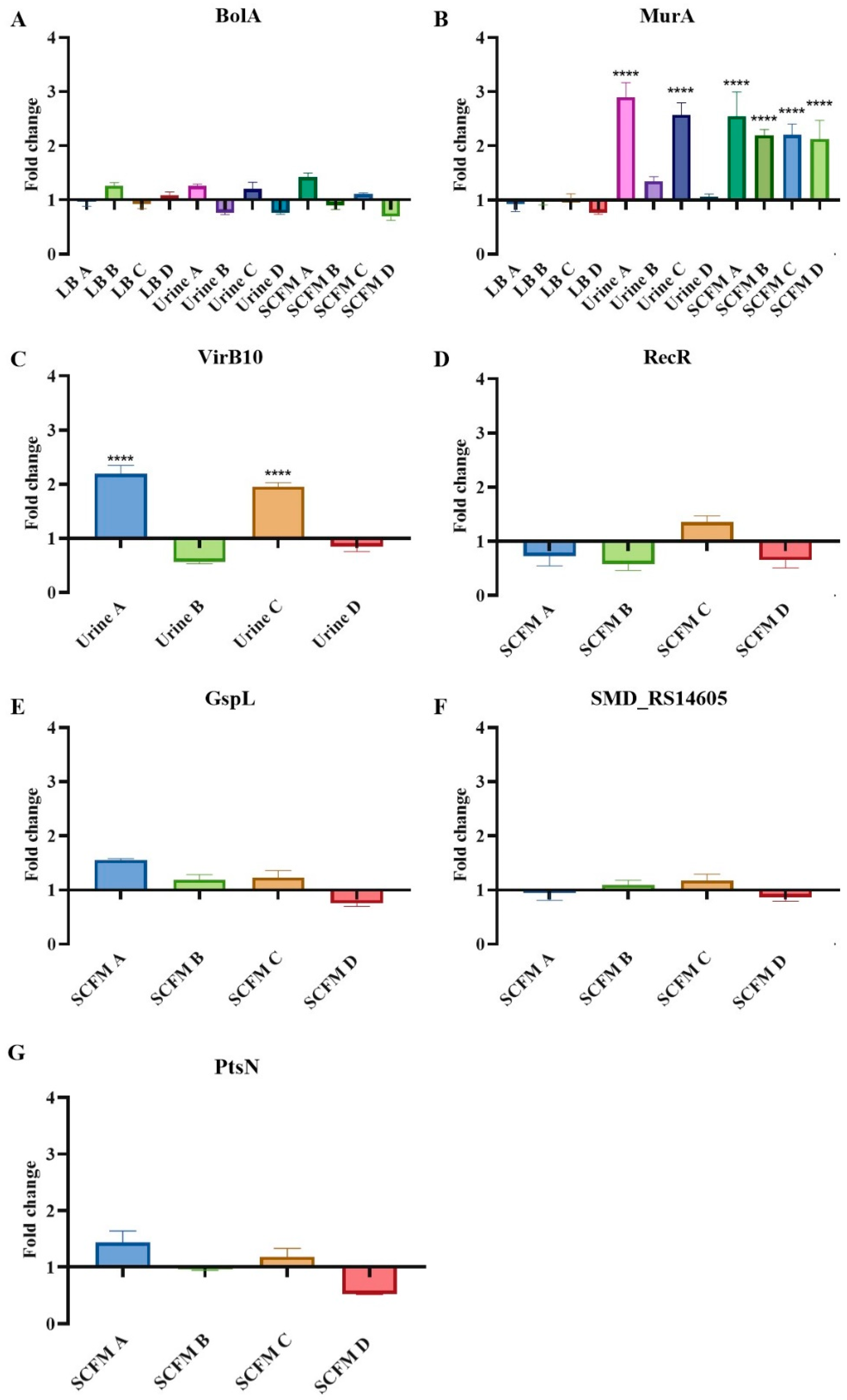
2.5. Expression Changes in Possible Resistance Determinants in the Fosfomycin-Evolved Populations
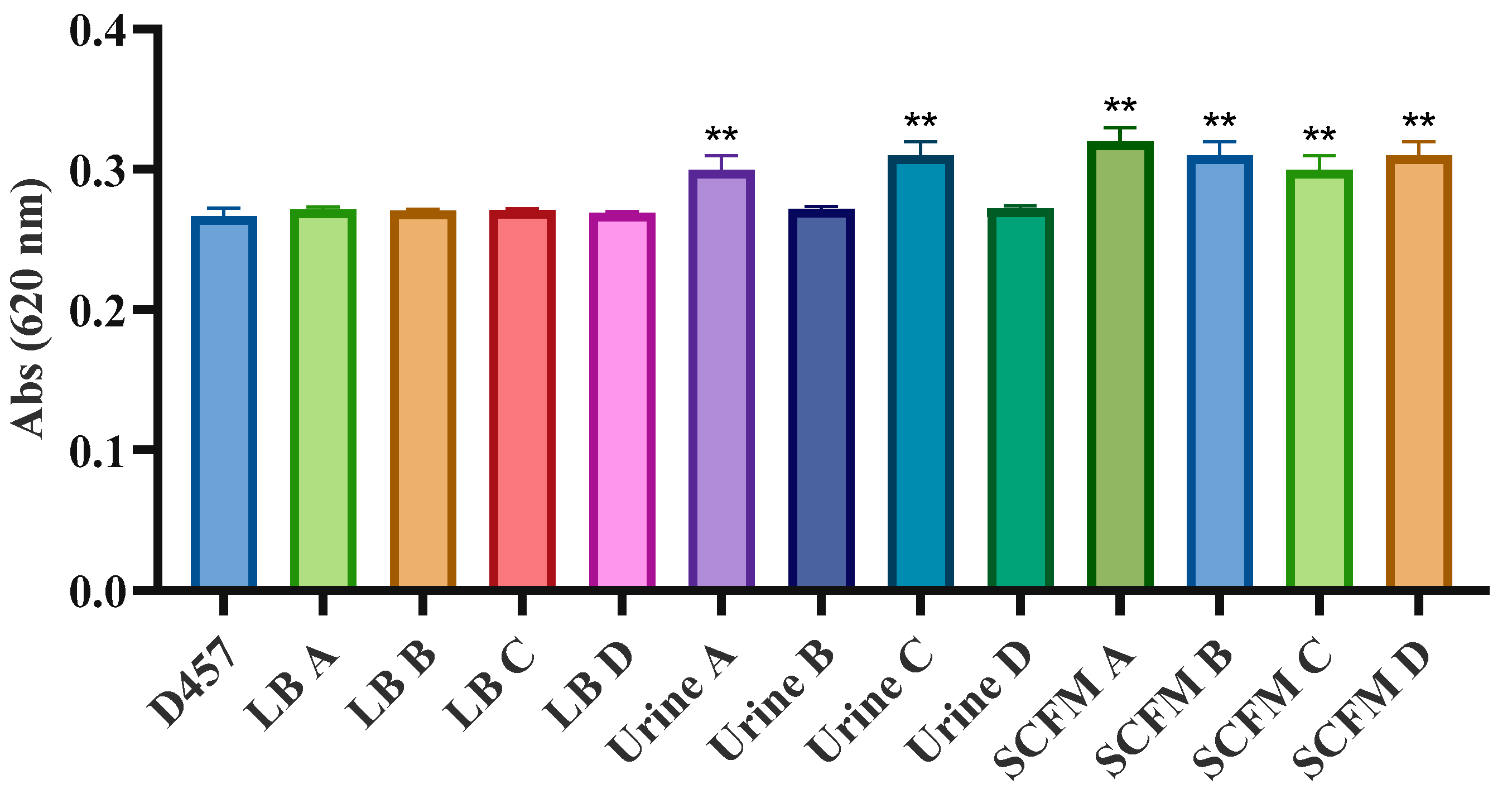
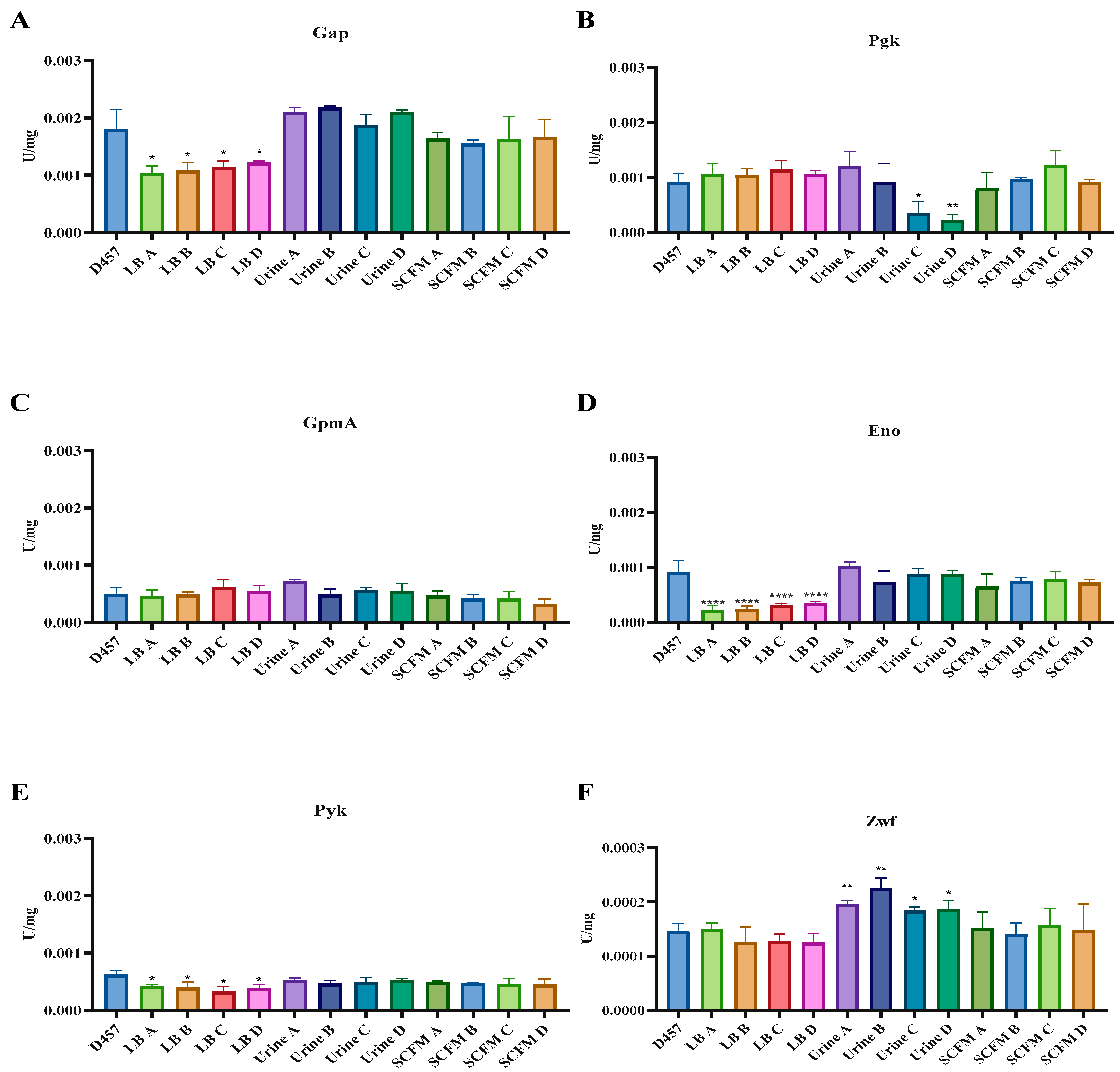
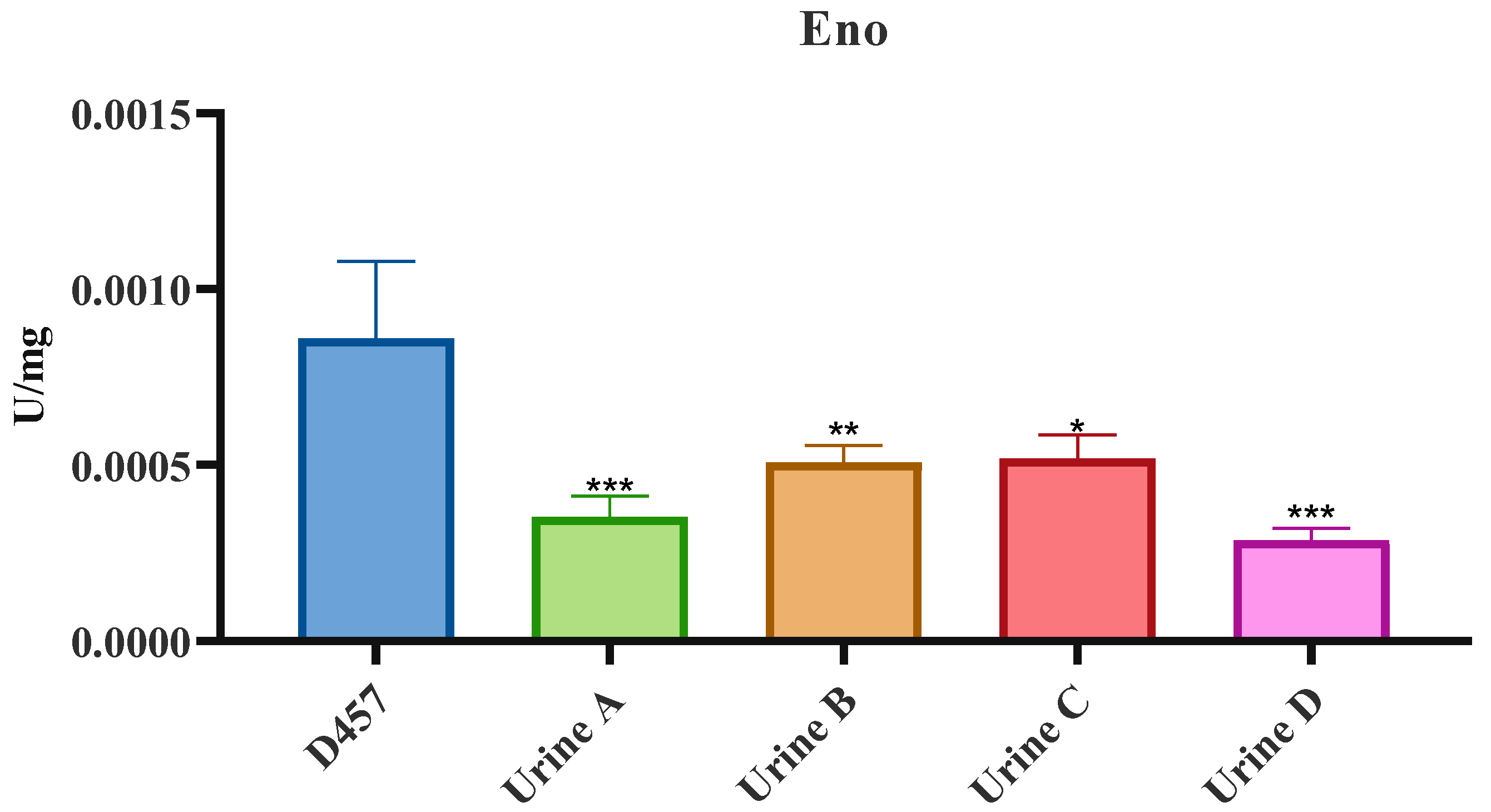
2.6. Activity of Central Metabolism Enzymes Is Associated with Fosfomycin Resistance in Some of the Fosfomycin-Evolved Populations
2.7. Fosfomycin Resistance Is not Related to an Impaired Entry of Fosfomycin Inside the Cell
3. Discussion
4. Materials and Methods
4.1. Growth Conditions and Antibiotic Susceptibility Assays
4.2. Experimental Evolutions
4.3. DNA Extraction, WGS and Identification of Mutations
4.4. Fitness Cost Determination
4.5. RNA Extraction and Quantitative Reverse Transcription PCR (qRT-PCR)
4.6. Assay of MurA Activity
4.7. In Vitro Activity Assays of the Enzymes of the Lower Glycolytic Pathway and Dehydrogenases
4.8. Quantification of Intracellular Fosfomycin
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kaye, K.S.; Pogue, J.M. Infections Caused by Resistant Gram-Negative Bacteria: Epidemiology and Management. Pharmacotherapy 2015, 35, 949–962. [Google Scholar] [CrossRef]
- Behera, B.; Mohanty, S.; Sahu, S.; Praharaj, A.K. In vitro Activity of Fosfomycin against Multidrug-Resistant Urinary and Nonurinary Gram-Negative Isolates. Indian J. Crit. Care Med. 2018, 22, 533–536. [Google Scholar] [PubMed]
- Brooke, J.S.; Di Bonaventura, G.; Berg, G.; Martinez, J.L. Editorial: A Multidisciplinary Look at Stenotrophomonas maltophilia: An Emerging Multi-Drug-Resistant Global Opportunistic Pathogen. Front. Microbiol. 2017, 8, 1511. [Google Scholar] [CrossRef] [PubMed]
- Brooke, J.S. Stenotrophomonas maltophilia: An emerging global opportunistic pathogen. Clin. Microbiol. Rev. 2012, 25, 2–41. [Google Scholar] [CrossRef] [PubMed]
- Vartivarian, S.E.; Papadakis, K.A.; Anaissie, E.J. Stenotrophomonas (Xanthomonas) maltophilia urinary tract infection. A disease that is usually severe and complicated. Arch. Intern. Med. 1996, 156, 433–435. [Google Scholar] [CrossRef] [PubMed]
- Gil-Gil, T.; Martínez, J.L.; Blanco, P. Mechanisms of antimicrobial resistance in Stenotrophomonas maltophilia: A review of current knowledge. Expert Rev. Anti-Infect. Ther. 2020, 18, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Green, H.; Jones, A.M. The microbiome and emerging pathogens in cystic fibrosis and non-cystic fibrosis bronchiectasis. Semin. Respir. Crit. Care Med. 2015, 36, 225–235. [Google Scholar] [CrossRef]
- Esposito, A.; Pompilio, A.; Bettua, C.; Crocetta, V.; Giacobazzi, E.; Fiscarelli, E.; Jousson, O.; Di Bonaventura, G. Evolution of Stenotrophomonas maltophilia in Cystic Fibrosis Lung over Chronic Infection: A Genomic and Phenotypic Population Study. Front. Microbiol. 2017, 8, 1590. [Google Scholar] [CrossRef]
- Pompilio, A.; Crocetta, V.; Ghosh, D.; Chakrabarti, M.; Gherardi, G.; Vitali, L.A.; Fiscarelli, E.; Di Bonaventura, G. Stenotrophomonas maltophilia Phenotypic and Genotypic Diversity during a 10-year Colonization in the Lungs of a Cystic Fibrosis Patient. Front. Microbiol. 2016, 7, 1551. [Google Scholar] [CrossRef]
- Falagas, M.E.; Giannopoulou, K.P.; Kokolakis, G.N.; Rafailidis, P.I. Fosfomycin: Use beyond urinary tract and gastrointestinal infections. Clin. Infect. Dis. 2008, 46, 1069–1077. [Google Scholar] [CrossRef]
- European Association of Urology. European Association of Urology Guidelines. In EAU Annual Congress, Milan 2021; 2021; ISBN 978-94-92671-13-4. [Google Scholar]
- Raz, R. Fosfomycin: An old—New antibiotic. Clin. Microbiol. Infect. 2012, 18, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Allen, R.C.; Pfrunder-Cardozo, K.R.; Hall, A.R. Collateral Sensitivity Interactions between Antibiotics Depend on Local Abiotic Conditions. mSystems 2021, 6, e0105521. [Google Scholar] [CrossRef] [PubMed]
- Scribner, M.R.; Santos-Lopez, A.; Marshall, C.W.; Deitrick, C.; Cooper, V.S. Parallel Evolution of Tobramycin Resistance across Species and Environments. mBio 2020, 11, e00932-20. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Lees, W.J.; Kempsell, K.E.; Lane, W.S.; Duncan, K.; Walsh, C.T. Characterization of a Cys115 to Asp substitution in the Escherichia coli cell wall biosynthetic enzyme UDP-GlcNAc enolpyruvyl transferase (MurA) that confers resistance to inactivation by the antibiotic fosfomycin. Biochemistry 1996, 35, 4923–4928. [Google Scholar] [CrossRef]
- Tsuruoka, T.; Yamada, Y. Characterization of spontaneous fosfomycin (phosphonomycin)-resistant cells of Escherichia coli B in vitro. J. Antibiot. 1975, 28, 906–911. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.M.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Arca, P.; Hardisson, C.; Suárez, J.E. Purification of a glutathione S-transferase that mediates fosfomycin resistance in bacteria. Antimicrob. Agents Chemother. 1990, 34, 844–848. [Google Scholar] [CrossRef]
- Gil-Gil, T.; Corona, F.; Martinez, J.L.; Bernardini, A. The Inactivation of Enzymes Belonging to the Central Carbon Metabolism Is a Novel Mechanism of Developing Antibiotic Resistance. mSystems 2020, 5, e00282-20. [Google Scholar] [CrossRef]
- Gil-Gil, T.; Ochoa-Sánchez, L.E.; Martínez, J.L. The Antibiotic Fosfomycin Mimics the Effects of the Intermediate Metabolites Phosphoenolpyruvate and Glyceraldehyde-3-Phosphate on the Stenotrophomonas maltophilia Transcriptome. Int. J. Mol. Sci. 2021, 23, 159. [Google Scholar] [CrossRef]
- Martinez, J.L.; Rojo, F. Metabolic regulation of antibiotic resistance. FEMS Microbiol. Rev. 2011, 35, 768–789. [Google Scholar] [CrossRef]
- Bayer-Santos, E.; Cenens, W.; Matsuyama, B.Y.; Oka, G.U.; Di Sessa, G.; Mininel, I.D.V.; Alves, T.L.; Farah, C.S. The opportunistic pathogen Stenotrophomonas maltophilia utilizes a type IV secretion system for interbacterial killing. PLoS Pathog. 2019, 15, e1007651. [Google Scholar] [CrossRef]
- Darbari, V.C.; Ciccone, J.; Patel, J.S.; Islam, B.; Agarwal, P.K.; Haider, S. Electrostatic Switching Controls Channel Dynamics of the Sensor Protein VirB10 in A. tumefaciens Type IV Secretion System. ACS Omega 2020, 5, 3271–3281. [Google Scholar] [CrossRef]
- Karaba, S.M.; White, R.C.; Cianciotto, N.P. Stenotrophomonas maltophilia encodes a type II protein secretion system that promotes detrimental effects on lung epithelial cells. Infect. Immun. 2013, 81, 3210–3219. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; White, K.A.; Polissi, A.; Georgopoulos, C.; Raetz, C.R. Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. J. Biol. Chem. 1998, 273, 12466–12475. [Google Scholar] [CrossRef] [PubMed]
- Doerrler, W.T.; Raetz, C.R. ATPase activity of the MsbA lipid flippase of Escherichia coli. J. Biol. Chem. 2002, 277, 36697–36705. [Google Scholar] [CrossRef]
- Doerrler, W.T.; Gibbons, H.S.; Raetz, C.R. MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J. Biol. Chem. 2004, 279, 45102–45109. [Google Scholar] [CrossRef] [PubMed]
- Mi, W.; Li, Y.; Yoon, S.H.; Ernst, R.K.; Walz, T.; Liao, M. Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 2017, 549, 233–237. [Google Scholar] [CrossRef]
- Polissi, A.; Georgopoulos, C. Mutational analysis and properties of the msbA gene of Escherichia coli, coding for an essential ABC family transporter. Mol. Microbiol. 1996, 20, 1221–1233. [Google Scholar] [CrossRef] [PubMed]
- Karow, M.; Georgopoulos, C. The essential Escherichia coli msbA gene, a multicopy suppressor of null mutations in the htrB gene, is related to the universally conserved family of ATP-dependent translocators. Mol. Microbiol. 1993, 7, 69–79. [Google Scholar] [CrossRef]
- Chiu, H.C.; Lin, T.L.; Yang, J.C.; Wang, J.T. Synergistic effect of imp/ostA and msbA in hydrophobic drug resistance of Helicobacter pylori. BMC Microbiol. 2009, 9, 136. [Google Scholar] [CrossRef]
- Jones, D.H.; Franklin, F.C.; Thomas, C.M. Molecular analysis of the operon which encodes the RNA polymerase sigma factor sigma 54 of Escherichia coli. Microbiology 1994, 140, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
- Powell, B.S.; Court, D.L.; Inada, T.; Nakamura, Y.; Michotey, V.; Cui, X.; Reizer, A.; Saier, M.H., Jr.; Reizer, J. Novel proteins of the phosphotransferase system encoded within the rpoN operon of Escherichia coli. Enzyme IIANtr affects growth on organic nitrogen and the conditional lethality of an erats mutant. J. Biol. Chem. 1995, 270, 4822–4839. [Google Scholar] [CrossRef]
- Pfluger-Grau, K.; Gorke, B. Regulatory roles of the bacterial nitrogen-related phosphotransferase system. Trends Microbiol. 2010, 18, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.R.; Park, Y.H.; Kim, M.; Kim, Y.R.; Park, S.; Peterkofsky, A.; Seok, Y.J. Reciprocal regulation of the autophosphorylation of enzyme INtr by glutamine and alpha-ketoglutarate in Escherichia coli. Mol. Microbiol. 2013, 88, 473–485. [Google Scholar] [CrossRef]
- Lee, C.R.; Cho, S.H.; Yoon, M.J.; Peterkofsky, A.; Seok, Y.J. Escherichia coli enzyme IIANtr regulates the K+ transporter TrkA. Proc. Natl. Acad. Sci. USA 2007, 104, 4124–4129. [Google Scholar] [CrossRef] [PubMed]
- Luttmann, D.; Heermann, R.; Zimmer, B.; Hillmann, A.; Rampp, I.S.; Jung, K.; Gorke, B. Stimulation of the potassium sensor KdpD kinase activity by interaction with the phosphotransferase protein IIA(Ntr) in Escherichia coli. Mol. Microbiol. 2009, 72, 978–994. [Google Scholar] [CrossRef] [PubMed]
- Cases, I.; Lopez, J.A.; Albar, J.P.; De Lorenzo, V. Evidence of multiple regulatory functions for the PtsN (IIA(Ntr)) protein of Pseudomonas putida. J. Bacteriol. 2001, 183, 1032–1037. [Google Scholar] [CrossRef]
- Mohamed, Y.F.; Abou-Shleib, H.M.; Khalil, A.M.; El-Guink, N.M.; El-Nakeeb, M.A. Membrane permeabilization of colistin toward pan-drug resistant Gram-negative isolates. Braz. J. Microbiol. 2016, 47, 381–388. [Google Scholar] [CrossRef]
- Guinote, I.B.; Moreira, R.N.; Freire, P.; Arraiano, C.M. Characterization of the BolA homolog IbaG: A new gene involved in acid resistance. J. Microbiol. Biotechnol. 2012, 22, 484–493. [Google Scholar] [CrossRef]
- Fleurie, A.; Zoued, A.; Alvarez, L.; Hines, K.M.; Cava, F.; Xu, L.; Davis, B.M.; Waldor, M.K. A Vibrio cholerae BolA-Like Protein Is Required for Proper Cell Shape and Cell Envelope Integrity. mBio 2019, 10, e00790-19. [Google Scholar] [CrossRef]
- Marquardt, J.L.; Siegele, D.A.; Kolter, R.; Walsh, C.T. Cloning and sequencing of Escherichia coli murZ and purification of its product, a UDP-N-acetylglucosamine enolpyruvyl transferase. J. Bacteriol. 1992, 174, 5748–5752. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef]
- Nilsson, A.I.; Berg, O.G.; Aspevall, O.; Kahlmeter, G.; Andersson, D.I. Biological costs and mechanisms of fosfomycin resistance in Escherichia coli. Antimicrob. Agents Chemother. 2003, 47, 2850–2858. [Google Scholar] [CrossRef]
- Falagas, M.E.; Vouloumanou, E.K.; Samonis, G.; Vardakas, K.Z. Fosfomycin. Clin. Microbiol. Rev. 2016, 29, 321–347. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J.; Sandlin, R.C.; Maurelli, A.T. In vitro and in vivo functional activity of Chlamydia MurA, a UDP-N-acetylglucosamine enolpyruvyl transferase involved in peptidoglycan synthesis and fosfomycin resistance. J. Bacteriol. 2003, 185, 1218–1228. [Google Scholar] [CrossRef] [PubMed]
- Sandoval, J.M.; Arenas, F.A.; Vasquez, C.C. Glucose-6-phosphate dehydrogenase protects Escherichia coli from tellurite-mediated oxidative stress. PLoS ONE 2011, 6, e25573. [Google Scholar] [CrossRef]
- Kim, J.; Jeon, C.O.; Park, W. Dual regulation of zwf-1 by both 2-keto-3-deoxy-6-phosphogluconate and oxidative stress in Pseudomonas putida. Microbiology 2008, 154, 3905–3916. [Google Scholar] [CrossRef]
- Martin-Gutierrez, G.; Docobo-Perez, F.; Rodriguez-Beltran, J.; Rodriguez-Martinez, J.M.; Aznar, J.; Pascual, A.; Blazquez, J. Urinary Tract Conditions Affect Fosfomycin Activity against Escherichia coli Strains Harboring Chromosomal Mutations Involved in Fosfomycin Uptake. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef]
- Alexander, M.K.; Miu, A.; Oh, A.; Reichelt, M.; Ho, H.; Chalouni, C.; Labadie, S.; Wang, L.; Liang, J.; Nickerson, N.N.; et al. Disrupting Gram-Negative Bacterial Outer Membrane Biosynthesis through Inhibition of the Lipopolysaccharide Transporter MsbA. Antimicrob. Agents Chemother. 2018, 62, e01142-18. [Google Scholar] [CrossRef] [PubMed]
- Reyes, C.L.; Ward, A.; Yu, J.; Chang, G. The structures of MsbA: Insight into ABC transporter-mediated multidrug efflux. FEBS Lett. 2006, 580, 1042–1048. [Google Scholar] [CrossRef] [PubMed]
- Leive, L.; Telesetsky, S.; Coleman, W.G., Jr.; Carr, D. Tetracyclines of various hydrophobicities as a probe for permeability of Escherichia coli outer membranes. Antimicrob. Agents Chemother. 1984, 25, 539–544. [Google Scholar] [CrossRef]
- Vuorio, R.; Vaara, M. The lipid A biosynthesis mutation lpxA2 of Escherichia coli results in drastic antibiotic supersusceptibility. Antimicrob. Agents Chemother. 1992, 36, 826–829. [Google Scholar] [CrossRef]
- Clements, J.M.; Coignard, F.; Johnson, I.; Chandler, S.; Palan, S.; Waller, A.; Wijkmans, J.; Hunter, M.G. Antibacterial activities and characterization of novel inhibitors of LpxC. Antimicrob. Agents Chemother. 2002, 46, 1793–1799. [Google Scholar] [CrossRef]
- Reuter, G.; Janvilisri, T.; Venter, H.; Shahi, S.; Balakrishnan, L.; van Veen, H.W. The ATP binding cassette multidrug transporter LmrA and lipid transporter MsbA have overlapping substrate specificities. J. Biol. Chem. 2003, 278, 35193–35198. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Hughes, D. Antibiotic resistance and its cost: Is it possible to reverse resistance? Nat. Rev. Microbiol. 2010, 8, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Hernando-Amado, S.; Sanz-Garcia, F.; Blanco, P.; Martinez, J.L. Fitness costs associated with the acquisition of antibiotic resistance. Essays Biochem. 2017, 61, 37–48. [Google Scholar] [PubMed]
- Levin, B.R.; Bull, J.J. Short-sighted evolution and the virulence of pathogenic microorganisms. Trends Microbiol. 1994, 2, 76–81. [Google Scholar] [CrossRef]
- Sanchez, P.; Alonso, A.; Martinez, J.L. Regulatory regions of smeDEF in Stenotrophomonas maltophilia strains expressing different amounts of the multidrug efflux pump SmeDEF. Antimicrob. Agents Chemother. 2004, 48, 2274–2276. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang le, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
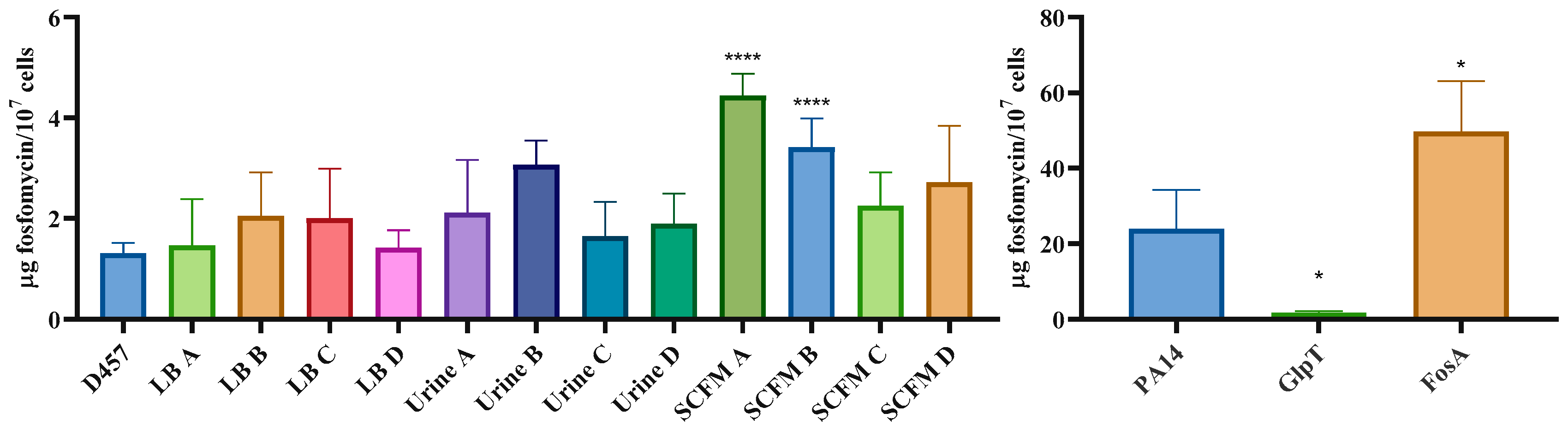
| Population | MIC (µg/mL) | |||
|---|---|---|---|---|
| MH | LB | Urine | SCFM | |
| D457 | 256 | 128 | 256 | 512 |
| LB control | 256 | 128 | 128 | 256 |
| LB A | 16,384 | >16,384 | 6250 | >16,384 |
| LB B | 16,384 | >16,384 | 6250 | >16,384 |
| LB C | 16,384 | 8192 | 3125 | >16,384 |
| LB D | 16,384 | 5892 | 3125 | >16,384 |
| Urine control | 256 | 128 | 128 | 256 |
| Urine A | >16,384 | >16,384 | >16,384 | 16,384 |
| Urine B | >16,384 | >16,384 | >16,384 | >16,384 |
| Urine C | >16,384 | >16,384 | >16,384 | >16,384 |
| Urine D | >16,384 | >16,384 | >16,384 | >16,384 |
| SCFM control | 256 | 128 | 128 | 256 |
| SCFM A | >16,384 | 8192 | 6250 | >16,384 |
| SCFM B | 8192 | 1024 | 3125 | 8192 |
| SCFM C | 8192 | 8192 | 6250 | >16,384 |
| SCFM D | 16,384 | 1024 | 1562 | 4092 |
| Medium | L | Gene | Product | Localization | Type | Nucleotide Change | Amino Acid Change | Frequency (%) | Domain | Provean Score |
|---|---|---|---|---|---|---|---|---|---|---|
| LB | A | eno | Phosphopyruvate hydratase | 1,828,585 | SNV | T → C | Leu106Pro | 93 | N-terminal | −6.9 |
| B | eno | Phosphopyruvate hydratase | 1,828,585 | SNV | T → C | Leu106Pro | 98 | N-terminal | −6.9 | |
| phaR | Polyhydroxyalkanoate synthesis repressor | 3,219,959 | SNV | C → T | Ser85Phe | 22 | PHB accumulation regulatory | −3.967 | ||
| C | eno | Phosphopyruvate hydratase | 1,828,585 | SNV | T → C | Leu106Pro | 98 | N-terminal | −6.9 | |
| D | eno | Phosphopyruvate hydratase | 1,828,585 | SNV | T → C | Leu106Pro | 94 | N-terminal | −6.9 | |
| Urine | A | bolA | BolA family transcriptional regulator | 1,158,049 | SNV | C → T | Ala52Ala | 28 | BolA-like | 0.0 |
| eno | Phosphopyruvate hydratase | 1,828,585 | SNV | T → C | Leu106Pro | 11 | N-terminal | −6.9 | ||
| virB10 | TrbI/VirB10 family protein | 2,939,074 | Ins | - → C | Asp173fs | ND | Bacterial conjugation TrbI | |||
| B | eno | Phosphopyruvate hydratase | 1,828,585 | SNV | T → C | Leu106Pro | 21 | N-terminal | −6.9 | |
| virB10 | TrbI/VirB10 family protein | 2,939,074 | Ins | - → C | Asp173fs | ND | Bacterial conjugation TrbI | |||
| C | bolA | BolA family transcriptional regulator | 1,158,049 | SNV | C → T | Ala52Ala | 12 | BolA-like | 0.0 | |
| eno | Phosphopyruvate hydratase | 1,828,585 | SNV | T → C | Leu106Pro | 9 | N-terminal | −6.9 | ||
| virB10 | TrbI/VirB10 family protein | 2,939,074 | Ins | - → C | Asp173fs | ND | Bacterial conjugation TrbI | |||
| D | eno | Phosphopyruvate hydratase | 1,828,585 | SNV | T → C | Leu106Pro | 13 | N-terminal | −6.9 | |
| virB10 | TrbI/VirB10 family protein | 2,939,074 | Ins | - → C | Asp173fs | ND | Bacterial conjugation TrbI | |||
| SCFM | A | gspL | General secretion pathway protein | 674,920 | SNV | G → T | Val247Phe | 77 | Fimbrial assembly pilN | −4.162 |
| ptsN | PTS sugar transporter subunit IIA | 1,152,021 | AGGGCCT → GCAGGCC | GlnAlaLeu126ArgProAla | 25 | PTS_EIIA_2 | L128A −4.542 | |||
| bolA | BolA family transcriptional regulator | 1,158,049 | SNV | C → T | Ala52Ala | 36 | BolA-like | 0.0 | ||
| SMD_RS14605 | ABC transporter ATP-binding protein | 3,119,746 | SNV | A → C | Thr419Pro | 19 | ABC transporter | −6.0 | ||
| recR | Recombination protein RecR | 1,087,048 | Ins | CAAGCGGGTGCCACAGAA | Upstream gene variant | ND | - | |||
| B | gspL | General secretion pathway protein | 674,920 | SNV | G → T | Val247Phe | 64 | Fimbrial assembly pilN | −4.162 | |
| bolA | BolA family transcriptional regulator | 1,158,049 | SNV | C → T | Ala52Ala | 91 | BolA-like | 0.0 | ||
| recR | Recombination protein RecR | 1,087,048 | Ins | CAAGCGGGTGCCACAGAA | Upstream gene variant | ND | - | |||
| C | bolA | BolA family transcriptional regulator | 1,158,049 | SNV | C → T | Ala52Ala | 53 | BolA-like | 0.0 | |
| recR | Recombination protein RecR | 1,087,048 | Ins | CAAGCGGGTGCCACAGAA | Upstream gene variant | ND | - | |||
| D | bolA | BolA family transcriptional regulator | 1,158,049 | SNV | C → T | Ala52Ala | 87 | BolA-like | 0.0 | |
| recR | Recombination protein RecR | 1,087,048 | Ins | CAAGCGGGTGCCACAGAA | Upstream gene variant | ND | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gil-Gil, T.; Martínez, J.L. Fosfomycin Resistance Evolutionary Pathways of Stenotrophomonas maltophilia in Different Growing Conditions. Int. J. Mol. Sci. 2022, 23, 1132. https://doi.org/10.3390/ijms23031132
Gil-Gil T, Martínez JL. Fosfomycin Resistance Evolutionary Pathways of Stenotrophomonas maltophilia in Different Growing Conditions. International Journal of Molecular Sciences. 2022; 23(3):1132. https://doi.org/10.3390/ijms23031132
Chicago/Turabian StyleGil-Gil, Teresa, and José L. Martínez. 2022. "Fosfomycin Resistance Evolutionary Pathways of Stenotrophomonas maltophilia in Different Growing Conditions" International Journal of Molecular Sciences 23, no. 3: 1132. https://doi.org/10.3390/ijms23031132
APA StyleGil-Gil, T., & Martínez, J. L. (2022). Fosfomycin Resistance Evolutionary Pathways of Stenotrophomonas maltophilia in Different Growing Conditions. International Journal of Molecular Sciences, 23(3), 1132. https://doi.org/10.3390/ijms23031132






