Biofilm and Small Colony Variants—An Update on Staphylococcus aureus Strategies toward Drug Resistance
Abstract
1. Introduction
2. The Development of DR or MDR
3. The Hotspots of MDR Relevant Recent Studies
4. Alternative Typical Phenotypic Switches of S. aureus
4.1. An Introduction of Biofilm Phenotype
4.1.1. Formation of S. aureus Biofilm
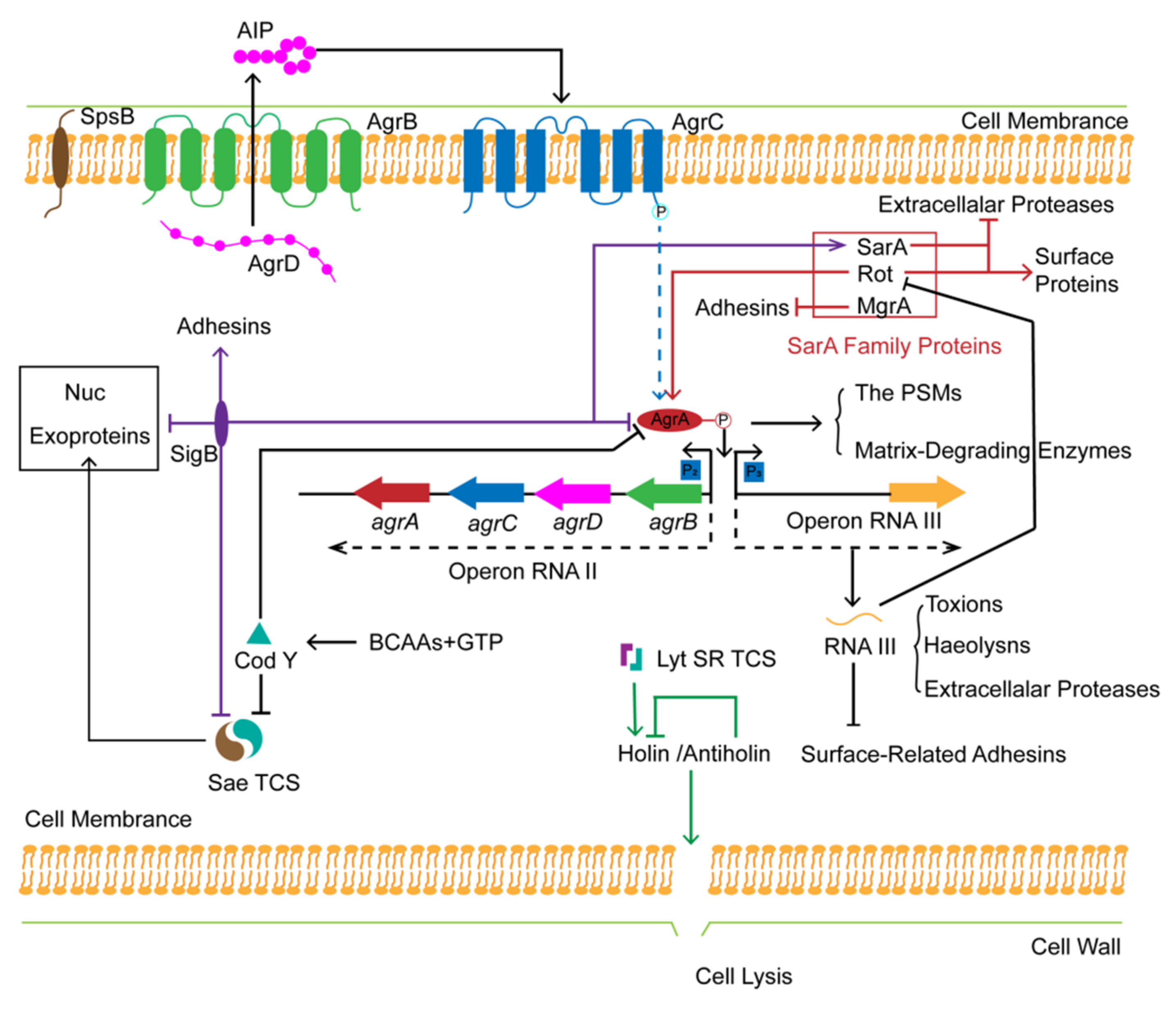
4.1.2. Global Regulation of S. aureus Biofilm Formation
- The Sae and the LytSR Two-Component Systems
- SarA Family Proteins
- Alternative Sigma Factor σB (SigB)
- The Transcriptional Repressor CodY
4.1.3. Drug-Resistance Mechanisms Induced by Biofilm of S. aureus
4.2. An Introduction of Biofilm Phenotype
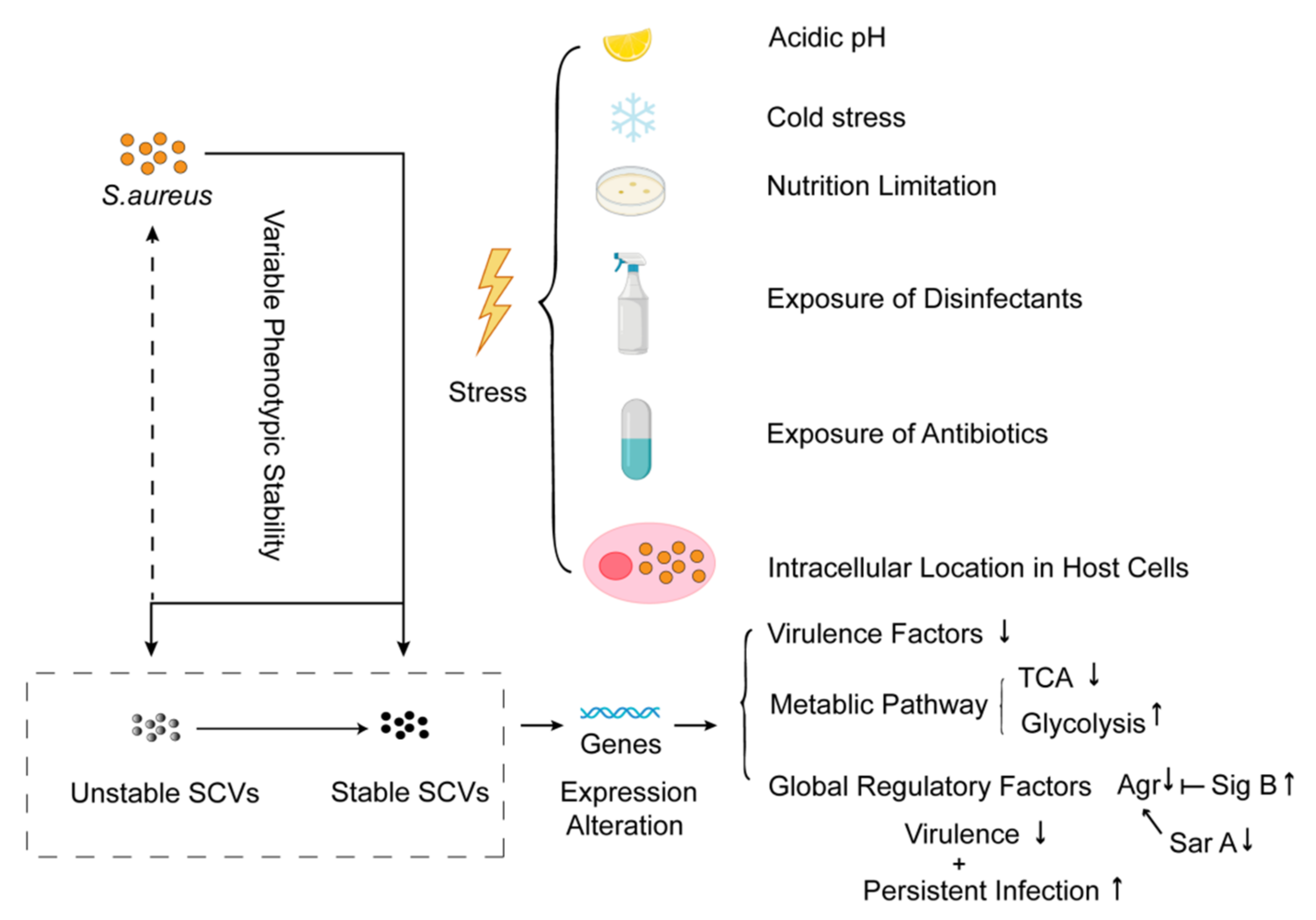
4.2.1. Formation of SCVs
4.2.2. Global Regulation of SCVs Formation
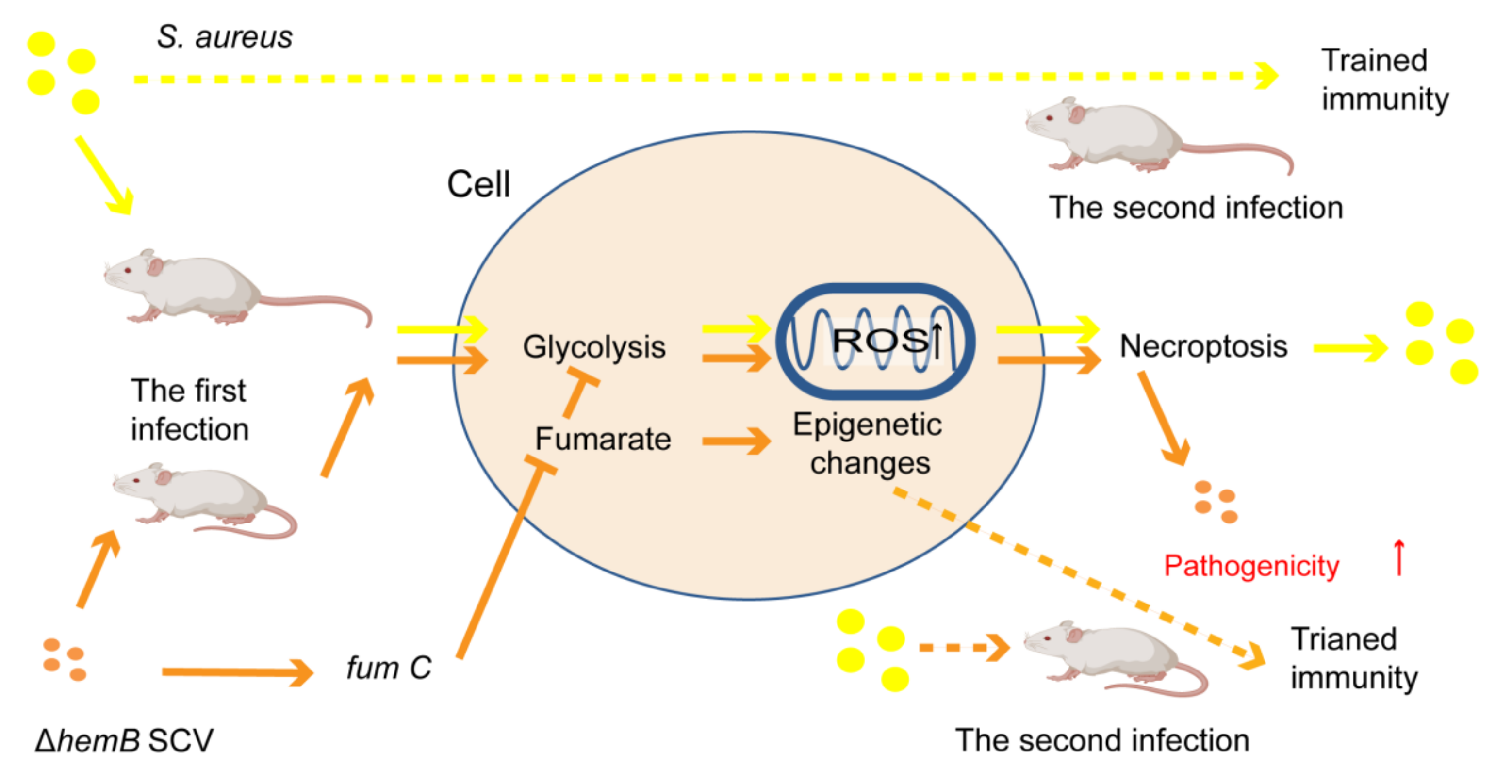
4.2.3. Drug-Resistance Mechanisms Induced by SCVs
5. Conclusions and Future Recommendations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lekshmi, M.; Ammini, P.; Kumar, S.; Varela, M.F. The Food Production Environment and the Development of Antimicrobial Resistance in Human Pathogens of Animal Origin. Microorganisms 2017, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Price, N.; Klein, J.L. Infectious Diseases; Oxford University Press (OUP): Oxford, UK, 2016. [Google Scholar]
- Zohra, T.; Numan, M.; Ikram, A.; Salman, M.; Khan, T.; Din, M.; Salman, M.; Farooq, A.; Amir, A.; Ali, M. Cracking the Challenge of Antimicrobial Drug Resistance with CRISPR/Cas9, Nanotechnology and other Strategies in ESKAPE Pathogens. Microorganisms 2021, 9, 954. [Google Scholar] [CrossRef]
- Levy, S.B.; Marshall, B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 2004, 10, S122–S129. [Google Scholar] [CrossRef] [PubMed]
- Rice, L.B. Progress and Challenges in Implementing the Research on ESKAPE Pathogens. Infect. Control Hosp. Epidemiol. 2010, 31, S7–S10. [Google Scholar] [CrossRef]
- Kahl, B.C.; Becker, K.; Loeffler, B. Clinical Significance and Pathogenesis of Staphylococcal Small Colony Variants in Persis-tent Infections. Clin. Microbiol. Rev. 2016, 29, 401–427. [Google Scholar] [CrossRef] [PubMed]
- Bui, L.M.; Conlon, B.P.; Kidd, S.P. Antibiotic tolerance and the alternative lifestyles of Staphylococcus aureus. Essays Biochem. 2017, 61, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.M. Phenotype Switching Is a Natural Consequence of Staphylococcus aureus Replication. J. Bacteriol. 2012, 194, 5404–5412. [Google Scholar] [CrossRef] [PubMed]
- Boles, B.R.; Thoendel, M.; Singh, P.K. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl. Acad. Sci. USA 2004, 101, 16630–16635. [Google Scholar] [CrossRef]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival Mechanisms of Clinically Relevant Microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef]
- Tuchscherr, L.; Geraci, J.; Löffler, B. Staphylococcus aureus Regulator Sigma B is Important to Develop Chronic Infections in Hematogenous Murine Osteomyelitis Model. Pathogens 2017, 6, 31. [Google Scholar] [CrossRef]
- Tuchscherr, L.; Loeffler, B. Staphylococcus aureus Dynamically Adapts Global Regulators and Virulence Factor Expression in the Course from Acute to Chronic Infection. Curr. Genet. 2016, 62, 15–17. [Google Scholar] [CrossRef]
- Tan, N.C.; Cooksley, C.M.; Roscioli, E.; Drilling, A.J.; Douglas, R.; Wormald, P.; Vreugde, S. Small-Colony Variants and Phenotype Switching of Intracellular Staphylococcus aureus in Chronic Rhinosinusitis. Allergy 2014, 69, 1364–1371. [Google Scholar] [CrossRef]
- Mbbs, B.S.P.; Cooksley, C.M.; Ramezanpour, M.; Vediappan, R.S.; Bassiouni, A.; Wormald, P.J.; Psaltis, A.J.; Vreugde, S. Staphylococcus aureus biofilm exoproteins are cytotoxic to human nasal epithelial barrier in chronic rhinosinusitis. Int. Forum Allergy Rhinol. 2020, 10, 871–883. [Google Scholar] [CrossRef]
- Xiao, N.; Yang, J.; Duan, N.; Lu, B.; Wang, L. Community-Associated Staphylococcus aureus PVL+ ST22 predominates in skin and soft tissue infections in Beijing, China. Infect. Drug Resist. 2019, 12, 2495–2503. [Google Scholar] [CrossRef]
- Besier, S.; Zander, J.; Siegel, E.; Saum, S.H.; Hunfeld, K.-P.; Ehrhart, A.; Brade, V.; Wichelhaus, T.A. Thymidine-Dependent Staphylococcus aureus Small-Colony Variants: Human Pathogens That Are Relevant Not Only in Cases of Cystic Fibrosis Lung Disease. J. Clin. Microbiol. 2008, 46, 3829–3832. [Google Scholar] [CrossRef][Green Version]
- Bates, D.M.; von Eiff, C.; McNamara, P.J.; Peters, G.; Yeaman, M.R.; Bayer, A.S.; Proctor, R.A. Staphylococcus aureus men D and hemB Mutants are as Infective as the Parent Strains, but the Menadione Biosynthetic Mutant Persists within the Kidney. J. Infect. Dis. 2003, 187, 1654–1661. [Google Scholar] [CrossRef]
- Herrmann, M. Antimicrobial Effects Promoting Biofilm Formation and Persistent Disease: The Role of a DNA-Binding Regu-lator, SarA, in Staphylococcal endocarditis. J. Infect. Dis. 2014, 209, 1153–1155. [Google Scholar] [CrossRef]
- Tuchscherr, L.; Pöllath, C.; Siegmund, A.; Deinhardt-Emmer, S.; Hoerr, V.; Svensson, C.-M.; Figge, M.T.; Monecke, S.; Löffler, B. Clinical, S. aureus Isolates Vary in Their Virulence to Promote Adaptation to the Host. Toxins 2019, 11, 135. [Google Scholar] [CrossRef]
- Rom, J.S.; Beenken, K.E.; Ramirez, A.M.; Walker, C.M.; Echols, E.J.; Smeltzer, M.S. Limiting protease production plays a key role in the pathogenesis of the divergent clinical isolates of Staphylococcus aureus LAC and UAMS-1. Virulence 2021, 12, 584–600. [Google Scholar] [CrossRef]
- Spanu, T.; Romano, L.; D’Inzeo, T.; Masucci, L.; Albanese, A.; Papacci, F.; Marchese, E.; Sanguinetti, M.; Fadda, G. Recurrent Ventriculoperitoneal Shunt Infection Caused by Small-Colony Variants of Staphylococcus aureus. Clin. Infect. Dis. 2005, 41, e48–e52. [Google Scholar] [CrossRef]
- Proctor, R.A.; von Eiff, C.; Kahl, B.C.; Becker, K.; McNamara, P.; Herrmann, M.; Peters, G. Small Colony Variants: A Patho-genic Form of Bacteria that Facilitates Persistent and Recurrent Infections. Nat. Rev. Microbiol. 2006, 4, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Xie, Y.; Yang, Z.; Cheng, D.; He, L.; Wang, H.; Liu, Q.; Li, M. The cytoplasmic loops of AgrC contribute to the quorum-sensing activity of Staphylococcus aureus. J. Microbiol. 2021, 59, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.; Yang, S.; Rao, X. Vancomycin resistant Staphylococcus aureus infections: A review of case updating and clinical features. J. Adv. Res. 2020, 21, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Chambers, H.F.; DeLeo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef]
- Kirby, W.M.M. Extraction of a Highly Potent Penicillin Inactivator from Penicillin Resistant Staphylococci. Science 1944, 99, 452–453. [Google Scholar] [CrossRef]
- Rountree, P.M.; Freeman, B.M. Infections caused by a particular phage type of Staphylococcus aureus. Med. J. Aust. 1955, 42, 157–161. [Google Scholar] [CrossRef]
- Barber, M. Methicillin-Resistant Staphylococci. J. Clin. Pathol. 1961, 14, 385–393. [Google Scholar] [CrossRef]
- Jevons, M.P.; Rolinson, G.N.; Knox, R. Celbenin-Resistant Staphylococci. Brit. Med. J. 1961, 2, 939. [Google Scholar] [CrossRef]
- Weigel, L.M.; Clewell, D.B.; Gill, S.R.; Clark, N.C.; McDougal, L.K.; Flannagan, S.E.; Kolonay, J.F.; Shetty, J.; Killgore, G.E.; Tenover, F.C. Genetic Analysis of a High-Level Vancomycin-Resistant Isolate of Staphylococcus aureus. Science 2003, 302, 1569–1571. [Google Scholar] [CrossRef]
- Dunn, S.J.; Connor, C.; McNally, A. The Evolution and Transmission of Multi-Drug Resistant Escherichia coil and Klebsiella Pneumoniae: The Complexity of Clones and Plasmids. Curr. Opin. Microbiol. 2019, 51, 51–56. [Google Scholar] [CrossRef]
- McInnes, R.S.; McCallum, G.E.; Lamberte, L.E.; van Schaik, W. Horizontal Transfer of Antibiotic Resistance Genes in the Human Gut Microbiome. Curr. Opin. Microbiol. 2020, 53, 35–43. [Google Scholar] [CrossRef]
- Munk, P.; Knudsen, B.E.; Lukjancenko, O.; Duarte, A.S.R.; Van Gompel, L.; Luiken, R.E.C.; Smit, L.A.M.; Schmitt, H.; Garcia, A.D.; Hansen, R.B.; et al. Abundance and diversity of the faecal resistome in slaughter pigs and broilers in nine European countries. Nat. Microbiol. 2018, 3, 898–908. [Google Scholar] [CrossRef]
- Sun, J.; Liao, X.; Souza, D.A.W.; Boolchandani, M.; Li, S.; Cheng, K.; Luis Martínez, J.; Li, L.; Feng, Y.; Fang, L.; et al. Environmental Remodeling of Human Gut Microbiota and Antibiotic Resistome in Livestock Farms. Nat. Commun. 2020, 11, 1427. [Google Scholar] [CrossRef]
- Danner, M.C.; Robertson, A.; Behrends, V.; Reiss, J. Antibiotic pollution in surface fresh waters: Occurrence and effects. Sci. Total Environ. 2019, 664, 793–804. [Google Scholar] [CrossRef] [PubMed]
- Naskar, A.; Kim, K.-S. Nanomaterials as Delivery Vehicles and Components of New Strategies to Combat Bacterial Infections: Advantages and Limitations. Microorganisms 2019, 7, 356. [Google Scholar] [CrossRef]
- Kumar, V.A.; Khan, S. Defining multidrug resistance in Gram-negative bacilli. Indian J. Med. Res. 2015, 141, 491–492. [Google Scholar] [CrossRef]
- Cassini, A.; Hogberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. At-tributable Deaths and Disability-Adjusted Life-Years Caused by Infections with Antibiotic-Resistant Bacteria in the EU and the European Economic Area in 2015: A Population-Level Modelling Analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- Stewardson, A.; Marimuthu, K.; Sengupta, S.; Allignol, A.; El-Bouseary, M.; Carvalho, M.J.; Hassan, B.; Delgado-Ramirez, M.A.; Arora, A.; Bagga, R.; et al. Effect of Carbapenem Resistance on Outcomes of Bloodstream Infection Caused by Enterobacteriaceae in Low-Income and Middle-Income Countries (PANORAMA): A Multinational Prospective Co-hort Study. Lancet Infect. Dis. 2019, 19, 601–610. [Google Scholar] [CrossRef]
- Gandra, S.; Tseng, K.K.; Arora, A.; Bhowmik, B.; Robinson, M.L.; Panigrahi, B.; Laxminarayan, R.; Klein, E.Y. The Mortality Burden of Multidrug-resistant Pathogens in India: A Retrospective, Observational Study. Clin. Infect. Dis. 2019, 69, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Li, H.; Ma, X.; Tang, Y.; Tang, H.; Hu, X.; Liu, Z. Small RNA AvrA Regulates IscR to Increase the Stress Tolerances in SmpB Deficiency of Aeromonas veronii. Front Cell Infect Mi. 2019, 9, 142. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.Y.; Ye, Y. A Study on Mining Bibliographic Records by Designed Software SATI: Case Study on Library and Information Science. J. Inf. Resour. Manag. 2012, 2, 50–58. [Google Scholar]
- Lee, J.; Zilm, P.S.; Kidd, S.P. Novel Research Models for Staphylococcus aureus Small Colony Variants (SCV) Development: Co-pathogenesis and Growth Rate. Front. Microbiol. 2020, 11, 321. [Google Scholar] [CrossRef]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef]
- Fraunholz, M.; Sinha, B. Intracellular Staphylococcus aureus: Live-in and let die. Front. Cell. Infect. Microbiol. 2012, 2, 43. [Google Scholar] [CrossRef]
- Strobel, M.; Pförtner, H.; Tuchscherr, L.; Völker, U.; Schmidt, F.; Kramko, N.; Schnittler, H.-J.; Fraunholz, M.; Löffler, B.; Peters, G.; et al. Post-invasion events after infection with Staphylococcus aureus are strongly dependent on both the host cell type and the infecting S. aureus strain. Clin. Microbiol. Infect. 2016, 22, 799–809. [Google Scholar] [CrossRef]
- Häffner, N.; Bär, J.; Haunreiter, V.D.; Shambat, S.M.; Seidl, K.; Crosby, H.A.; Horswill, A.R.; Zinkernagel, A.S. Intracellular Environment and agr System Affect Colony Size Heterogeneity of Staphylococcus aureus. Front. Microbiol. 2020, 11, 1415. [Google Scholar] [CrossRef]
- Balaban, N.Q.; Merrin, J.; Chait, R.; Kowalik, L.; Leibler, S. Bacterial Persistence as a Phenotypic Switch. Science 2004, 305, 1622–1625. [Google Scholar] [CrossRef]
- Wood, T.; Knabel, S.J.; Kwan, B.W. Bacterial Persister Cell Formation and Dormancy. Appl. Environ. Microbiol. 2013, 79, 7116–7121. [Google Scholar] [CrossRef]
- Keren, I.; Kaldalu, N.; Spoering, A.; Wang, Y.; Lewis, K. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 2004, 230, 13–18. [Google Scholar] [CrossRef]
- Singh, R.; Ray, P.; Das, A.; Sharma, M. Role of persisters and small-colony variants in antibiotic resistance of planktonic and biofilm-associated Staphylococcus aureus: An in vitro study. J. Med. Microbiol. 2009, 58, 1067–1073. [Google Scholar] [CrossRef]
- Chen, L.; Wen, Y. The role of bacterial biofilm in persistent infections and control strategies. Int. J. Oral Sci. 2011, 3, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Craft, K.M.; Nguyen, J.M.; Berg, L.J.; Townsend, S.D. Methicillin-Resistant Staphylococcus aureus (MRSA): Antibiotic-Resistance and the Biofilm Phenotype. Medchemcomm 2019, 1, 1231–1241. [Google Scholar] [CrossRef] [PubMed]
- Schilcher, K.; Horswill, A.R. Staphylococcal Biofilm Development: Structure, Regulation, and Treatment Strategies. Microbiol. Mol. Biol. Rev. 2020, 84, e00026-19. [Google Scholar] [CrossRef]
- Landini, P.; Antoniani, D.; Burgess, J.G.; Nijland, R. Molecular Mechanisms of Compounds Affecting Bacterial Biofilm For-mation and Dispersal. Appl. Microbiol. Biot. 2010, 86, 813–823. [Google Scholar] [CrossRef]
- Kohler, C.; Von Eiff, C.; Liebeke, M.; McNamara, P.J.; Lalk, M.; Proctor, R.A.; Hecker, M.; Engelmann, S. A Defect in Menadione Biosynthesis Induces Global Changes in Gene Expression in Staphylococcus aureus. J. Bacteriol. 2008, 190, 6351–6364. [Google Scholar] [CrossRef]
- Idrees, M.; Sawant, S.; Karodia, N.; Rahman, A. Staphylococcus aureus Biofilm: Morphology, Genetics, Pathogenesis and Treatment Strategies. Int. J. Environ. Res. Public Health 2021, 18, 7602. [Google Scholar] [CrossRef]
- Reffuveille, F.; Josse, J.; Vallé, Q.; Gangloff, C.M.; Gangloff, S.C. Staphylococcus aureus Biofilms and Their Impact on the Medical Field. In The Rise of Virulence and Antibiotic Resistance in Staphylococcus aureus; InTech: Rijeka, Croatia, 2017; p. 187. [Google Scholar]
- Nguyen, H.T.T.; Nguyen, T.H.; Otto, M. The Staphylococcal Exopolysaccharide PIA—Biosynthesis and Role in Biofilm Formation, Colonization, and Infection. Comput. Struct. Biotec. 2020, 18, 3324–3334. [Google Scholar] [CrossRef]
- Corrigan, R.; Rigby, D.; Handley, P.; Foster, T.J. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 2007, 153, 2435–2446. [Google Scholar] [CrossRef]
- Hauck, C.R.; Ohlsen, K. Sticky Connections: Extracellular Matrix Protein Recognition and Integrin-Mediated Cellular Invasion by Staphylococcus aureus Christof R Hauck and Knut Ohlsen. Curr. Opin. Microbiol. 2006, 9, 5–11. [Google Scholar] [CrossRef]
- Foster, T.J.; Geoghegan, J.A.; Ganesh, V.K.; Höök, M. Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 2014, 12, 49–62. [Google Scholar] [CrossRef]
- Idrees, M.; Mohammad, A.R.; Karodia, N.; Rahman, A. Multimodal Role of Amino Acids in Microbial Control and Drug Development. Antibiotics 2020, 9, 330. [Google Scholar] [CrossRef]
- Taglialegna, A.; Lasa, I.; Valle, J. Amyloid Structures as Biofilm Matrix Scaffolds. J. Bacteriol. 2016, 198, 2579–2588. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Lin, S.; Soteyome, T.; Peters, B.M.; Li, Y.; Chen, H.; Su, J.; Li, L.; Li, B.; Xu, Z.; et al. Biofilm Formation of Staphylococcus aureus under Food Heat Processing Conditions: First Report on CML Production within Biofilm. Sci. Rep. 2019, 9, 1312. [Google Scholar] [CrossRef] [PubMed]
- Kong, K.-F.; Vuong, C.; Otto, M. Staphylococcus quorum sensing in biofilm formation and infection. Int. J. Med. Microbiol. 2006, 296, 133–139. [Google Scholar] [CrossRef]
- Lu, L.; Hu, W.; Tian, Z.; Yuan, D.; Yi, G.; Zhou, Y.; Cheng, Q.; Zhu, J.; Li, M. Developing Natural Products as Potential Anti-Biofilm Agents. Chin. Med. UK 2019, 14, 11. [Google Scholar] [CrossRef]
- Dunman, P.M.; Murphy, E.; Haney, S.; Palacios, D.; Tucker-Kellogg, G.; Wu, S.; Brown, E.L.; Zagursky, R.J.; Shlaes, D.; Projan, S.J. Transcription Profiling-Based Identification of Staphylococcus aureus Genes Regulated by the agr and/or sarA Loci. J. Bacteriol. 2001, 183, 7341–7353. [Google Scholar] [CrossRef]
- Harraghy, N.; Kerdudou, S.; Herrmann, M. Quorum-sensing systems in Staphylococci as therapeutic targets. Anal. Bioanal. Chem. 2006, 387, 437–444. [Google Scholar] [CrossRef]
- Gordon, C.P.; Olson, S.D.; Lister, J.L.; Kavanaugh, J.S.; Horswill, A.R. Truncated Autoinducing Peptides as Antagonists of Staphylococcus lugdunensis Quorum Sensing. J. Med. Chem. 2016, 59, 8879–8888. [Google Scholar] [CrossRef]
- Novick, R.P.; Ross, H.F.; Projan, S.J.; Kornblum, J.; Kreiswirth, B.; Moghazeh, S. Synthesis of Staphylococcal Virulence Factors is Controlled by a Regulatory RNA Molecule. Embo J. 1993, 12, 3967–3975. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Wang, R.; Khan, B.A.; Sturdevant, D.E.; Otto, M. Role of the Accessory Gene Regulator Agr in Community-Associated Methicillin-Resistant Staphylococcus aureus Pathogenesis. Infect. Immun. 2011, 79, 1927–1935. [Google Scholar] [CrossRef]
- Le, K.Y.; Villaruz, A.E.; Zheng, Y.; He, L.; Fisher, E.L.; Nguyen, T.H.; Ho, T.V.; Yeh, A.J.; Joo, H.-S.; Cheung, G.Y.; et al. Role of Phenol-Soluble Modulins in Staphylococcus epidermidis Biofilm Formation and Infection of Indwelling Medical Devices. J. Mol. Biol. 2019, 431, 3015–3027. [Google Scholar] [CrossRef]
- Boles, B.R.; Horswill, A.R. Agr-Mediated Dispersal of Staphylococcus aureus Biofilms. PLoS Pathog. 2008, 4, e1000052. [Google Scholar] [CrossRef]
- Lister, J.L.; Horswill, A.R. Staphylococcus aureus biofilms: Recent developments in biofilm dispersal. Front. Cell. Infect. Microbiol. 2014, 4, 178. [Google Scholar] [CrossRef]
- Giraudo, A.T.; Calzolari, A.; Cataldi, A.A.; Bogni, C.; Nagel, R. The Sae Locus of Staphylococcus aureus Encodes a Two-Component Regulatory System. Fems Microbiol. Lett. 1999, 180, 117. [Google Scholar] [PubMed]
- Olson, M.E.; Nygaard, T.K.; Ackermann, L.; Watkins, R.L.; Zurek, O.W.; Pallister, K.B.; Griffith, S.; Kiedrowski, M.R.; Flack, C.E.; Kavanaugh, J.S.; et al. Staphylococcus aureus Nuclease is an SaeRS-Dependent Virulence Factor. Infect. Immun. 2013, 81, 1316–1324. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.Q.; Willard, J.; Yeaman, M.R.; Cheung, A.L.; Bayer, A.S. Regulation of Staphylococcus aureus Alpha-Toxin Gene (Hla) Expression by Agr, sarA, and Sae in vitro and in Experimental Infective Endocarditis. J. Infect. Dis. 2006, 194, 1267–1275. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Yu, C.; Sun, J.; Liu, H.; Landwehr, C.; Holmes, D.; Ji, Y. Inactivation of a Two-Component Signal Transduction System, SaeRS, Eliminates Adherence and Attenuates Virulence of Staphylococcus aureus. Infect. Immun. 2006, 74, 4655–4665. [Google Scholar] [CrossRef] [PubMed]
- Rogasch, K.; Rühmling, V.; Pané-Farré, J.; Höper, D.; Weinberg, C.; Fuchs, S.; Schmudde, M.; Bröker, B.; Wolz, C.; Hecker, M.; et al. Influence of the Two-Component System SaeRS on Global Gene Expression in Two Different Staphylococcus aureus Strains. J. Bacteriol. 2006, 188, 7742–7758. [Google Scholar] [CrossRef]
- Flack, C.E.; Zurek, O.W.; Meishery, D.D.; Pallister, K.B.; Malone, C.L.; Horswill, A.R.; Voyich, J.M. Differential regulation of staphylococcal virulence by the sensor kinase SaeS in response to neutrophil-derived stimuli. Proc. Natl. Acad. Sci. USA 2014, 111, E2037–E2045. [Google Scholar] [CrossRef]
- Sadykov, M.R.; Bayles, K.W. The control of death and lysis in staphylococcal biofilms: A coordination of physiological signals. Curr. Opin. Microbiol. 2012, 15, 211–215. [Google Scholar] [CrossRef]
- Rice, K.C.; Firek, B.A.; Nelson, J.B.; Yang, S.-J.; Patton, T.G.; Bayles, K.W. The Staphylococcus aureus cidAB Operon: Evaluation of Its Role in Regulation of Murein Hydrolase Activity and Penicillin Tolerance. J. Bacteriol. 2003, 185, 2635–2643. [Google Scholar] [CrossRef]
- Groicher, K.H.; Firek, B.A.; Fujimoto, D.F.; Bayles, K.W. The Staphylococcus aureus lrgAB Operon Modulates Murein Hydro-lase Activity and Penicillin Tolerance. J. Bacteriol. 2000, 182, 1794–1801. [Google Scholar] [CrossRef]
- Ranjit, D.K.; Endres, J.L.; Bayles, K.W. Staphylococcus aureus CidA and LrgA Proteins Exhibit Holin-Like Properties. J. Bacteriol. 2011, 193, 2468–2476. [Google Scholar] [CrossRef]
- Brunskill, E.W.; Bayles, K.W. Identification and Molecular Characterization of a Putative Regulatory Locus that Affects Autolysis in Staphylococcus aureus. J. Bacteriol. 1996, 178, 611–618. [Google Scholar] [CrossRef]
- Schmidt, K.A.; Manna, A.C.; Cheung, A.L. SarT influences sarS expression in Staphylococcus aureus. Infect. Immun. 2003, 71, 5139–5148. [Google Scholar] [CrossRef]
- Heinrichs, J.H.; Bayer, M.G.; Cheung, A.L. Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. J. Bacteriol. 1996, 178, 418–423. [Google Scholar] [CrossRef]
- Zielinska, A.K.; Beenken, K.E.; Mrak, L.N.; Spencer, H.J.; Post, G.R.; Skinner, R.A.; Tackett, A.J.; Horswill, A.R.; Smeltzer, M.S. SarA-mediated Repression of Protease Production Plays a Key Role in the Pathogenesis of Staphylococcus aureus USA300 Isolates. Mol. Microbiol. 2012, 86, 1183–1196. [Google Scholar] [CrossRef]
- McNamara, P.J.; Milligan-Monroe, K.C.; Khalili, S.; Proctor, R.A. Identification, Cloning, and Initial Characterization of rot, a Locus Encoding a Regulator of Virulence Factor Expression in Staphylococcus aureus. J. Bacteriol. 2000, 182, 3197–3203. [Google Scholar] [CrossRef]
- Saïd-Salim, B.; Dunman, P.M.; McAleese, F.M.; Macapagal, D.; Murphy, E.; McNamara, P.J.; Arvidson, S.; Foster, T.J.; Projan, S.J.; Kreiswirth, B.N. Global Regulation of Staphylococcus aureus Genes by Rot. J. Bacteriol. 2003, 185, 610–619. [Google Scholar] [CrossRef]
- Tuffs, S.W.; Herfst, C.A.; Baroja, M.L.; Podskalniy, V.A.; DeJong, E.N.; Coleman, C.E.M.; McCormick, J.K. Regulation of Toxic Shock Syndrome Toxin-1 by the Accessory Gene Regulator in Staphylococcus aureus is Mediated by the Repressor of Toxins. Mol. Microbiol. 2019, 112, 1163–1177. [Google Scholar] [CrossRef]
- Geisinger, E.; Adhikari, R.P.; Jin, R.; Ross, H.F.; Novick, R.P. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol. Microbiol. 2006, 61, 1038–1048. [Google Scholar] [CrossRef]
- Boisset, S.; Geissmann, T.; Huntzinger, E.; Fechter, P.; Bendridi, N.; Possedko, M.; Chevalier, C.; Helfer, A.C.; Benito, Y.; Jacquier, A.; et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007, 21, 1353–1366. [Google Scholar] [CrossRef]
- Crosby, H.; Schlievert, P.; Merriman, J.A.; King, J.M.; Salgado-Pabon, W.; Horswill, A.R. The Staphylococcus aureus Global Regulator MgrA Modulates Clumping and Virulence by Controlling Surface Protein Expression. PLoS Pathog. 2016, 12, e1005604. [Google Scholar] [CrossRef]
- Pane-Farre, J.; Jonas, B.; Foerstner, K.; Engelmann, S.; Hecker, M. The sigma(B) Regulon in Staphylococcus aureus and its Regulation. Int. J. Med. Microbiol. 2006, 296, 237–258. [Google Scholar] [CrossRef] [PubMed]
- Atwood, D.N.; Loughran, A.J.; Courtney, A.P.; Anthony, A.C.; Meeker, D.G.; Spencer, H.J.; Gupta, R.K.; Lee, C.Y.; Beenken, K.E.; Smeltzer, M.S. Comparative impact of diverse regulatory loci on Staphylococcus aureus biofilm formation. MicrobiologyOpen 2015, 4, 436–451. [Google Scholar] [CrossRef] [PubMed]
- Kullik, I.; Giachino, P.; Fuchs, T. Deletion of the Alternative Sigma Factor sigma(B) in Staphylococcus aureus Reveals its Function as a Global Regulator of Virulence Genes. J. Bacteriol. 1998, 180, 4814–4820. [Google Scholar] [CrossRef] [PubMed]
- Donegan, N.P.; Cheung, A.L. Regulation of the mazEF Toxin-Antitoxin Module in Staphylococcus aureus and Its Impact on sigB Expression. J. Bacteriol. 2009, 191, 2795–2805. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, M.; Dunman, P.; Kormanec, J.; Macapagal, D.; Murphy, E.; Mounts, W.; Berger-Bachi, B.; Projan, S. Microar-ray-Based Analysis of the Staphylococcus aureus sigma(B) Regulon. J. Bacteriol. 2004, 186, 4085–4099. [Google Scholar] [CrossRef]
- Nair, S.P.; Bischoff, M.; Senn, M.M.; Berger-Bachi, B. The sigma(B) Regulon Influences Internalization of Staphylococcus aureus by Osteoblasts. Infect. Immun. 2003, 71, 4167–4170. [Google Scholar] [CrossRef]
- Bischoff, M.; Entenza, J.M.; Giachino, P. Influence of a Functional sigB Operon on the Global Regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 2001, 183, 4079–4089. [Google Scholar] [CrossRef]
- Horsburgh, M.J.; Aish, J.L.; White, I.J.; Shaw, L.; Lithgow, J.K.; Foster, S.J. Sigma(B) Modulates Virulence Determinant Expression and Stress Resistance: Characterization of a Functional rsbU Strain Derivedfrom Staphylococcus aureus 8325–4. J. Bacteriol. 2002, 184, 5457–5467. [Google Scholar] [CrossRef] [PubMed]
- Geiger, T.; Goerke, C.; Mainiero, M.; Kraus, D.; Wolz, C. The Virulence Regulator Sae of Staphylococcus aureus: Promoter Activities and Response to Phagocytosis-Related Signals. J. Bacteriol. 2008, 190, 3419–3428. [Google Scholar] [CrossRef] [PubMed]
- Stenz, L.; Francois, P.; Whiteson, K.; Wolz, C.; Linder, P.; Schrenzel, J. The CodY pleiotropic repressor controls virulence in gram-positive pathogens. FEMS Immunol. Med. Microbiol. 2011, 62, 123–139. [Google Scholar] [CrossRef] [PubMed]
- Pohl, K.; Francois, P.; Stenz, L.; Schlink, F.; Geiger, T.; Herbert, S.; Goerke, C.; Schrenzel, J.; Wolz, C. CodY in Staphylococcus aureus: A Regulatory Link between Metabolism and Virulence Gene Expression. J. Bacteriol. 2009, 191, 2953–2963. [Google Scholar] [CrossRef]
- Majerczyk, C.D.; Dunman, P.M.; Luong, T.T.; Lee, C.Y.; Sadykov, M.R.; Somerville, G.; Bodi, K.; Sonenshein, A.L. Direct Targets of CodY in Staphylococcus aureus. J. Bacteriol. 2010, 192, 2861–2877. [Google Scholar] [CrossRef]
- Waters, N.R.; Samuels, D.J.; Behera, R.K.; Livny, J.; Rhee, K.Y.; Sadykov, M.R.; Brinsmade, S.R. A Spectrum of CodY Activities Drives Metabolic Reorganization and Virulence Gene Expression in Staphylococcus aureus. Mol. Microbiol. 2016, 101, 495–514. [Google Scholar] [CrossRef]
- Majerczyk, C.D.; Sadykov, M.R.; Luong, T.T.; Lee, C.; Somerville, G.A.; Sonenshein, A.L. Staphylococcus aureus CodY Nega-tively Regulates Virulence Gene Expression. J. Bacteriol. 2008, 190, 2257–2265. [Google Scholar] [CrossRef]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal Biofilms. Microbiol. Spectr. 2018, 6, 10. [Google Scholar] [CrossRef]
- Verma, J.; Bag, S.; Saha, B.; Kumar, P.; Ghosh, T.S.; Dayal, M.; Senapati, T.; Mehra, S.; Dey, P.; Desigamani, A.; et al. Genomic plasticity associated with antimicrobial resistance inVibrio cholerae. Proc. Natl. Acad. Sci. USA 2019, 116, 6226–6231. [Google Scholar] [CrossRef]
- Hoiby, N.; Johansen, H.K.; Moser, C.; Song, Z.J.; Ciofu, O.; Kharazmi, A. Pseudomonas aeruginosa and the in vitro and in vivo Biofilm Mode of Growth. Microbes Infect. 2001, 3, 23–35. [Google Scholar] [CrossRef]
- Bassetti, M.; Vena, A.; Croxatto, A.; Righi, E.; Guery, B. How to manage Pseudomonas aeruginosa infections. Drugs Context 2018, 7, 212527. [Google Scholar] [CrossRef]
- Cramton, S.E.; Gerke, C.; Schnell, N.F.; Nichols, W.W.; Gotz, F. The Intercellular Adhesion (Ica) Locus is Present in Staphylococcus aureus and is Required for Biofilm Formation. Infect. Immun. 1999, 67, 5427–5433. [Google Scholar] [CrossRef]
- Gotz, F. Staphylococcus and Biofilms. Mol. Microbiol. 2002, 43, 1367–1378. [Google Scholar] [CrossRef]
- Donlan, R.M. Role of Biofilms in Antimicrobial Resistance. Asaio J. 2000, 46, S47–S52. [Google Scholar] [CrossRef]
- Ou, C.; Shang, D.; Yang, J.; Chen, B.; Chang, J.; Jin, F.; Shi, C. Prevalence of Multidrug-Resistant Staphylococcus aureus Isolates with Strong Biofilm Formation Ability Among Animal-Based Food in Shanghai. Food Control. 2020, 112, 107106. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial Biofilms: A Common Cause of Persistent Infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Bjarnsholt, T.; Jensen, P.O.; Fiandaca, M.J.; Pedersen, J.; Hansen, C.R.; Andersen, C.B.; Pressler, T.; Givskov, M.; Hoiby, N. Pseudomonas aeruginosa Biofilms in the Respiratory Tract of Cystic Fibrosis Patients. Pediatr. Pulm. 2009, 44, 547–558. [Google Scholar] [CrossRef]
- Wu, H.; Moser, C.; Wang, H.-Z.; Høiby, N.; Song, Z.-J. Strategies for combating bacterial biofilm infections. Int. J. Oral Sci. 2015, 7, 1–7. [Google Scholar] [CrossRef]
- Miyaue, S.; Suzuki, E.; Komiyama, Y.; Kondo, Y.; Morikawa, M.; Maeda, S. Bacterial Memory of Persisters: Bacterial Persister Cells Can Retain Their Phenotype for Days or Weeks After Withdrawal from Colony–Biofilm Culture. Front. Microbiol. 2018, 9, 1396. [Google Scholar] [CrossRef]
- Wood, T.K. Strategies for combating persister cell and biofilm infections. Microb. Biotechnol. 2017, 10, 1054–1056. [Google Scholar] [CrossRef] [PubMed]
- Conlon, B.P.; Rowe, S.E.; Lewis, K. Persister Cells in Biofilm Associated Infections. In Advances in Experimental Medicine and Biology; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2014; Volume 831, pp. 1–9. [Google Scholar]
- Anderl, J.N.; Zahller, J.; Roe, F.; Stewart, P.S. Role of Nutrient Limitation and Stationary-Phase Existence in Klebsiella Pneumoniae Biofilm Resistance to Ampicillin and Ciprofloxacin. Antimicrob. Agents Chemother. 2003, 47, 1251–1256. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.R.W.; Allison, D.G.; Gilbert, P. Resistance of bacterial biofilms to antibiotics a growth-rate related effect? J. Antimicrob. Chemother. 1988, 22, 777–780. [Google Scholar] [CrossRef] [PubMed]
- Waters, E.; Rowe, S.E.; O’Gara, J.P.; Conlon, B.P. Convergence of Staphylococcus aureus Persister and Biofilm Research: Can Biofilms Be Defined as Communities of Adherent Persister Cells? PLoS Pathog. 2016, 12, e1006012. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Bryers, J.D. Non-invasive determination of conjugative transfer of plasmids bearing antibiotic-resistance genes in biofilm-bound bacteria: Effects of substrate loading and antibiotic selection. Appl. Microbiol. Biotechnol. 2012, 97, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Proctor, R.A.; Van Langevelde, P.; Kristjansson, M.; Maslow, J.N.; Arbeit, R.D. Persistent and Relapsing Infections Associated with Small-Colony Variants of Staphylococcus aureus. Clin. Infect. Dis. 1995, 20, 95–102. [Google Scholar] [CrossRef]
- Kim, N.-H.; Kang, Y.M.; Han, W.D.; Park, K.U.; Park, K.-H.; Yoo, J.I.; Lee, D.-G.; Park, C.; Song, K.-H.; Kim, E.S.; et al. Small-Colony Variants in Persistent and Recurrent Staphylococcus aureus Bacteremia. Microb. Drug Resist. 2016, 22, 538–544. [Google Scholar] [CrossRef]
- Kussmann, M.; Karer, M.; Obermueller, M.; Schmidt, K.; Barousch, W.; Moser, D.; Nehr, M.; Ramharter, M.; Poeppl, W.; Makristathis, A.; et al. Emergence of a Dalbavancin Induced Glycopep-tide/Lipoglycopeptide Non-Susceptible Staphylococcus aureus During Treatment of a Cardiac Device-Related Endocarditis. Emerg. Microbes Infec. 2018, 7, 202. [Google Scholar] [CrossRef]
- Schwerdt, M.; Neumann, C.; Schwartbeck, B.; Kampmeier, S.; Herzog, S.; Görlich, D.; Dübbers, A.; Große-Onnebrink, J.; Kessler, C.; Küster, P.; et al. Staphylococcus aureus in the airways of cystic fibrosis patients—A retrospective long-term study. Int. J. Med. Microbiol. 2018, 308, 631–639. [Google Scholar] [CrossRef]
- VonEiff, C.; Bettin, D.; Proctor, R.A.; Rolauffs, B.; Lindner, N.; Winkelmann, W.; Peters, G. Recovery of Small Colony Variants of Staphylococcus aureus Following Gentamicin Bead Placement for Osteomyelitis. Clin. Infect. Dis. 1997, 25, 1250–1251. [Google Scholar] [CrossRef]
- Besier, S.; Smaczny, C.; von Mallinckrodt, C.; Krahl, A.; Ackermann, H.; Brade, V.; Wichelhaus, T.A. Prevalence and Clinical Significance of Staphylococcus aureus Small-Colony Variants in Cystic Fibrosis Lung Disease. J. Clin. Microbiol. 2007, 45, 168–172. [Google Scholar] [CrossRef]
- Kahl, B.C.; Belling, G.; Reichelt, R.; Herrmann, M.; Proctor, R.A.; Peters, G. Thymidine-Dependent Small-Colony Variants of Staphylococcus aureus Exhibit Gross Morphological and Ultrastructural Changes Consistent with Impaired Cell Separation. J. Clin. Microbiol. 2003, 41, 410–413. [Google Scholar] [CrossRef]
- Kahl, B.; Herrmann, M.; Everding, A.S.; Koch, H.G.; Becker, K.; Harms, E.; Proctor, R.A.; Peters, G. Persistent Infection with Small Colony Variant Strains of Staphylococcus aureus in Patients with Cystic Fibrosis. J. Infect. Dis. 1998, 177, 1023–1029. [Google Scholar] [CrossRef]
- Leimer, N.; Rachmühl, C.; Marques, M.P.; Bahlmann, A.S.; Furrer, A.; Eichenseher, F.; Seidl, K.; Matt, U.; Loessner, M.J.; Schuepbach, R.A.; et al. Nonstable Staphylococcus aureus Small-Colony Variants are Induced by Low pH and Sensitized to Antimicrobial Therapy by Phagolysosomal Alkalinization. J. Infect. Dis. 2016, 213, 305–313. [Google Scholar] [CrossRef]
- Tuchscherr, L.; Loeffler, B.; Proctor, R.A. Persistence of Staphylococcus aureus: Multiple Metabolic Pathways Impact the Expression of Virulence Factors in Small-Colony Variants (SCVs). Front. Microbiol. 2020, 11, 1028. [Google Scholar] [CrossRef]
- Kahl, B.C. Small colony variants (SCVs) of Staphylococcus aureus—A bacterial survival strategy. Infect. Genet. Evol. 2014, 21, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Sendi, P.; Proctor, R.A. Staphylococcus aureus as an intracellular pathogen: The role of small colony variants. Trends Microbiol. 2009, 17, 54–58. [Google Scholar] [CrossRef]
- Proctor, R. Respiration and Small Colony Variants of Staphylococcus aureus. Microbiol. Spectr. 2019, 7, 22. [Google Scholar] [CrossRef]
- Oun, S.; Redder, P.; Didier, J.-P.; François, P.; Corvaglia, A.-R.; Buttazzoni, E.; Giraud, C.; Girard, M.; Schrenzel, J.; Linder, P. The CshA DEAD-box RNA helicase is important for quorum sensing control in Staphylococcus aureus. RNA Biol. 2013, 10, 157–165. [Google Scholar] [CrossRef]
- Roux, C.M.; DeMuth, J.P.; Dunman, P.M. Characterization of Components of the Staphylococcus aureus mRNA Degradosome Holoenzyme-Like Complex. J. Bacteriol. 2011, 193, 5520–5526. [Google Scholar] [CrossRef]
- Tuchscherr, L.; Heitmann, V.; Hussain, M.; Viemann, D.; Roth, J.; von Eiff, C.; Peters, G.; Becker, K.; Loeffler, B. Staphylococcus aureus Small-Colony Variants are Adapted Phenotypes for Intracellular Persistence. J. Infect. Dis. 2010, 202, 1031–1040. [Google Scholar] [CrossRef] [PubMed]
- Tuchscherr, L.; Bischoff, M.; Lattar, S.M.; Noto Llana, M.; Pförtner, H.; Niemann, S.; Geraci, J.; Van De Vyver, H.; Fraunholz, M.J.; Cheung, A.L.; et al. Sigma Factor SigB Is Crucial to Mediate Staphylococcus aureus Adaptation during Chronic Infections. PLoS Pathog. 2015, 11, e1004870. [Google Scholar] [CrossRef] [PubMed]
- Deora, R.; Tseng, T.; Misra, T.K. Alternative transcription factor sigmaSB of Staphylococcus aureus: Characterization and role in transcription of the global regulatory locus sar. J. Bacteriol. 1997, 179, 6355–6359. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.L.; Zhang, G. Global regulation of virulence determinants in Staphylococcus aureus by the SarA protein family. Front. Biosci. Landmark 2002, 7, D1825–D1842. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.M.; Park, B.S.; Kim, J.-I.; Kim, S.E.; Lee, J.; Oh, S.C.; Enkhbayar, P.; Matsushima, N.; Lee, H.; Yoo, O.J.; et al. Crystal Structure of the TLR4-MD-2 Complex with Bound Endotoxin Antagonist Eritoran. Cell 2007, 130, 906–917. [Google Scholar] [CrossRef] [PubMed]
- Samuelsen, O.; Haukland, H.H.; Kahl, B.C.; von Eiff, C.; Proctor, R.A.; Ulvatne, H.; Sandvik, K.; Vorland, L.H. Staphylococcus aureus Small Colony Variants are Resistant to the Antimicrobial Peptide Lactoferricin B. J. Antimicrob. Chemoth. 2005, 56, 1126–1129. [Google Scholar] [CrossRef]
- Werth, N.; Beerlage, C.; Rosenberger, C.; Yazdi, A.S.; Edelmann, M.; Amr, A.; Bernhardt, W.; Von Eiff, C.; Becker, K.; Schäfer, A.; et al. Activation of Hypoxia Inducible Factor 1 Is a General Phenomenon in Infections with Human Pathogens. PLoS ONE 2010, 5, e11576. [Google Scholar] [CrossRef]
- Tuchscherr, L.; Medina, E.; Hussain, M.; Völker, W.; Heitmann, V.; Niemann, S.; Holzinger, D.; Roth, J.; Proctor, R.A.; Becker, K.; et al. Staphylococcus aureus phenotype switching: An effective bacterial strategy to escape host immune response and establish a chronic infection. EMBO Mol. Med. 2011, 3, 129–141. [Google Scholar] [CrossRef]
- Arts, R.J.; Joosten, L.A.; Netea, M.G. Immunometabolic circuits in trained immunity. Semin. Immunol. 2016, 28, 425–430. [Google Scholar] [CrossRef]
- Lung, T.W.F.; Monk, I.R.; Acker, K.P.; Mu, A.; Wang, N.; Riquelme, S.A.; Pires, S.; Noguera, L.P.; Dach, F.; Gabryszewski, S.J.; et al. Staphylococcus aureus small colony variants impair host immunity by activating host cell glycolysis and inducing necroptosis. Nat. Microbiol. 2020, 5, 141–153. [Google Scholar] [CrossRef]
- Arts, R.J.W.; Novakovic, B.; ter Horst, R.; Carvalho, A.; Bekkering, S.; Lachmandas, E.; Rodrigues, F.; Silvestre, R.; Cheng, S.; Wang, S.; et al. Glutaminolysis and Fumarate Accumulation Integrate Immunometabolic and Epigenetic Programs in Trained Immunity. Cell Metab. 2016, 24, 807–819. [Google Scholar] [CrossRef]
- Lechner, S.; Lewis, K.; Bertram, R. Staphylococcus aureus Persisters Tolerant to Bactericidal Antibiotics. J. Mol. Microbiol. Biotechnol. 2012, 22, 235–244. [Google Scholar] [CrossRef]
- Proctor, R.; Vesga, O.; Otten, M.; Koo, S.-P.; Yeaman, M.; Sahl, H.-G.; Bayer, A. Staphylococcus aureus Small Colony Variants Cause Persistent and Resistant Infections. Chemotherapy 1996, 42, 47–52. [Google Scholar] [CrossRef]
- Melter, O.; Radojevič, B. Small colony variants of Staphylococcus aureus—Review. Folia. Microbiol. 2010, 55, 548–558. [Google Scholar] [CrossRef]
- Garcia, L.G.; Lemaire, S.; Kahl, B.C.; Becker, K.; Proctor, R.A.; Denis, O.; Tulkens, P.M.; Van Bambeke, F. Antibiotic activity against small-colony variants of Staphylococcus aureus: Review of in vitro, animal and clinical data. J. Antimicrob. Chemother. 2013, 68, 1455–1464. [Google Scholar] [CrossRef]
- McNamara, P.J.; Proctor, A.R. Staphylococcus aureus small colony variants, electron transport and persistent infections. Int. J. Antimicrob. Agents 2000, 14, 117–122. [Google Scholar] [CrossRef]
- Darouiche, O.R.; Hamill, R.J. Antibiotic penetration of and bactericidal activity within endothelial cells. Antimicrob. Agents Chemother. 1994, 38, 1059–1064. [Google Scholar] [CrossRef][Green Version]
- Mishra, R.; Panda, A.K.; De Mandal, S.; Shakeel, M.; Bisht, S.S.; Khan, J. Natural Anti-Biofilm Agents: Strategies to Control Biofilm-Forming Pathogens. Front. Microbiol. 2020, 11, 566325. [Google Scholar] [CrossRef]
- An, A.Y.; Choi, K.-Y.G.; Baghela, A.S.; Hancock, R.E.W. An Overview of Biological and Computational Methods for Designing Mechanism-InforMed. Anti-biofilm Agents. Front. Microbiol. 2021, 12, 640787. [Google Scholar] [CrossRef]
- Malaekeh-Nikouei, B.; Bazzaz, B.S.F.; Mirhadi, E.; Tajani, A.S.; Khameneh, B. The role of nanotechnology in combating biofilm-based antibiotic resistance. J. Drug Deliv. Sci. Technol. 2020, 60, 101880. [Google Scholar] [CrossRef]
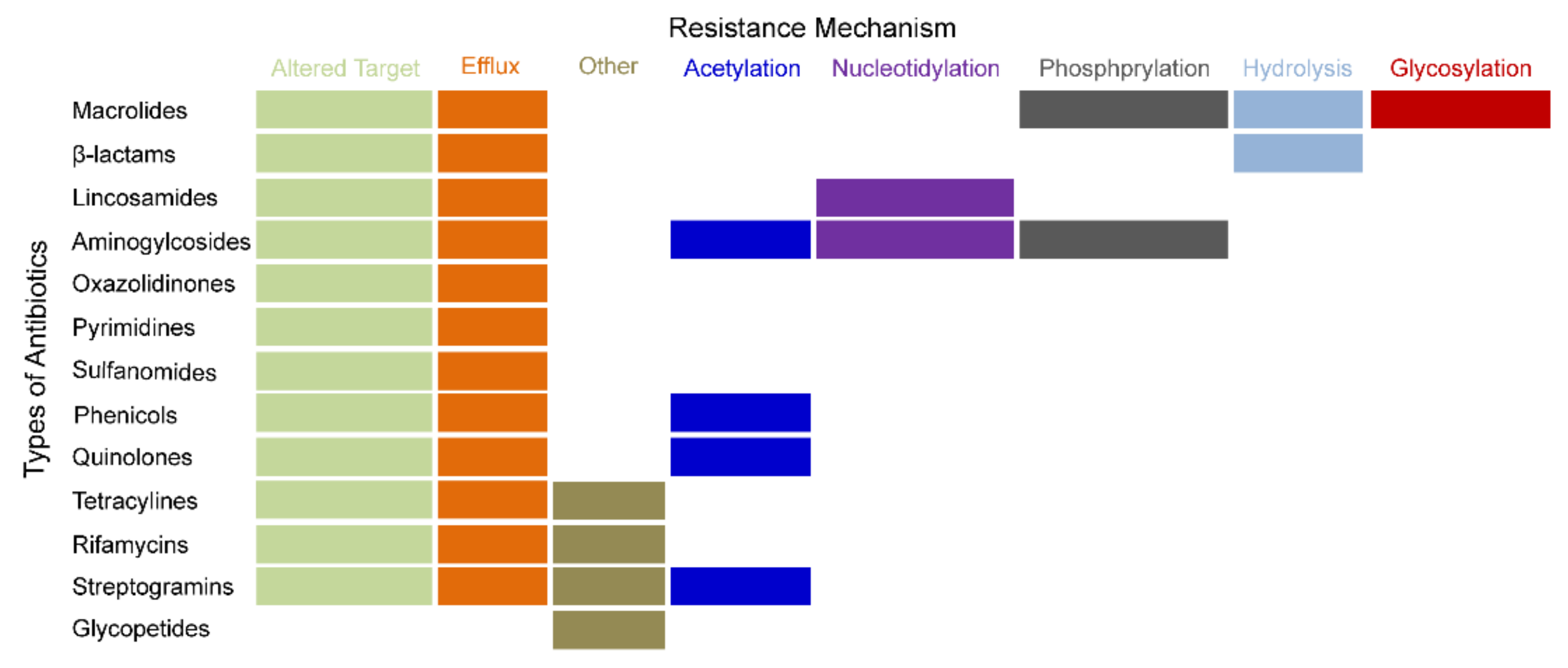

| Resistant Stage | Occurrence Time | Mechanism | Drug Resistance | Typical Strain | Reference |
|---|---|---|---|---|---|
| Penicillin-resistant strains | In the mid- 1940s | Plasmid-encoded penicillinase hydrolysing the β-lactam ring of penicillin | Penicillin | Phage type 80/81 S. aureus | [26,27] |
| Methicillin-resistant S. aureus (MRSA) | 1961 | Gene mecA and mecC encoding the low-affinity penicillin-binding protein PBP2A or PBP2ALGA | Entire β-lactam class of antibiotics | MRSA COMRSAL | [28,29] |
| Vancomycin-resistant S. aureus (VRSA) | At the end of the 20th century | Mediated by vanA gene cluster | Vancomycin | N/A | [24,30] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, H.; Tong, Y.; Cheng, J.; Abbas, Z.; Li, Z.; Wang, J.; Zhou, Y.; Si, D.; Zhang, R. Biofilm and Small Colony Variants—An Update on Staphylococcus aureus Strategies toward Drug Resistance. Int. J. Mol. Sci. 2022, 23, 1241. https://doi.org/10.3390/ijms23031241
Guo H, Tong Y, Cheng J, Abbas Z, Li Z, Wang J, Zhou Y, Si D, Zhang R. Biofilm and Small Colony Variants—An Update on Staphylococcus aureus Strategies toward Drug Resistance. International Journal of Molecular Sciences. 2022; 23(3):1241. https://doi.org/10.3390/ijms23031241
Chicago/Turabian StyleGuo, Henan, Yucui Tong, Junhao Cheng, Zaheer Abbas, Zhongxuan Li, Junyong Wang, Yichen Zhou, Dayong Si, and Rijun Zhang. 2022. "Biofilm and Small Colony Variants—An Update on Staphylococcus aureus Strategies toward Drug Resistance" International Journal of Molecular Sciences 23, no. 3: 1241. https://doi.org/10.3390/ijms23031241
APA StyleGuo, H., Tong, Y., Cheng, J., Abbas, Z., Li, Z., Wang, J., Zhou, Y., Si, D., & Zhang, R. (2022). Biofilm and Small Colony Variants—An Update on Staphylococcus aureus Strategies toward Drug Resistance. International Journal of Molecular Sciences, 23(3), 1241. https://doi.org/10.3390/ijms23031241







