Cognate RNA-Binding Modes by the Alternative-Splicing Regulator MBNL1 Inferred from Molecular Dynamics
Abstract
1. Introduction
2. Results and Discussion
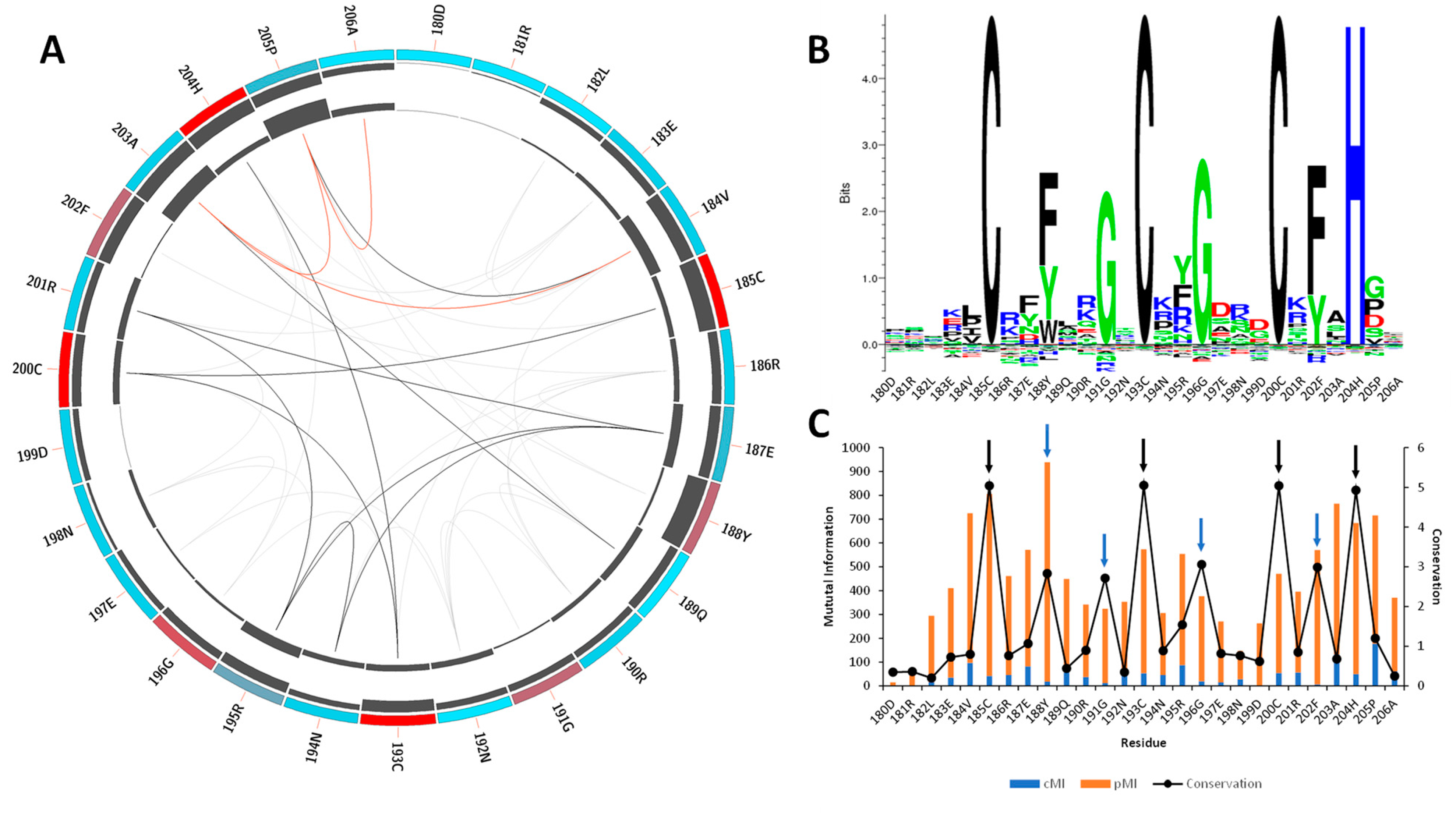
2.1. Sequence Coevolution of the CCCH Domain
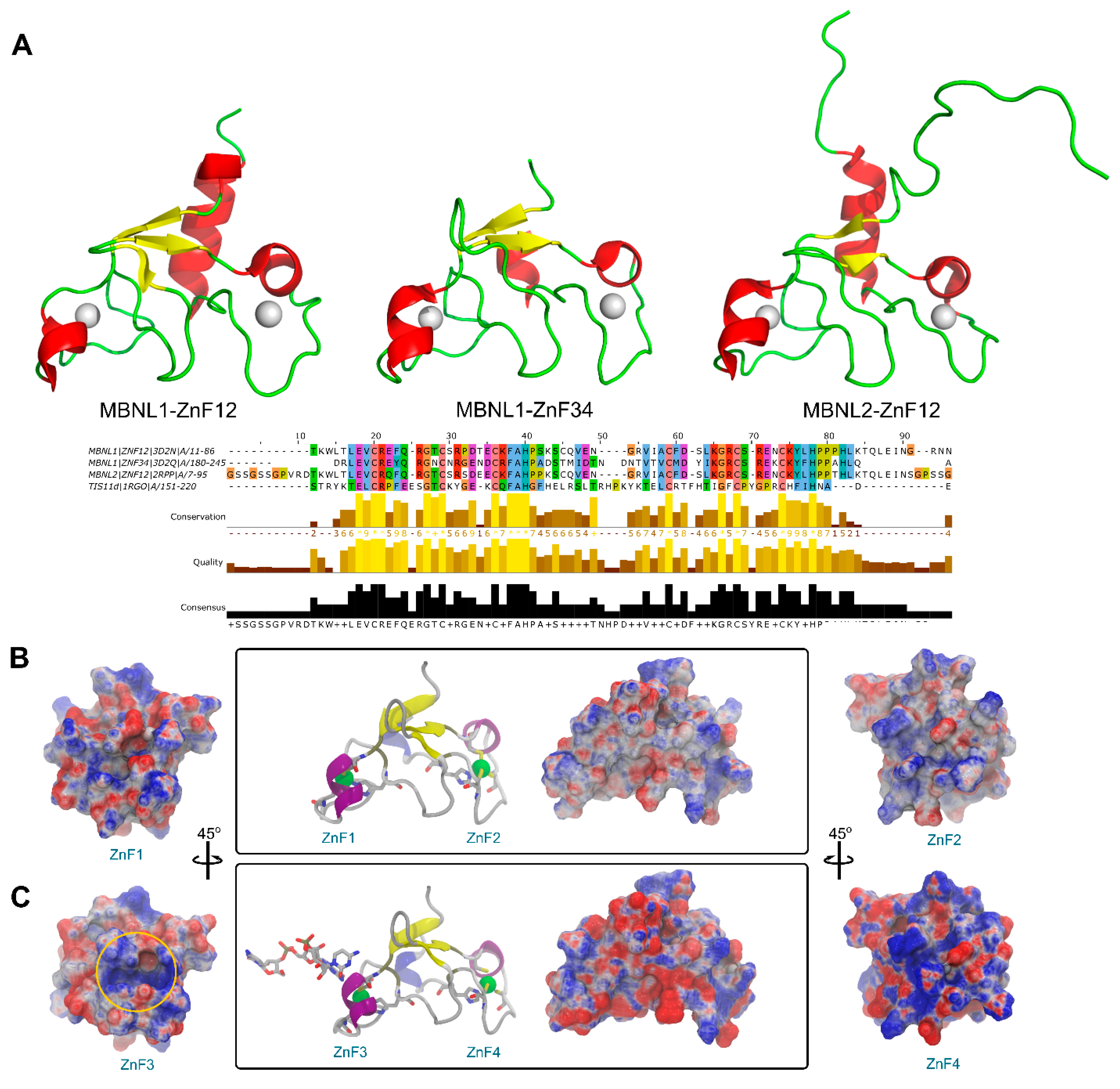
2.2. MBNL Domains from a Structural Perspective
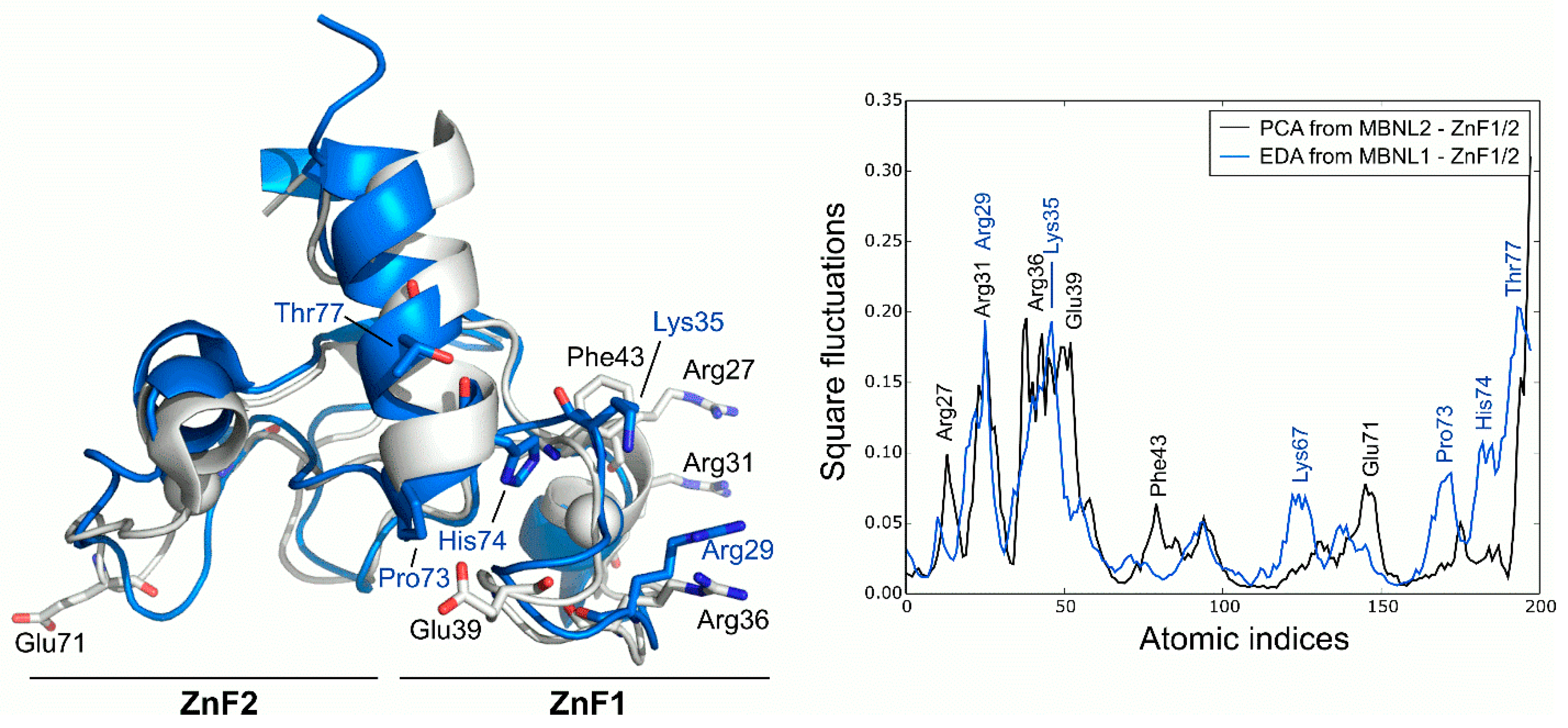
2.3. ZnF1/2 from MBNL1 and MBNL2 Exhibit Equivalent Large-Scale Motions
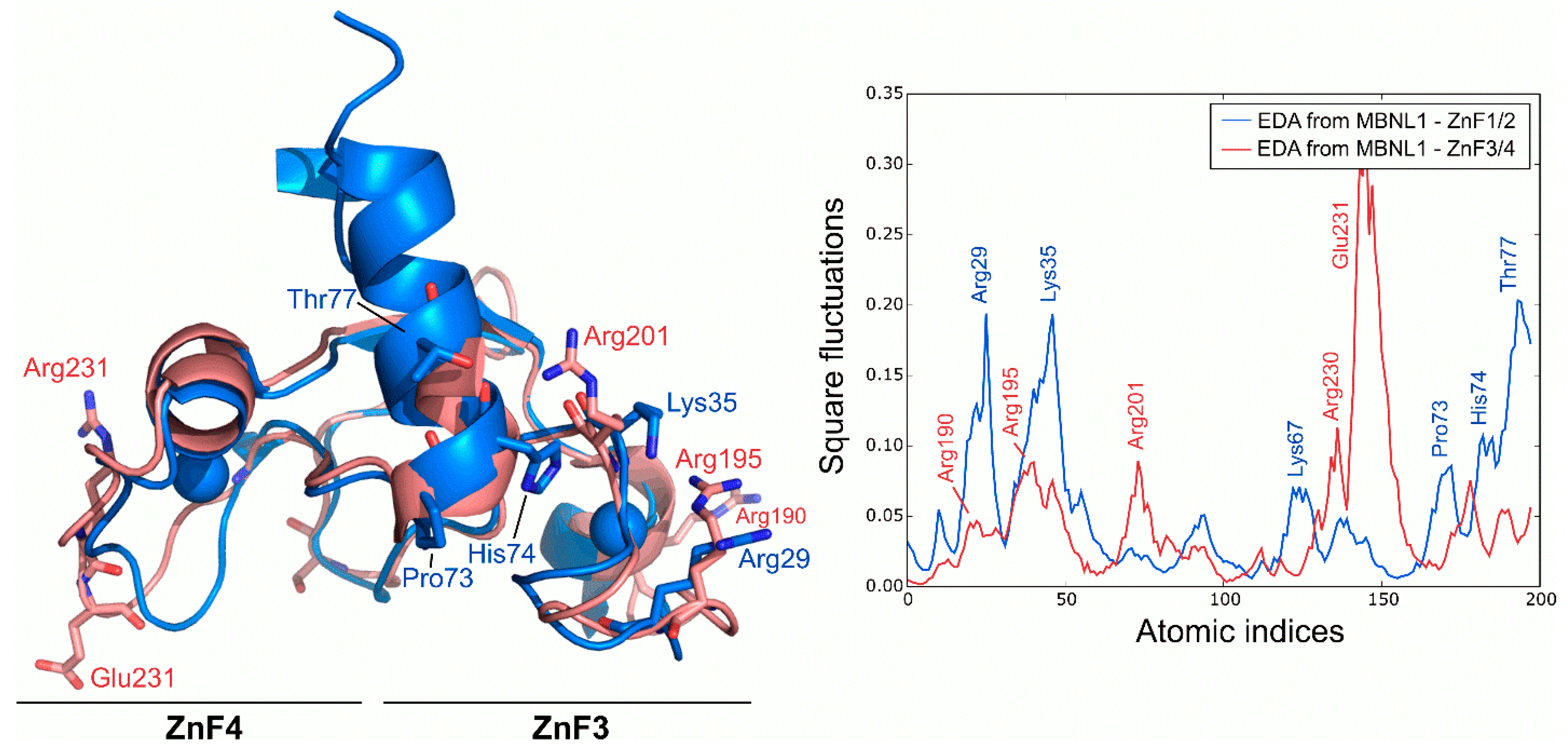
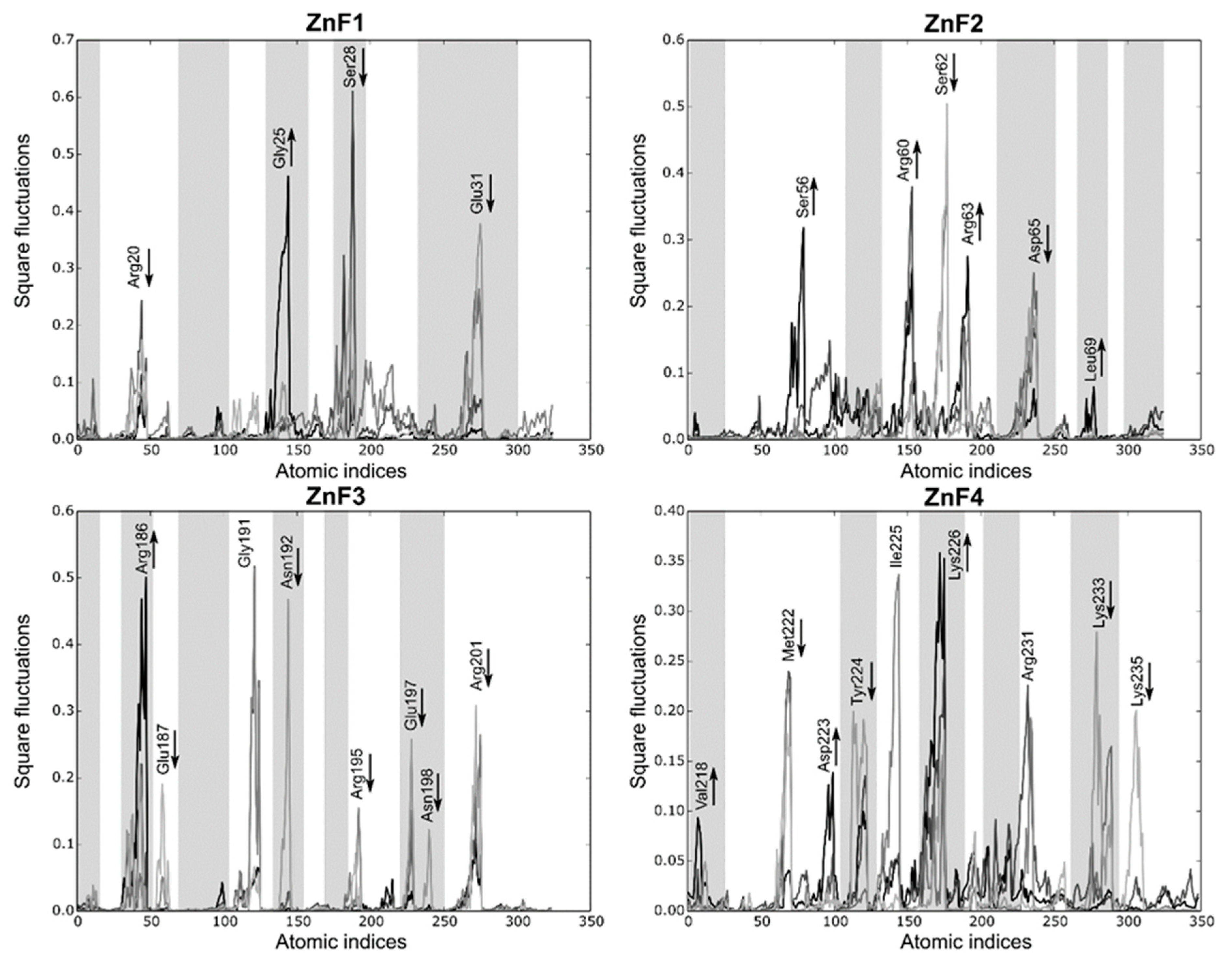
2.4. Intrinsic Fluctuations of ZnF3/4 Differ from Those of ZnF1/2
2.5. Effect of RNA Binding over Local Fluctuations
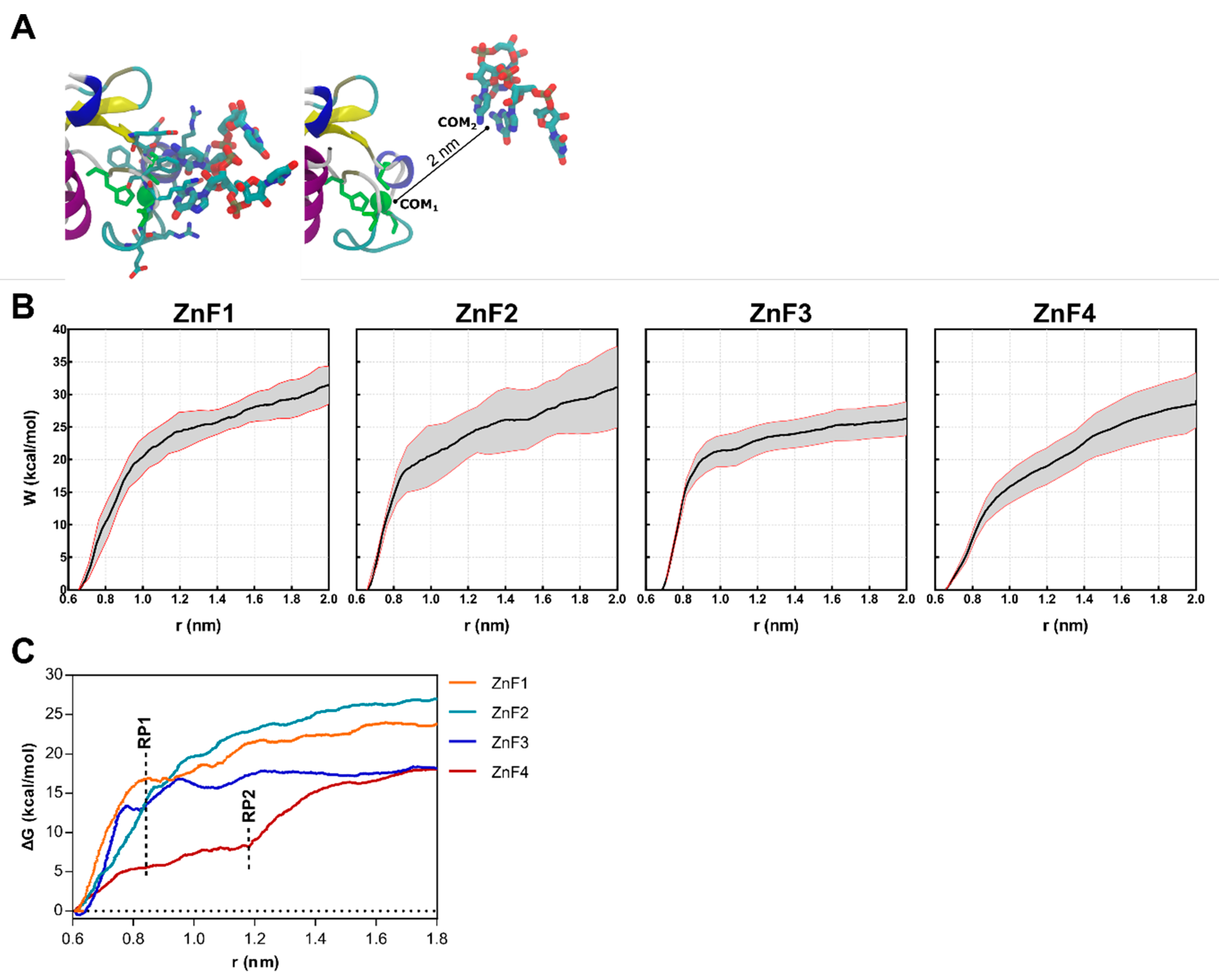
2.6. Both ZnFs of MBNL1 Have Differentiated Affinity for RNA
3. Materials and Methods
3.1. Conservation and Coevolution Analyses
3.2. System Preparation
3.3. Molecular Dynamics
3.4. Essential Dynamics and Principal Components Analysis
3.5. Steered Molecular Dynamics
3.6. Pocket Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Matlin, A.J.; Clark, F.; Smith, C.W.J. Understanding Alternative Splicing: Towards a Cellular Code. Nat. Rev. Mol. Cell Biol. 2005, 6, 386–398. [Google Scholar] [CrossRef]
- Black, D.L. Mechanisms of Alternative Pre-Messenger RNA Splicing. Annu. Rev. Biochem. 2003, 72, 291–336. [Google Scholar] [CrossRef]
- Baralle, F.E.; Giudice, J. Alternative Splicing as a Regulator of Development and Tissue Identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef]
- Romero, P.R.; Zaidi, S.; Fang, Y.Y.; Uversky, V.N.; Radivojac, P.; Oldfield, C.J.; Cortese, M.S.; Sickmeier, M.; LeGall, T.; Obradovic, Z.; et al. Alternative Splicing in Concert with Protein Intrinsic Disorder Enables Increased Functional Diversity in Multicellular Organisms. Proc. Natl. Acad. Sci. USA 2006, 103, 8390–8395. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Chen, Y.; Donahue, C.P.; Wolfe, M.S.; Varani, G. Structural Basis for Stabilization of the Tau Pre-MRNA Splicing Regulatory Element by Novantrone (Mitoxantrone). Chem. Biol. 2009, 16, 557–566. [Google Scholar] [CrossRef]
- Barbany, M.; Morata, J.; Meyer, T.; Lois, S.; Orozco, M.; de la Cruz, X. Characterization of the Impact of Alternative Splicing on Protein Dynamics: The Cases of Glutathione S-Transferase and Ectodysplasin-A Isoforms. Proteins Struct. Funct. Bioinform. 2012, 80, 2235–2249. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, S.X.; Rao, X.; Liu, Y. Modulator-Dependent RBPs Changes Alternative Splicing Outcomes in Kidney Cancer. Front. Genet. 2020, 11, 265. [Google Scholar] [CrossRef]
- Malhotra, S.; Sowdhamini, R. Sequence Search and Analysis of Gene Products Containing RNA Recognition Motifs in the Human Genome. BMC Genom. 2014, 15, 1159. [Google Scholar] [CrossRef]
- Barbany, M.; Meyer, T.; Hospital, A.; Faustino, I.; D’Abramo, M.; Morata, J.; Orozco, M.; de la Cruz, X. Molecular Dynamics Study of Naturally Existing Cavity Couplings in Proteins. PLoS ONE 2015, 10, e0119978. [Google Scholar] [CrossRef]
- Warf, M.B.; Berglund, J.A. MBNL Binds Similar RNA Structures in the CUG Repeats of Myotonic Dystrophy and Its Pre-MRNA Substrate Cardiac Troponin T. RNA 2007, 13, 2238–2251. [Google Scholar] [CrossRef]
- Teplova, M.; Patel, D.J. Structural Insights into RNA Recognition by the Alternative-Splicing Regulator Muscleblind-like MBNL1. Nat. Struct. Mol. Biol. 2008, 15, 1343–1351. [Google Scholar] [CrossRef] [PubMed]
- Edge, C.; Gooding, C.; Smith, C.W. Dissecting Domains Necessary for Activation and Repression of Splicing by Muscleblind-like Protein 1. BMC Mol. Biol. 2013, 14, 29. [Google Scholar] [CrossRef] [PubMed]
- Klinck, R.; Fourrier, A.; Thibault, P.; Toutant, J.; Durand, M.; Lapointe, E.; Caillet-Boudin, M.-L.; Sergeant, N.; Gourdon, G.; Meola, G.; et al. RBFOX1 Cooperates with MBNL1 to Control Splicing in Muscle, Including Events Altered in Myotonic Dystrophy Type 1. PLoS ONE 2014, 9, e107324. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.-H.; Richardson, S.L.; Ho, Y.-J.; Lucas, A.M.H.; Tuccinardi, T.; Baranger, A.M.; Zimmerman, S.C. Investigating the Binding Mode of an Inhibitor of the MBNL1⋅RNA Complex in Myotonic Dystrophy Type 1 (DM1) Leads to the Unexpected Discovery of a DNA-Selective Binder. ChemBioChem 2012, 13, 2505–2509. [Google Scholar] [CrossRef]
- Goers, E.S.; Purcell, J.; Voelker, R.B.; Gates, D.P.; Berglund, J.A. MBNL1 Binds GC Motifs Embedded in Pyrimidines to Regulate Alternative Splicing. Nucleic Acids Res. 2010, 38, 2467–2484. [Google Scholar] [CrossRef]
- Laurent, F.-X.; Sureau, A.; Klein, A.F.; Trouslard, F.; Gasnier, E.; Furling, D.; Marie, J. New Function for the RNA Helicase P68/DDX5 as a Modifier of MBNL1 Activity on Expanded CUG Repeats. Nucleic Acids Res. 2012, 40, 3159–3171. [Google Scholar] [CrossRef]
- Warf, M.B.; Diegel, J.V.; von Hippel, P.H.; Berglund, J.A. The Protein Factors MBNL1 and U2AF65 Bind Alternative RNA Structures to Regulate Splicing. Proc. Natl. Acad. Sci. USA 2009, 106, 9203–9208. [Google Scholar] [CrossRef]
- Childs-Disney, J.L.; Hoskins, J.; Rzuczek, S.G.; Thornton, C.A.; Disney, M.D. Rationally Designed Small Molecules Targeting the RNA That Causes Myotonic Dystrophy Type 1 Are Potently Bioactive. ACS Chem. Biol. 2012, 7, 856–862. [Google Scholar] [CrossRef]
- López-Martínez, A.; Soblechero-Martín, P.; de-la-Puente-Ovejero, L.; Nogales-Gadea, G.; Arechavala-Gomeza, V. An Overview of Alternative Splicing Defects Implicated in Myotonic Dystrophy Type I. Genes 2020, 11, 1109. [Google Scholar] [CrossRef]
- Philips, A.V.; Timchenko, L.T.; Cooper, T.A. Disruption of Splicing Regulated by a CUG-Binding Protein in Myotonic Dystrophy. Science 1998, 280, 737–741. [Google Scholar] [CrossRef]
- Michalowski, S.; Miller, J.W.; Urbinati, C.R.; Paliouras, M.; Swanson, M.S.; Griffith, J. Visualization of Double-Stranded RNAs from the Myotonic Dystrophy Protein Kinase Gene and Interactions with CUG-Binding Protein. Nucleic Acids Res. 1999, 27, 3534–3542. [Google Scholar] [CrossRef]
- Grammatikakis, I.; Goo, Y.-H.; Echeverria, G.V.; Cooper, T.A. Identification of MBNL1 and MBNL3 Domains Required for Splicing Activation and Repression. Nucleic Acids Res. 2011, 39, 2769–2780. [Google Scholar] [CrossRef]
- Tran, H.; Gourrier, N.; Lemercier-Neuillet, C.; Dhaenens, C.-M.; Vautrin, A.; Fernandez-Gomez, F.J.; Arandel, L.; Carpentier, C.; Obriot, H.; Eddarkaoui, S.; et al. Analysis of Exonic Regions Involved in Nuclear Localization, Splicing Activity, and Dimerization of Muscleblind-like-1 Isoforms. J. Biol. Chem. 2011, 286, 16435–16446. [Google Scholar] [CrossRef] [PubMed]
- Purcell, J.; Oddo, J.C.; Wang, E.T.; Berglund, J.A. Combinatorial Mutagenesis of MBNL1 Zinc Fingers Elucidates Distinct Classes of Regulatory Events. Mol. Cell Biol. 2012, 32, 4155–4167. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Konieczny, P.; Stepniak-Konieczna, E.; Sobczak, K. MBNL Proteins and Their Target RNAs, Interaction and Splicing Regulation. Nucleic Acids Res. 2014, 42, 10873–10887. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Bahar, I. Sequence Evolution Correlates with Structural Dynamics. Mol. Biol. Evol. 2012, 29, 2253–2263. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Ramisetty, S.R.; Hussain, N.; Baranger, A.M. MBNL1-RNA Recognition: Contributions of MBNL1 Sequence and RNA Conformation. ChemBioChem 2012, 13, 112–119. [Google Scholar] [CrossRef]
- Hale, M.A.; Richardson, J.I.; Day, R.C.; McConnell, O.L.; Arboleda, J.; Wang, E.T.; Berglund, J.A. An Engineered RNA Binding Protein with Improved Splicing Regulation. Nucleic Acids Res. 2018, 46, 3152–3168. [Google Scholar] [CrossRef]
- Jarzynski, C. A Nonequilibrium Equality for Free Energy Differences. Phys. Rev. Lett. 1996, 78, 2690. [Google Scholar] [CrossRef]
- Simonetti, F.L.; Teppa, E.; Chernomoretz, A.; Nielsen, M.; Marino Buslje, C. MISTIC: Mutual Information Server to Infer Coevolution. Nucleic Acids Res. 2013, 41, W8–W14. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Dang, W.; Abe, C.; Tsuda, K.; Inoue, M.; Watanabe, S.; Kobayashi, N.; Kigawa, T.; Matsuda, T.; Yabuki, T.; et al. Solution Structure of the RNA Binding Domain in the Human Muscleblind-like Protein 2. Protein Sci. 2008, 18, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Babin, V.; Berryman, J.T.; Betz, R.M.; Cai, Q.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Gohlke, H.; et al. AMBER 14. University of California: San Francisco, CA, USA, 2014. [Google Scholar]
- Pang, Y.-P. Novel Zinc Protein Molecular Dynamics Simulations: Steps Toward Antiangiogenesis for Cancer Treatment. J. Mol. Model 1999, 5, 196–202. [Google Scholar] [CrossRef]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J.C. Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of n-Alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Toukmaji, A.; Sagui, C.; Board, J.; Darden, T. Efficient Particle-Mesh Ewald Based Approach to Fixed and Induced Dipolar Interactions. J. Chem. Phys. 2000, 113, 10913–10927. [Google Scholar] [CrossRef]
- Sagui, C.; Pedersen, L.G.; Darden, T.A. Towards an Accurate Representation of Electrostatics in Classical Force Fields: Efficient Implementation of Multipolar Interactions in Biomolecular Simulations. J. Chem. Phys. 2004, 120, 73–87. [Google Scholar] [CrossRef]
- Meireles, L.; Gur, M.; Bakan, A.; Bahar, I. Pre-Existing Soft Modes of Motion Uniquely Defined by Native Contact Topology Facilitate Ligand Binding to Proteins. Protein Sci. 2011, 20, 1645–1658. [Google Scholar] [CrossRef]
- Bakan, A.; Meireles, L.M.; Bahar, I. ProDy: Protein Dynamics Inferred from Theory and Experiments. Bioinformatics 2011, 27, 1575–1577. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Durrant, J.D.; Votapka, L.; Sørensen, J.; Amaro, R.E. POVME 2.0: An Enhanced Tool for Determining Pocket Shape and Volume Characteristics. J. Chem. Theory Comput. 2014, 10, 5047–5056. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González, À.L.; Fernández-Remacha, D.; Borrell, J.I.; Teixidó, J.; Estrada-Tejedor, R. Cognate RNA-Binding Modes by the Alternative-Splicing Regulator MBNL1 Inferred from Molecular Dynamics. Int. J. Mol. Sci. 2022, 23, 16147. https://doi.org/10.3390/ijms232416147
González ÀL, Fernández-Remacha D, Borrell JI, Teixidó J, Estrada-Tejedor R. Cognate RNA-Binding Modes by the Alternative-Splicing Regulator MBNL1 Inferred from Molecular Dynamics. International Journal of Molecular Sciences. 2022; 23(24):16147. https://doi.org/10.3390/ijms232416147
Chicago/Turabian StyleGonzález, Àlex L., Daniel Fernández-Remacha, José Ignacio Borrell, Jordi Teixidó, and Roger Estrada-Tejedor. 2022. "Cognate RNA-Binding Modes by the Alternative-Splicing Regulator MBNL1 Inferred from Molecular Dynamics" International Journal of Molecular Sciences 23, no. 24: 16147. https://doi.org/10.3390/ijms232416147
APA StyleGonzález, À. L., Fernández-Remacha, D., Borrell, J. I., Teixidó, J., & Estrada-Tejedor, R. (2022). Cognate RNA-Binding Modes by the Alternative-Splicing Regulator MBNL1 Inferred from Molecular Dynamics. International Journal of Molecular Sciences, 23(24), 16147. https://doi.org/10.3390/ijms232416147






