Muscle Regeneration in Holothurians without the Upregulation of Muscle Genes
Abstract
1. Introduction
2. Results and Discussion
2.1. De Novo Transcriptome Assembly
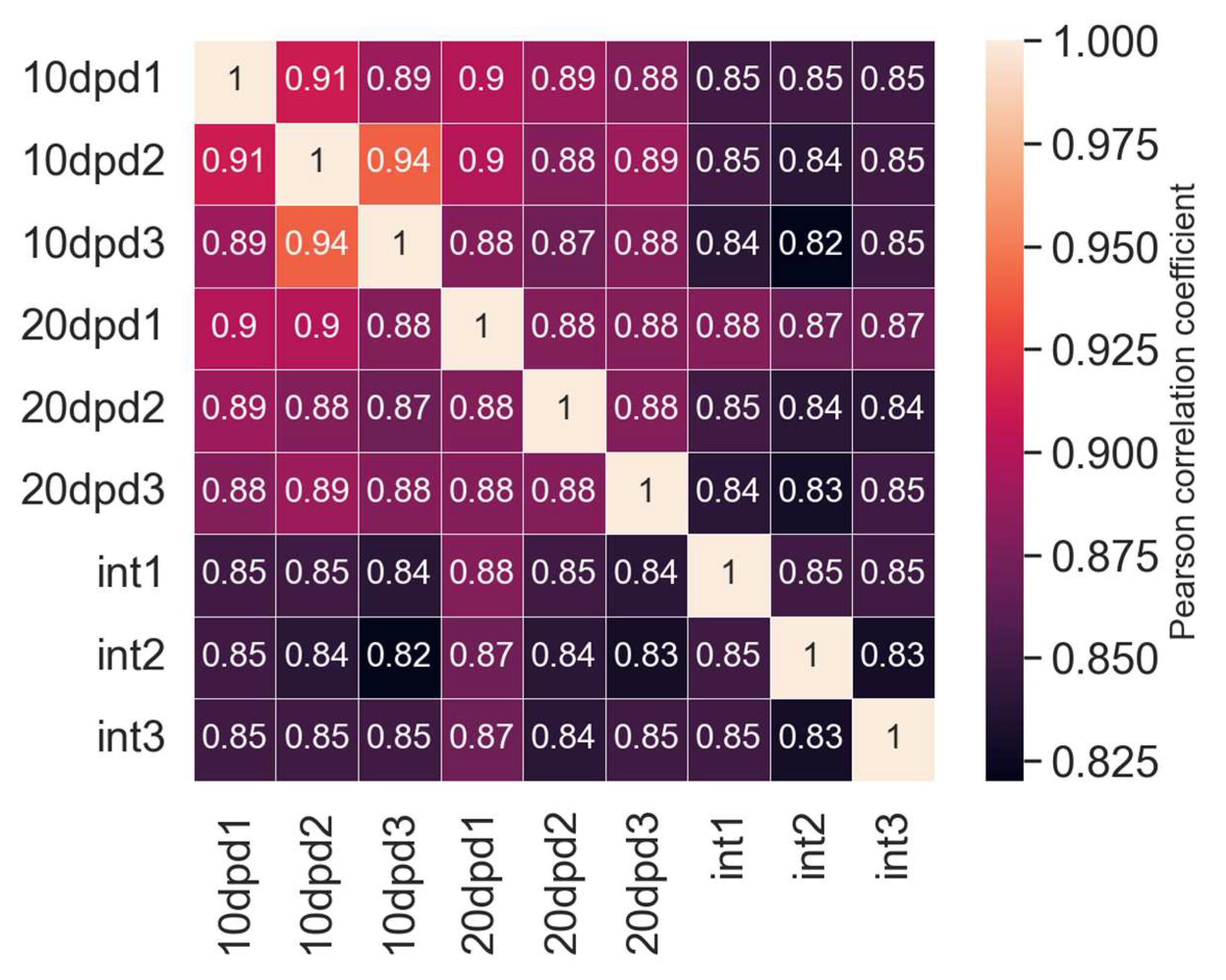
2.2. Differential Expression Analysis
2.3. Annotation
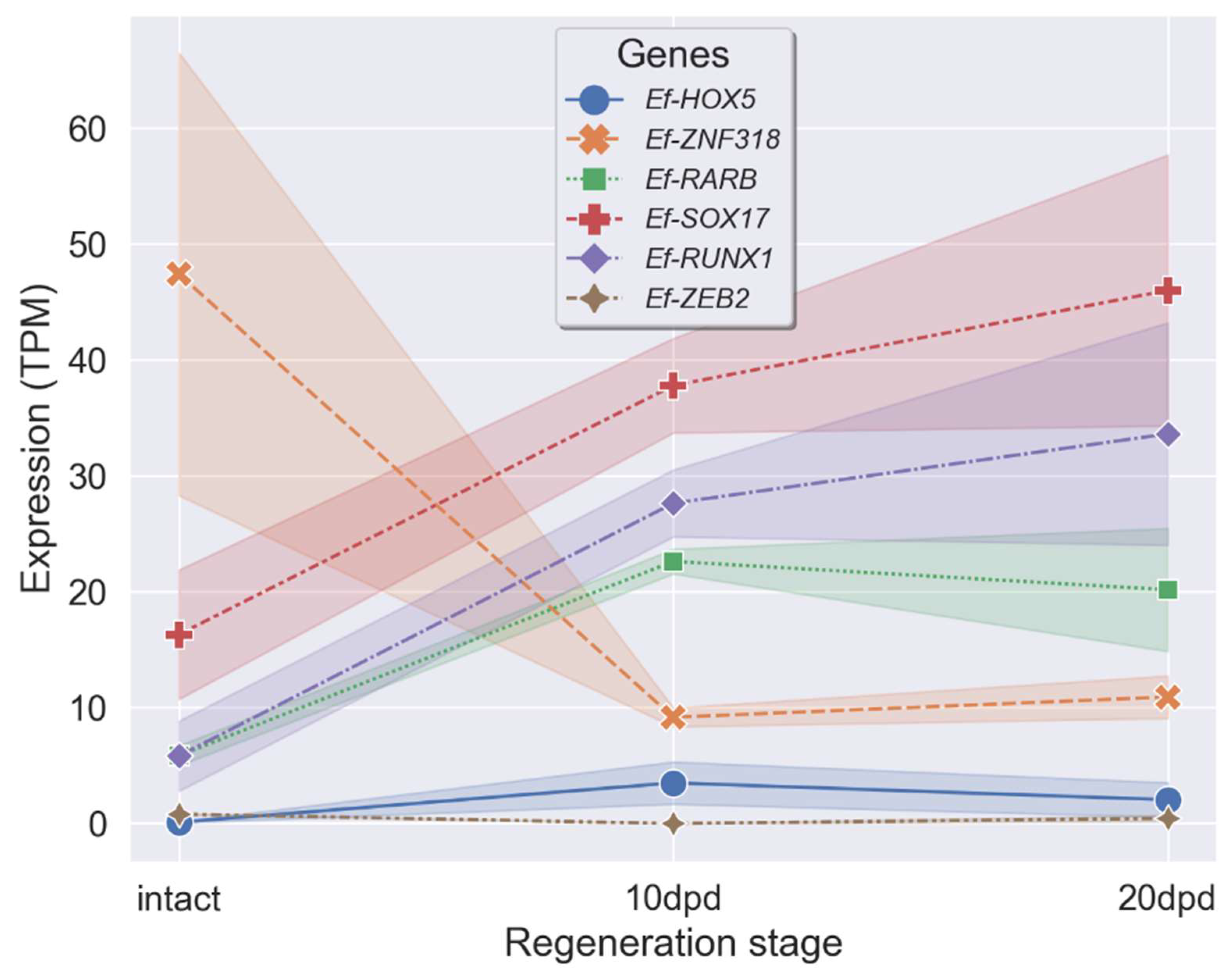
2.4. Search for Genes of Transcription Factors with Differential Expression during LMB Regeneration
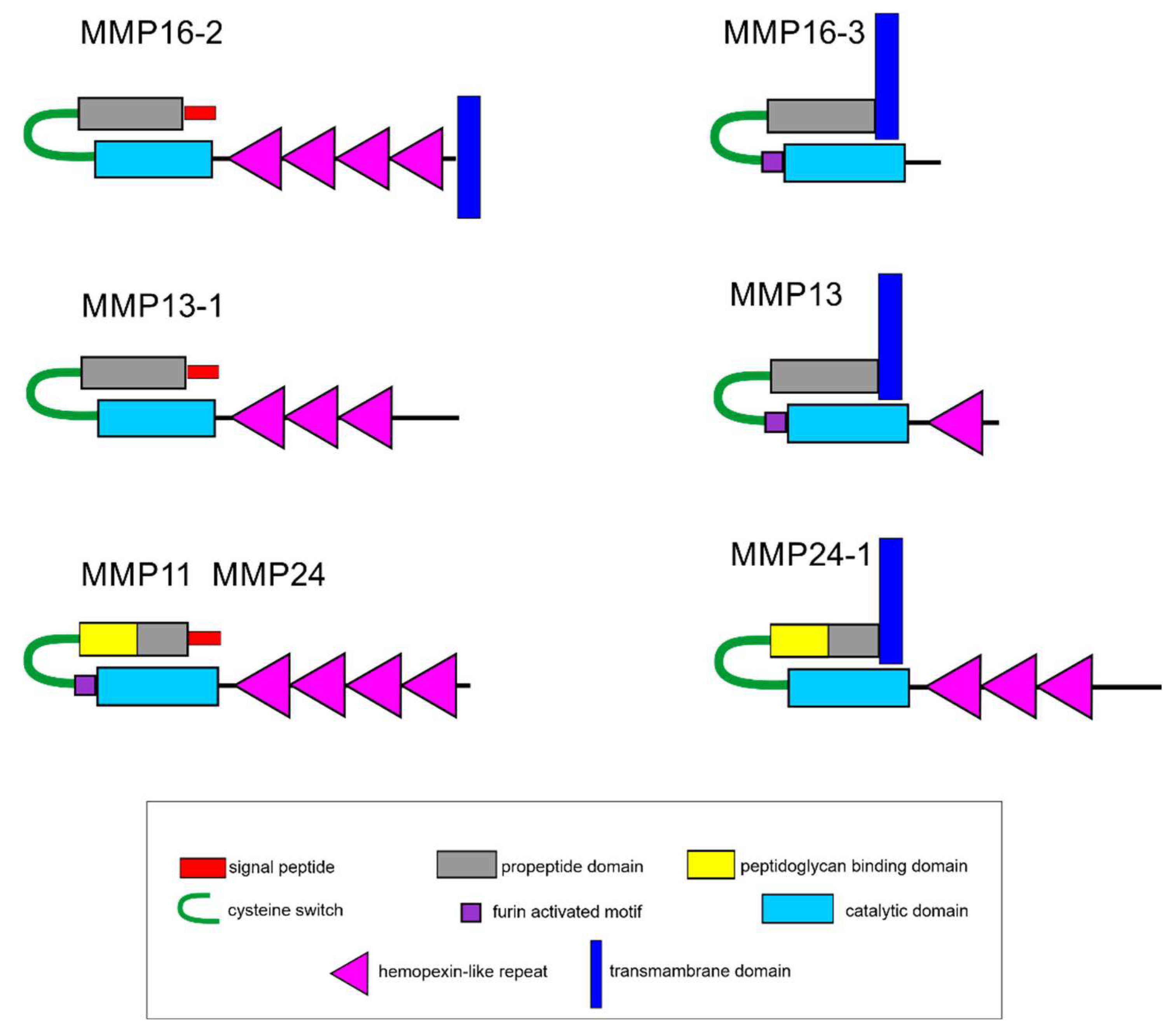
2.5. Search for Genes of MMPs and ECM Proteins with Differential Expression during LMB Regeneration
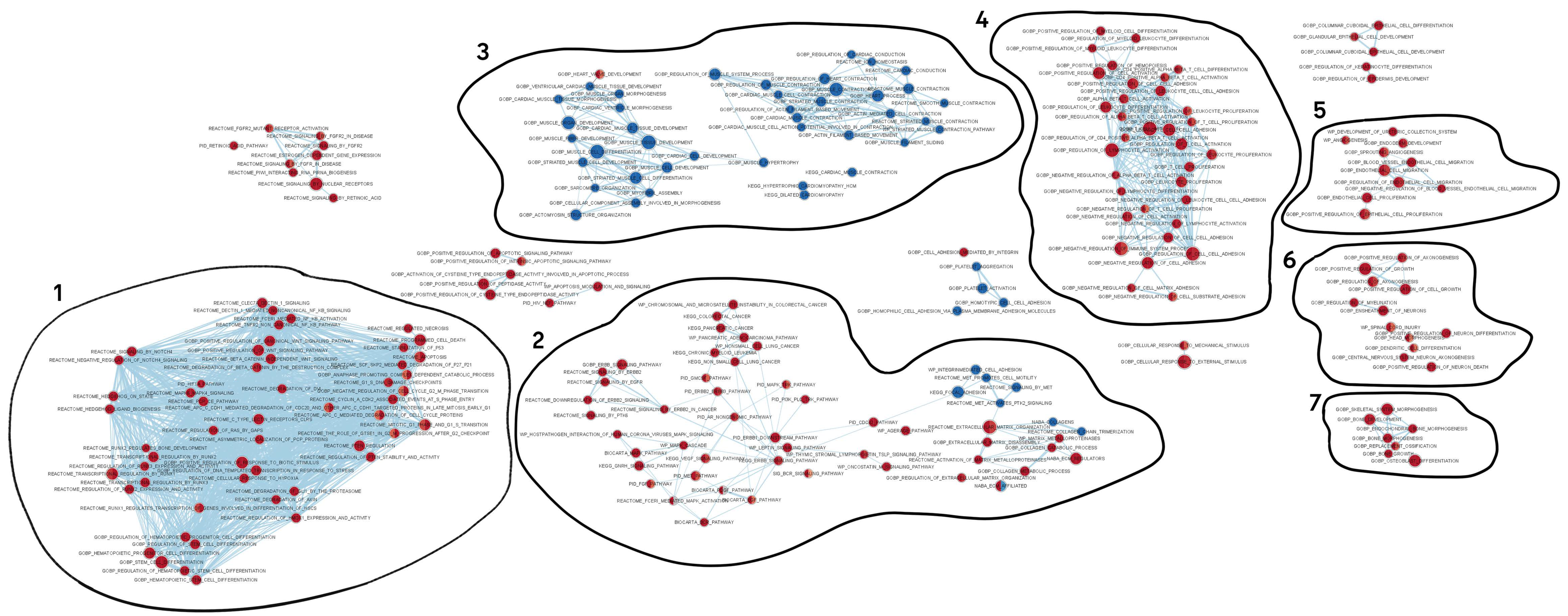
2.6. Network of Enrichment Biological Processes and Pathways
3. Materials and Methods
3.1. Animals
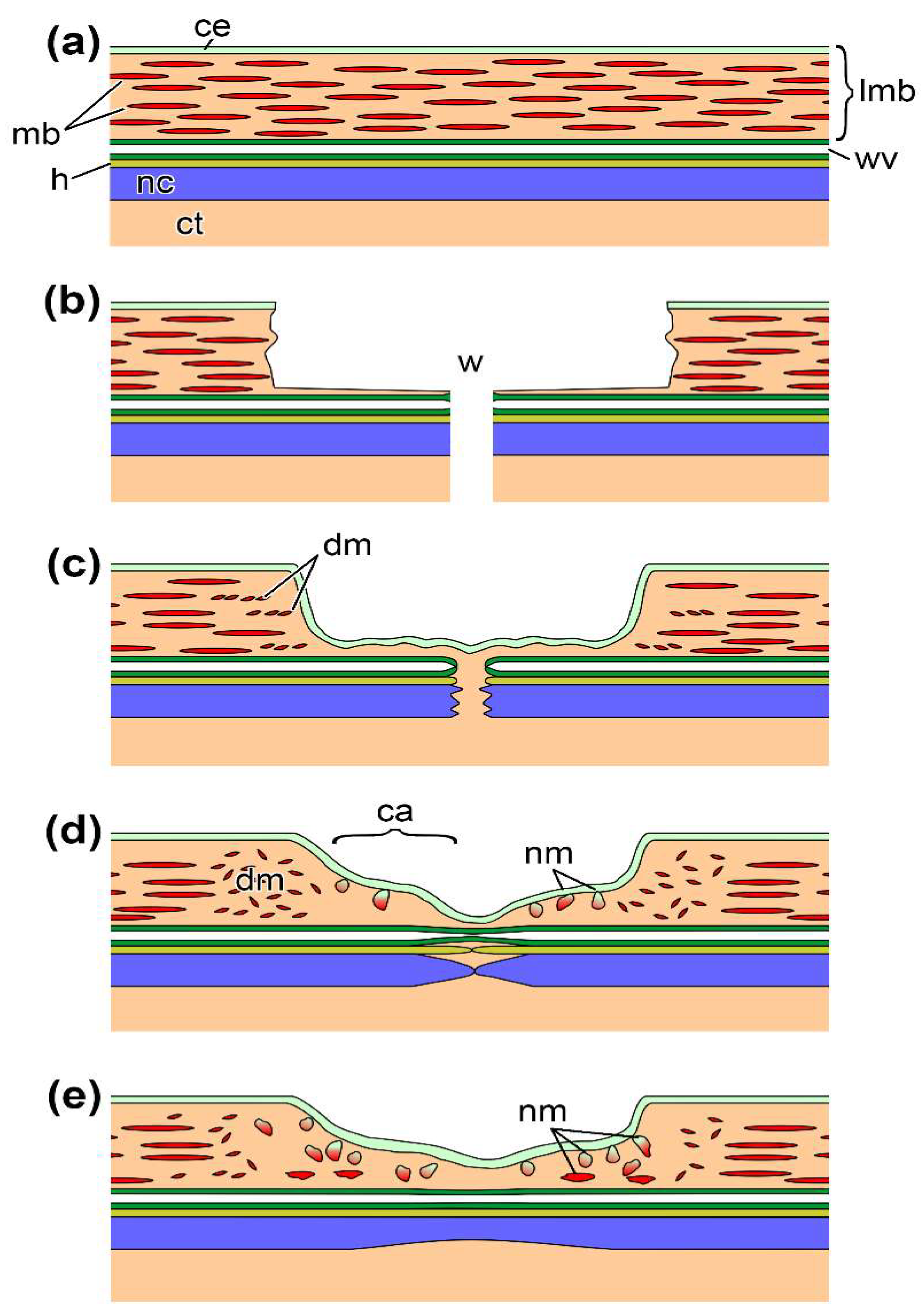
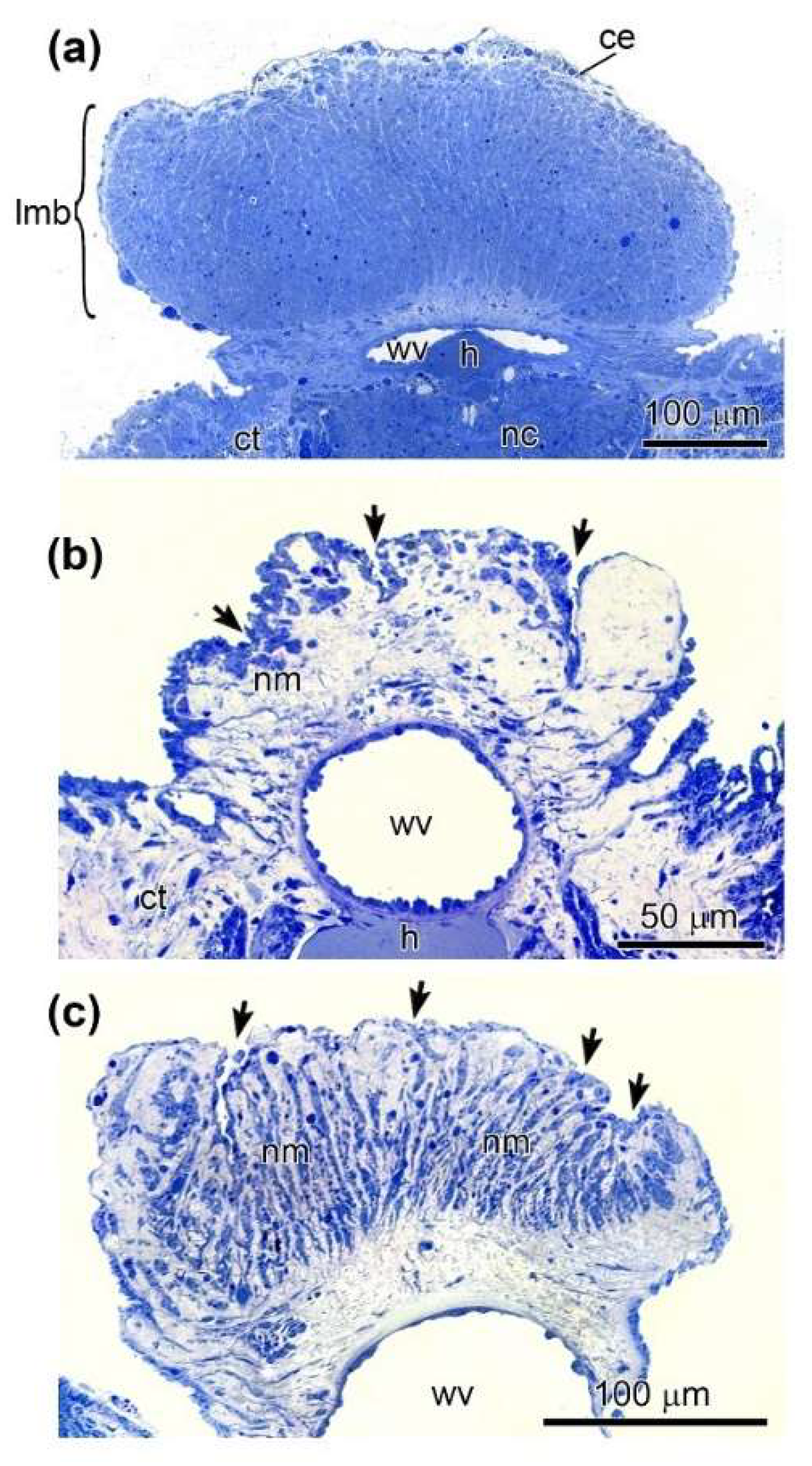
3.2. Light Microscopy
3.3. Sample Collection and RNA Extraction
3.4. Transcriptome Sequencing
3.5. De Novo Transcriptome Assembly
3.6. Differential Expression Analysis
3.7. Annotation
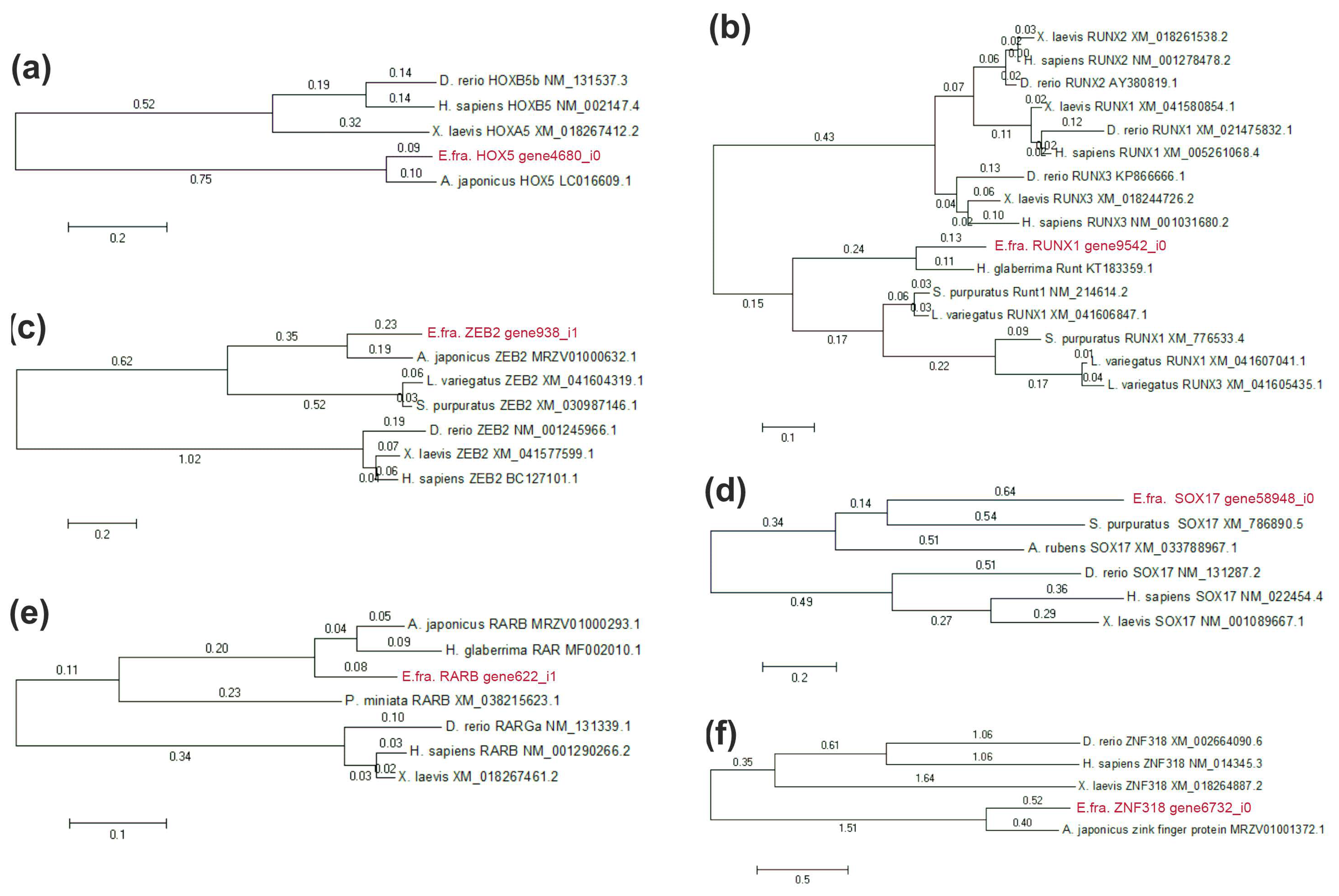
3.8. Phylogenetic Tree Construction
3.9. MMP Domain Structure
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Carlson, B.M. Muscle regeneration in amphibians and mammals: Passing the torch. Dev. Dyn. 2003, 226, 167–181. [Google Scholar] [CrossRef]
- Galliot, B.; Tanaka, E.; Simon, A. Molecular and cellular basis of regeneration and tissue repair: Regeneration and tissue repair: Themes and variations. Cell. Mol. Life Sci. 2008, 65, 1. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.; Sleep, E.; Raya, M.; Martí, M.; Raya, A.; Belmonte, J.C.I. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 2010, 464, 606–609. [Google Scholar] [CrossRef] [PubMed]
- Beffagna, G. Zebrafish as a smart model to understand regeneration after heart injury: How fish could help humans. Front. Cardiovasc. Med. 2019, 6, 107. [Google Scholar] [CrossRef]
- Brockes, J.P. Amphibian limb regeneration: Rebuilding a complex structure. Science 1997, 276, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-C.J.; Grigoriev, N.G.; Spencer, A.N. Wound healing in jellyfish striated muscle involves rapid switching between two modes of cell motility and a change in the source of regulatory calcium. Dev. Biol. 2000, 225, 87–100. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Leclère, L.; Röttinger, E. Diversity of cnidarian muscles: Function, anatomy, development and regeneration. Front. Cell Dev. Biol. 2017, 4, 157. [Google Scholar] [CrossRef]
- Zullo, L.; Bozzo, M.; Daya, A.; Di Clemente, A.; Mancini, F.P.; Megighian, A.; Nesher, N.; Röttinger, E.; Shomrat, T.; Tiozzo, S.; et al. The diversity of muscles and their regenerative potential across animals. Cells 2020, 9, 1925. [Google Scholar] [CrossRef]
- Dolmatov, I.Y. Regeneration in echinoderms. Russ. J. Mar. Biol. 1999, 22, 225–233. [Google Scholar]
- Dolmatov, I.Y.; Ginanova, T.T. Muscle regeneration in holothurians. Microsc. Res. Tech. 2001, 55, 452–463. [Google Scholar] [CrossRef]
- Candia Carnevali, M.D.; Burighel, P. Regeneration in echinoderms and ascidians. In eLS; Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2010. [Google Scholar] [CrossRef]
- Brusca, R.C.; Brusca, G.J. Invertebrates, 2nd ed.; Sinauer Associates: Sunderland, MA, USA, 2003; 936p. [Google Scholar]
- Garcia-Arraras, J.; Dolmatov, I. Echinoderms: Potential model systems for studies on muscle regeneration. Curr. Pharm. Des. 2010, 16, 942–955. [Google Scholar] [CrossRef]
- Dolmatov, I.Y.; Eliseikina, M.G.; Ginanova, T.T.; Lamash, N.E.; Korchagin, V.P.; Bulgakov, A.A. Muscle regeneration in the holothurian Apostichopus japonicus. Rouxs Arch. Dev. Biol. 1996, 205, 486–493. [Google Scholar] [CrossRef]
- Shulga, A.P.; Lamash, N.E. Proteinases with gelatinase activity and their role in ambulacrum regeneration in holothurians Eupentacta fraudatrix (D’yakonov and Baranova, 1958) and Cucumaria japonica (Semper, 1868) (Echinodermata: Holothuroidea). Russ. J. Mar. Biol. 2020, 46, 461–471. [Google Scholar] [CrossRef]
- Dolmatov, I.Y.; Shulga, A.P.; Ginanova, T.T.; Eliseikina, M.G.; Lamash, N.E. Metalloproteinase inhibitor GM6001 delays regeneration in holothurians. Tissue Cell 2019, 59, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Pineda, P.A.; Ramírez-Gómez, F.; Pérez-Ortiz, J.; González-Díaz, S.; Santiago-De Jesús, F.; Hernández-Pasos, J.; Del Valle-Avila, C.; Rojas-Cartagena, C.; Suárez-Castillo, E.C.; Tossas, K.; et al. Gene expression profiling of intestinal regeneration in the sea cucumber. BMC Genom. 2009, 10, 262. [Google Scholar] [CrossRef]
- Quispe-Parra, D.J.; Medina-Feliciano, J.G.; Cruz-González, S.; Ortiz-Zuazaga, H.; García-Arrarás, J.E. Transcriptomic analysis of early stages of intestinal regeneration in Holothuria glaberrima. Sci. Rep. 2021, 11, 346. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinforma. Oxf. Engl. 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Bürglin, T.R.; Affolter, M. Homeodomain proteins: An update. Chromosoma 2016, 125, 497–521. [Google Scholar] [CrossRef]
- Hajirnis, N.; Mishra, R.K. Homeotic genes: Clustering, modularity, and diversity. Front. Cell Dev. Biol. 2021, 9, 718308. [Google Scholar] [CrossRef]
- Rux, D.R.; Wellik, D.M. Hox genes in the adult skeleton: Novel functions beyond embryonic development: Hox genes in the adult skeleton. Dev. Dyn. 2017, 246, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Hombría, J.C.-G.; Lovegrove, B. Beyond homeosis—Hox function in morphogenesis and organogenesis. Differentiation 2003, 71, 461–476. [Google Scholar] [CrossRef]
- Poliacikova, G.; Maurel-Zaffran, C.; Graba, Y.; Saurin, A.J. Hox proteins in the regulation of muscle development. Front. Cell Dev. Biol. 2021, 9, 731996. [Google Scholar] [CrossRef]
- Wu, Y.; Moser, M.; Bautch, V.L.; Patterson, C. HoxB5 is an upstream transcriptional switch for differentiation of the vascular endothelium from precursor cells. Mol. Cell. Biol. 2003, 23, 5680–5691. [Google Scholar] [CrossRef] [PubMed]
- Kam, M.K.M.; Lui, V.C.H. Roles of hoxb5 in the development of vagal and trunk neural crest cells. Dev. Growth Differ. 2015, 57, 158–168. [Google Scholar] [CrossRef]
- Thorndyke, M.C.; Chen, W.-C.; Beesley, P.W.; Patruno, M. Molecular approach to echinoderm regeneration. Microsc. Res. Tech. 2001, 55, 474–485. [Google Scholar] [CrossRef]
- Gheldof, A.; Hulpiau, P.; van Roy, F.; De Craene, B.; Berx, G. Evolutionary functional analysis and molecular regulation of the ZEB transcription factors. Cell. Mol. Life Sci. 2012, 69, 2527–2541. [Google Scholar] [CrossRef] [PubMed]
- Vandewalle, C. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res. 2005, 33, 6566–6578. [Google Scholar] [CrossRef] [PubMed]
- Brabletz, S.; Brabletz, T. The ZEB/MiR-200 feedback loop—A motor of cellular plasticity in development and cancer? EMBO Rep. 2010, 11, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, G.; Manabe, I.; Tsushima, K.; Fujiu, K.; Oishi, Y.; Imai, Y.; Maemura, K.; Miyagishi, M.; Higashi, Y.; Kondoh, H.; et al. ΔEF1 mediates TGF-β signaling in vascular smooth muscle cell differentiation. Dev. Cell 2006, 11, 93–104. [Google Scholar] [CrossRef]
- Siles, L.; Ninfali, C.; Cortés, M.; Darling, D.S.; Postigo, A. ZEB1 protects skeletal muscle from damage and is required for its regeneration. Nat. Commun. 2019, 10, 1364. [Google Scholar] [CrossRef]
- Di Filippo, E.S.; Costamagna, D.; Giacomazzi, G.; Cortés-Calabuig, Á.; Stryjewska, A.; Huylebroeck, D.; Fulle, S.; Sampaolesi, M. Zeb2 regulates myogenic differentiation in pluripotent stem cells. Int. J. Mol. Sci. 2020, 21, 2525. [Google Scholar] [CrossRef] [PubMed]
- Ghyselinck, N.B.; Duester, G. Retinoic acid signaling pathways. Development 2019, 146, dev167502. [Google Scholar] [CrossRef]
- Zhao, L.; Son, J.S.; Wang, B.; Tian, Q.; Chen, Y.; Liu, X.; de Avila, J.M.; Zhu, M.-J.; Du, M. Retinoic acid signalling in fibro/adipogenic progenitors robustly enhances muscle regeneration. EBioMedicine 2020, 60, 103020. [Google Scholar] [CrossRef]
- Viera-Vera, J.; García-Arrarás, J.E. Retinoic acid signaling is associated with cell proliferation, muscle cell dedifferentiation, and overall rudiment size during intestinal regeneration in the sea cucumber, Holothuria glaberrima. Biomolecules 2019, 9, 873. [Google Scholar] [CrossRef] [PubMed]
- Osato, M. Point mutations in the RUNX1/AML1 gene: Another actor in RUNX leukemia. Oncogene 2004, 23, 4284–4296. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.M.; Johnson, S.A.; Sims, N.A.; Trivett, M.K.; Slavin, J.L.; Rubin, B.P.; Waring, P.; McArthur, G.A.; Walkley, C.R.; Holloway, A.J.; et al. Terminal osteoblast differentiation, mediated by Runx2 and P27KIP1, is disrupted in osteosarcoma. J. Cell Biol. 2004, 167, 925–934. [Google Scholar] [CrossRef]
- Kagoshima, H.; Shigesada, K.; Kohara, Y. RUNX regulates stem cell proliferation and differentiation: Insights from studies of C. elegans. J. Cell. Biochem. 2007, 100, 1119–1130. [Google Scholar] [CrossRef]
- Sullivan, J.C.; Sher, D.; Eisenstein, M.; Shigesada, K.; Reitzel, A.M.; Marlow, H.; Levanon, D.; Groner, Y.; Finnerty, J.R.; Gat, U. The evolutionary origin of the Runx/CBFbeta transcription factors—Studies of the most basal metazoans. BMC Evol. Biol. 2008, 8, 228. [Google Scholar] [CrossRef]
- Seo, W.; Taniuchi, I. The roles of RUNX family proteins in development of immune cells. Mol. Cells 2020, 43, 107–113. [Google Scholar] [CrossRef]
- Hall, A.; Choi, K.; Liu, W.; Rose, J.; Zhao, C.; Yu, Y.; Na, Y.; Cai, Y.; Coover, R.A.; Lin, Y.; et al. RUNX represses Pmp22 to drive neurofibromagenesis. Sci. Adv. 2019, 5, eaau8389. [Google Scholar] [CrossRef]
- Chuang, L.; Ito, Y. The multiple interactions of RUNX with the Hippo–YAP pathway. Cells 2021, 10, 2925. [Google Scholar] [CrossRef] [PubMed]
- Coffman, J.A.; Dickey-Sims, C.; Haug, J.S.; McCarthy, J.J.; Robertson, A.J. Evaluation of developmental phenotypes produced by morpholino antisense targeting of a sea urchin Runx gene. BMC Biol. 2004, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Dickey-Sims, C.; Robertson, A.J.; Rupp, D.E.; McCarthy, J.J.; Coffman, J.A. Runx-dependent expression of PKC is critical for cell survival in the sea urchin embryo. BMC Biol. 2005, 3, 18. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Robertson, A.J.; Dickey-Sims, C.; Ransick, A.; Rupp, D.E.; McCarthy, J.J.; Coffman, J.A. CBFβ is a facultative Runx partner in the sea urchin embryo. BMC Biol. 2006, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.J.; Coluccio, A.; Knowlton, P.; Dickey-Sims, C.; Coffman, J.A. Runx expression is mitogenic and mutually linked to Wnt activity in blastula-stage sea urchin embryos. PLoS ONE 2008, 3, e3770. [Google Scholar] [CrossRef]
- She, Z.-Y.; Yang, W.-X. SOX family transcription factors involved in diverse cellular events during development. Eur. J. Cell Biol. 2015, 94, 547–563. [Google Scholar] [CrossRef]
- Alonso-Martin, S.; Auradé, F.; Mademtzoglou, D.; Rochat, A.; Zammit, P.S.; Relaix, F. SOXF factors regulate murine satellite cell self-renewal and function through inhibition of β-Catenin activity. eLife 2018, 7, e26039. [Google Scholar] [CrossRef]
- Dolmatov, I.Y.; Kalacheva, N.V.; Tkacheva, E.S.; Shulga, A.P.; Zavalnaya, E.G.; Shamshurina, E.V.; Girich, A.S.; Boyko, A.V.; Eliseikina, M.G. Expression of Piwi, MMP, TIMP, and Sox during gut regeneration in holothurian Eupentacta fraudatrix (Holothuroidea, Dendrochirotida). Genes 2021, 12, 1292. [Google Scholar] [CrossRef]
- Dolmatov, I.Y.; Nizhnichenko, V.A.; Dolmatova, L.S. Matrix metalloproteinases and tissue inhibitors of metalloproteinases in echinoderms: Structure and possible functions. Cells 2021, 10, 2331. [Google Scholar] [CrossRef]
- Fanjul-Fernández, M.; Folgueras, A.R.; Cabrera, S.; López-Otín, C. Matrix metalloproteinases: Evolution, gene regulation and functional analysis in mouse models. Biochim. Biophys. Acta BBA—Mol. Cell Res. 2010, 1803, 3–19. [Google Scholar] [CrossRef]
- Nagase, H.; Visse, R.; Murphy, G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc. Res. 2006, 69, 562–573. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, Y. Role of matrix metalloproteinases in skeletal muscle: Migration, differentiation, regeneration and fibrosis. Cell Adhes. Migr. 2009, 3, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.; Leong, D.; Smith, L.R.; Barton, E.R. Matrix metalloproteinase 13 is a new contributor to skeletal muscle regeneration and critical for myoblast migration. Am. J. Physiol.-Cell Physiol. 2013, 305, C529–C538. [Google Scholar] [CrossRef]
- Smith, L.R.; Kok, H.J.; Zhang, B.; Chung, D.; Spradlin, R.A.; Rakoczy, K.D.; Lei, H.; Boesze-Battaglia, K.; Barton, E.R. Matrix metalloproteinase 13 from satellite cells is required for efficient muscle growth and regeneration. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2020, 54, 333–353. [Google Scholar] [CrossRef]
- Dolmatov, I.Y.; Afanasyev, S.V.; Boyko, A.V. Molecular mechanisms of fission in echinoderms: Transcriptome analysis. PLoS ONE 2018, 13, e0195836. [Google Scholar] [CrossRef] [PubMed]
- Boyko, A.V.; Girich, A.S.; Tkacheva, E.S.; Dolmatov, I.Y. The Eupentacta fraudatrix transcriptome provides insights into regulation of cell transdifferentiation. Sci. Rep. 2020, 10, 1522. [Google Scholar] [CrossRef] [PubMed]
- Dolmatov, I.Y. Molecular aspects of regeneration mechanisms in holothurians. Genes 2021, 12, 250. [Google Scholar] [CrossRef]
- Nakano, A.; Nakano, H.; Smith, K.A.; Palpant, N.J. The developmental origins and lineage contributions of endocardial endothelium. Biochim. Biophys. Acta 2016, 1863, 1937–1947. [Google Scholar] [CrossRef]
- O’Donnell, A.; Yutzey, K.E. Mechanisms of heart valve development and disease. Dev. Camb. Engl. 2020, 147, dev183020. [Google Scholar] [CrossRef]
- Wallace, D.M. Large- and small-scale phenol extractions. Methods Enzymol. 1987, 152, 33–41. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinforma. Oxf. Engl. 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. J. Comput. Mol. Cell Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Boyko, A.V.; Girich, A.S.; Eliseikina, M.G.; Maslennikov, S.I.; Dolmatov, I.Y. Reference assembly and gene expression analysis of Apostichopus japonicus larval development. Sci. Rep. 2019, 9, 1131. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011, 12, 323. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Zerbino, D.R.; Achuthan, P.; Akanni, W.; Amode, M.R.; Barrell, D.; Bhai, J.; Billis, K.; Cummins, C.; Gall, A.; Girón, C.G.; et al. Ensembl 2018. Nucleic Acids Res. 2018, 46, D754–D761. [Google Scholar] [CrossRef]
- Kudtarkar, P.; Cameron, R.A. Echinobase: An expanding resource for echinoderm genomic information. Database 2017, bax074. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Merico, D.; Isserlin, R.; Stueker, O.; Emili, A.; Bader, G.D. Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PLoS ONE 2010, 5, e13984. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nizhnichenko, V.A.; Boyko, A.V.; Ginanova, T.T.; Dolmatov, I.Y. Muscle Regeneration in Holothurians without the Upregulation of Muscle Genes. Int. J. Mol. Sci. 2022, 23, 16037. https://doi.org/10.3390/ijms232416037
Nizhnichenko VA, Boyko AV, Ginanova TT, Dolmatov IY. Muscle Regeneration in Holothurians without the Upregulation of Muscle Genes. International Journal of Molecular Sciences. 2022; 23(24):16037. https://doi.org/10.3390/ijms232416037
Chicago/Turabian StyleNizhnichenko, Vladimir A., Alexey V. Boyko, Talia T. Ginanova, and Igor Yu. Dolmatov. 2022. "Muscle Regeneration in Holothurians without the Upregulation of Muscle Genes" International Journal of Molecular Sciences 23, no. 24: 16037. https://doi.org/10.3390/ijms232416037
APA StyleNizhnichenko, V. A., Boyko, A. V., Ginanova, T. T., & Dolmatov, I. Y. (2022). Muscle Regeneration in Holothurians without the Upregulation of Muscle Genes. International Journal of Molecular Sciences, 23(24), 16037. https://doi.org/10.3390/ijms232416037






