Immunoproteasome Inhibition Ameliorates Aged Dystrophic Mouse Muscle Environment
Abstract
:1. Introduction
2. Results
2.1. Immunohistological Analysis of Skeletal Muscle Reveals Higher Percentage of Small Fibers and Metabolic Switch in Mdx Mice Treated with IP Inhibitor
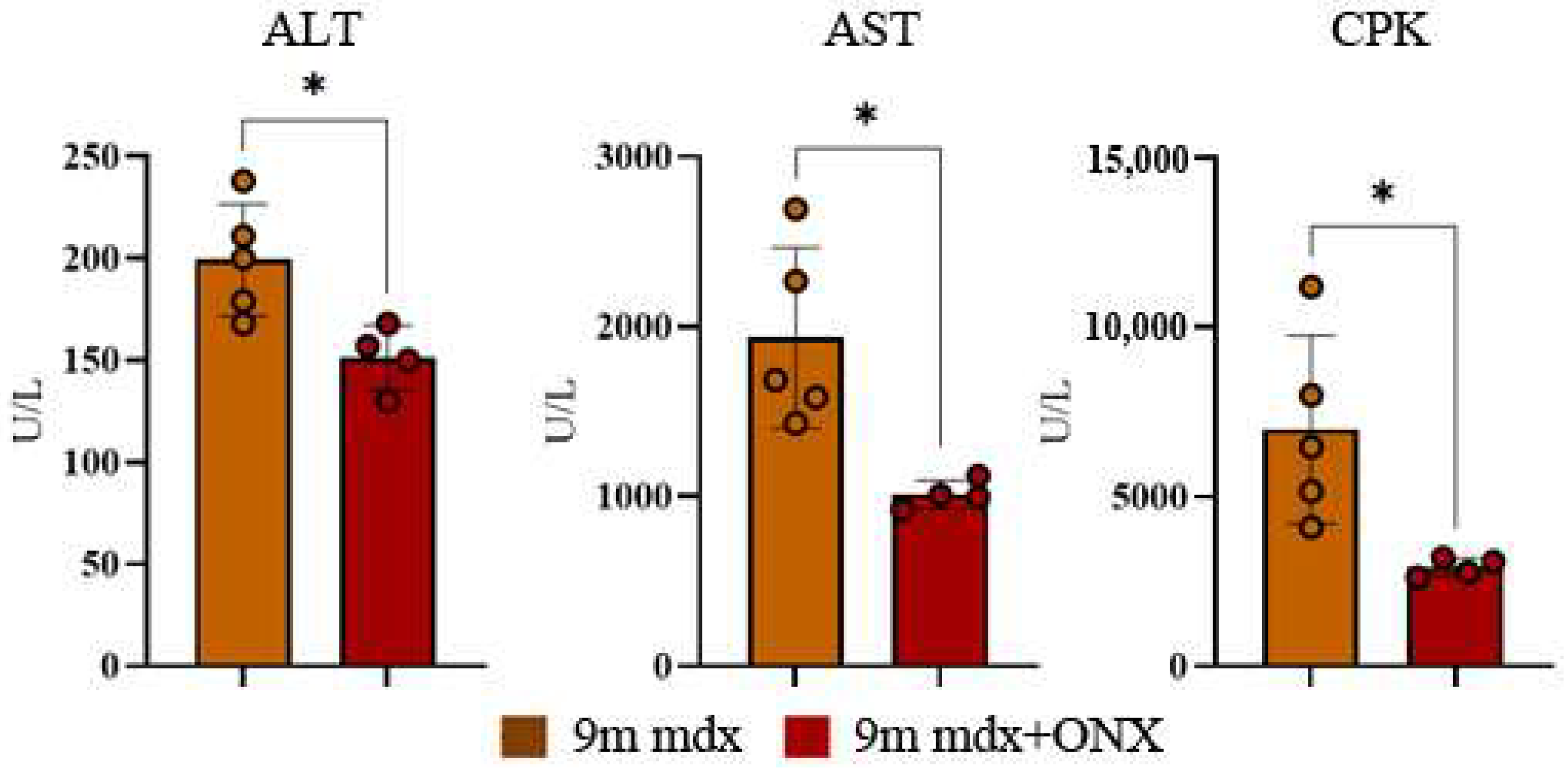
2.2. Serum Markers of Muscle Cell Damage Are Lowered by ONX-0914 Administration
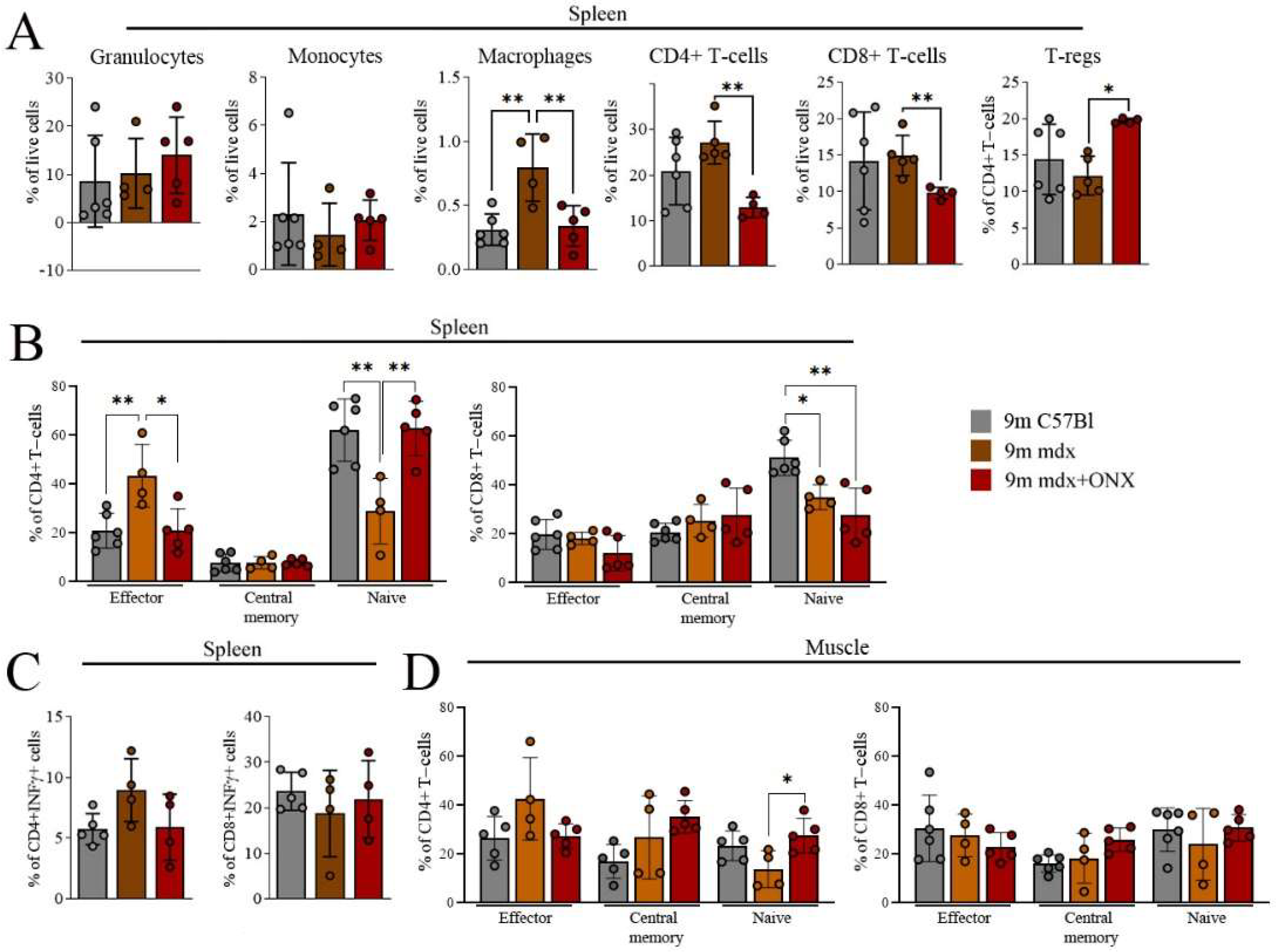
2.3. ONX-0914 Modulates Skeletal Muscle and Spleen Immune Cell Population
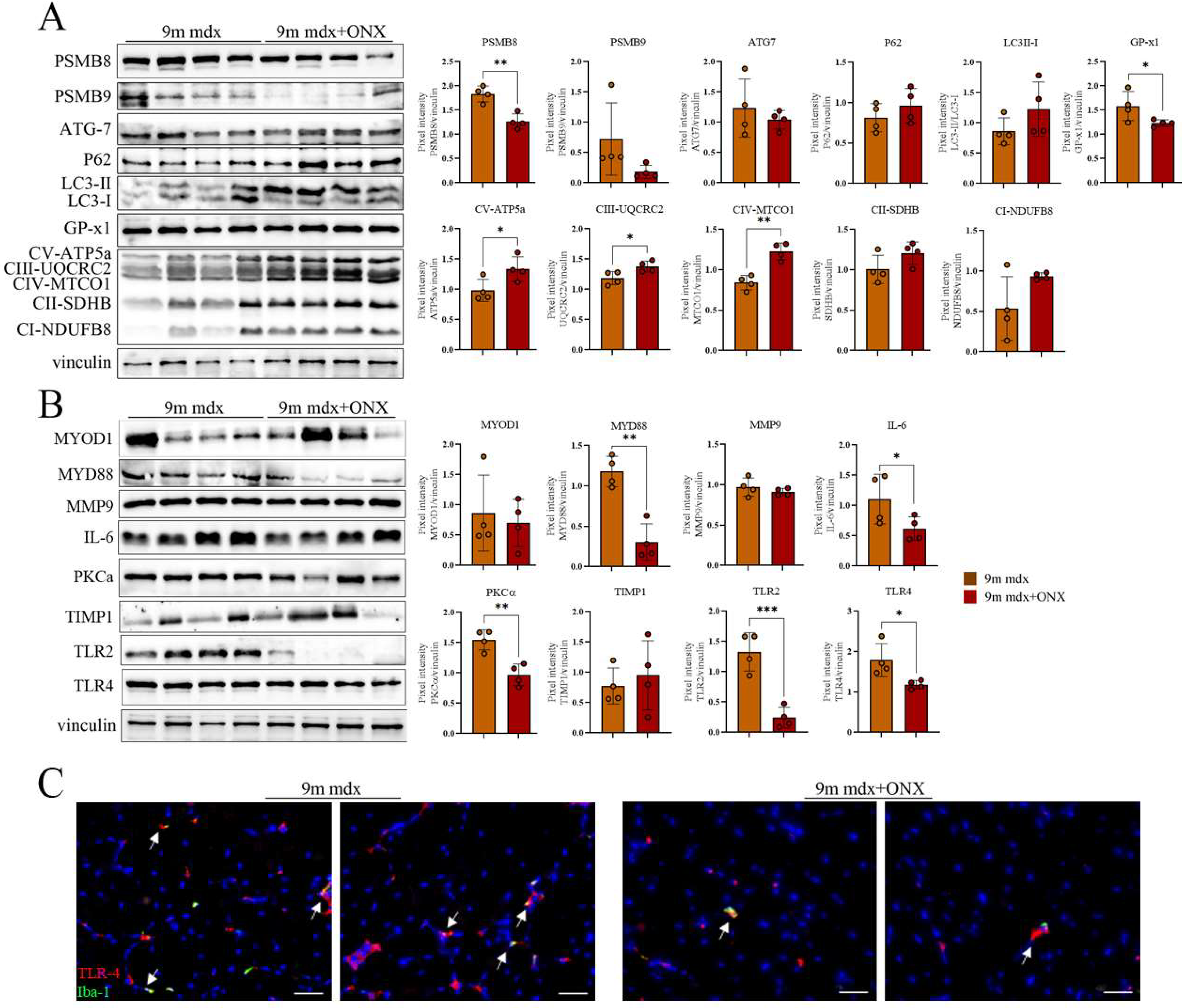
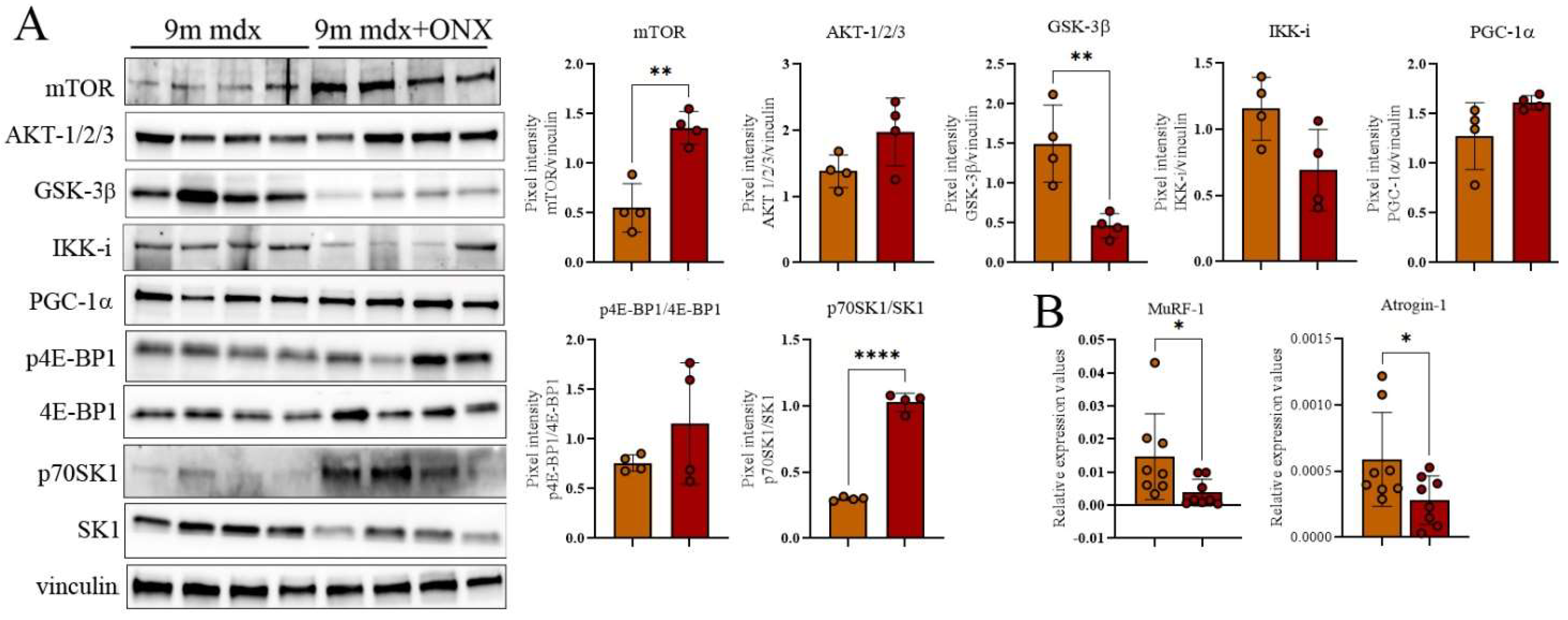
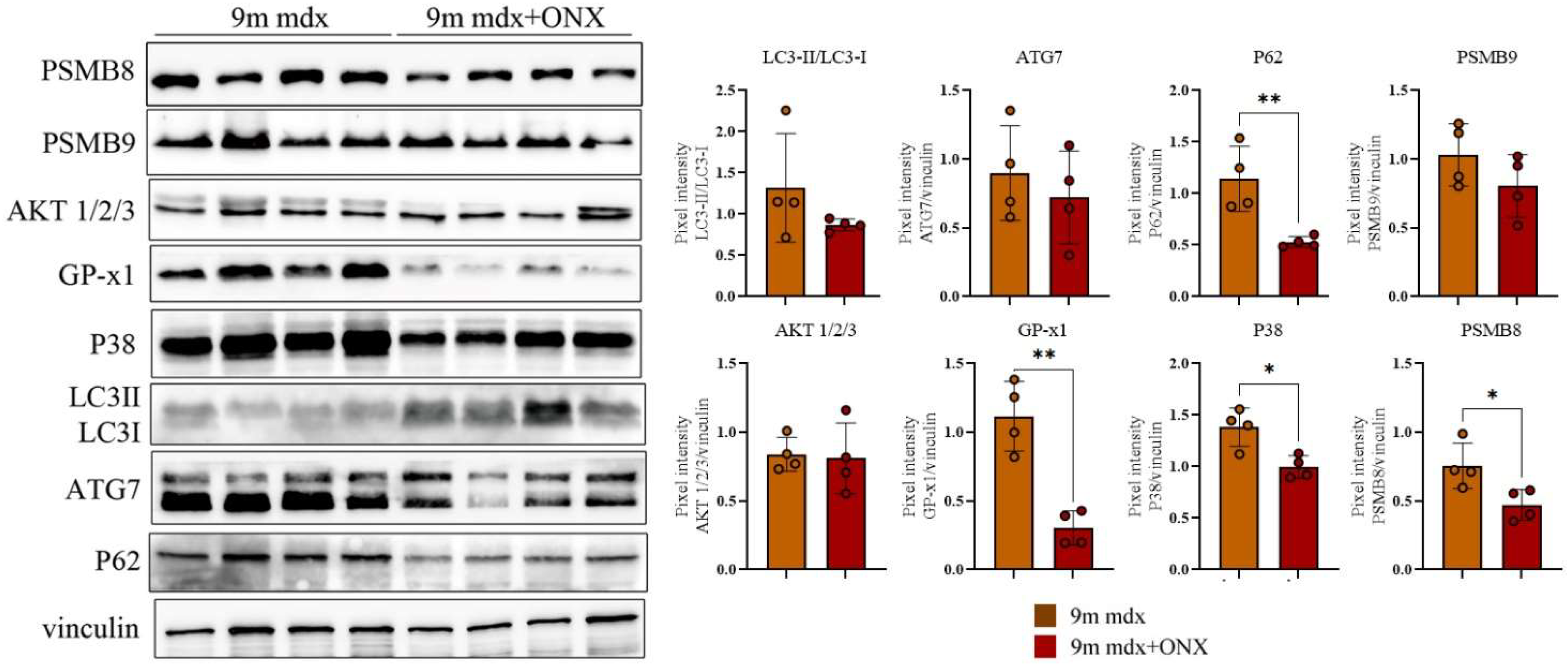
2.4. Proteomic Markers of Inflammation, Fibrosis and Oxidative Stress in Skeletal and Cardiac Muscle Are Modulated by ONX-0914 Administration
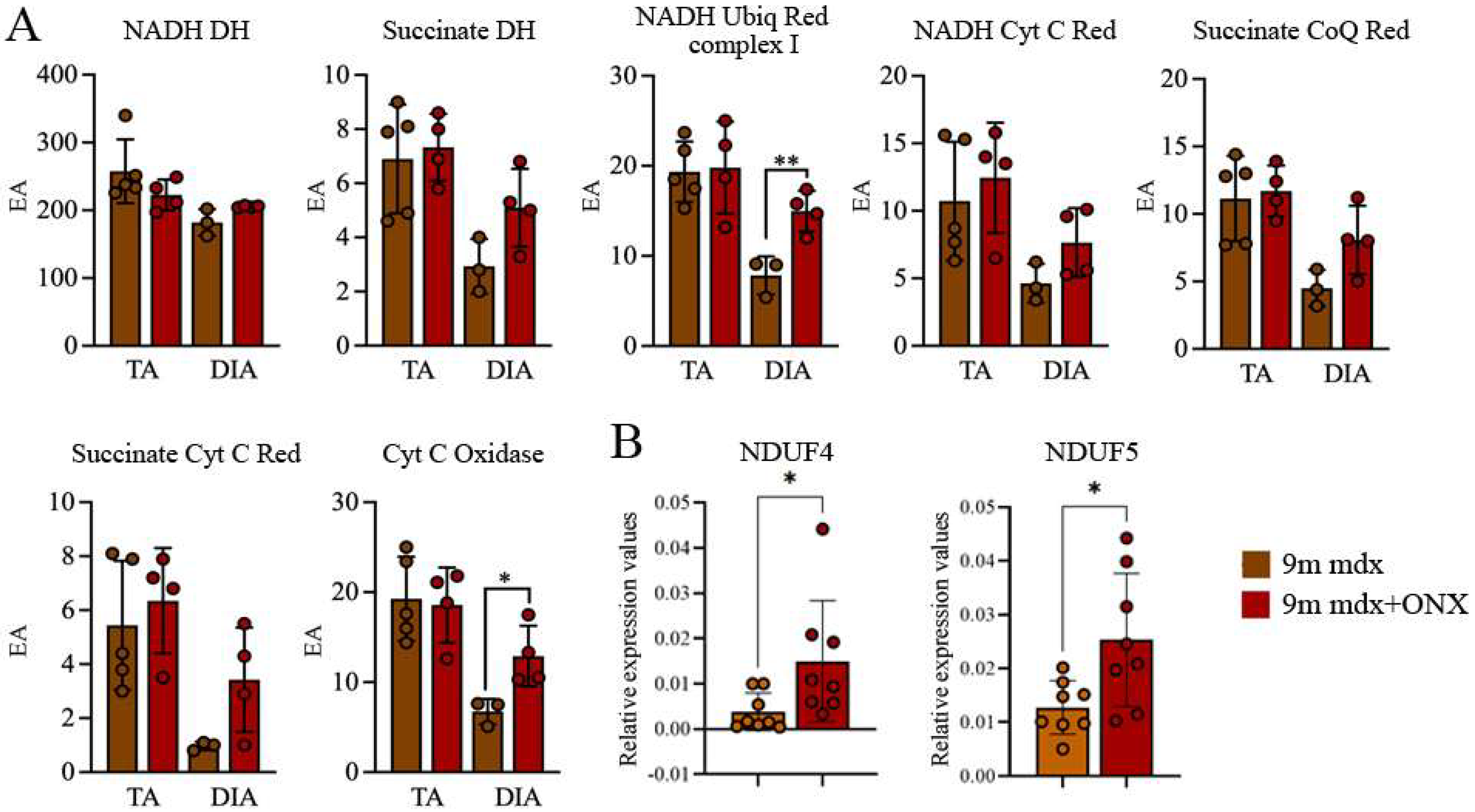
2.5. In Diaphragms of Aged mdx Mice
3. Discussion
4. Materials and Methods
4.1. Ethical Statement
4.2. Serum Analysis
4.3. Flow Cytometry Analysis
4.4. WB Analysis
4.5. Histological and Immunofluorescence Analysis of Muscle Sections
4.6. Quantitative (RT-qPCR) Experiments
4.7. Mitochondrial Enzymes
4.8. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Villalta, S.A.; Rosenberg, A.S.; Bluestone, J.A. The immune system in Duchenne muscular dystrophy: Friend or foe. Rare Dis. 2015, 3, e1010966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merlini, L.; Vagheggini, A.; Cocchi, D. Sarcopenia and sarcopenic obesity in patients with muscular dystrophy. Front. Aging Neurosci. 2014, 6, 274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ubaida-Mohien, C.; Lyashkov, A.; Gonzalez-Freire, M.; Tharakan, R.; Shardell, M.; Moaddel, R.; Semba, R.D.; Chia, C.W.; Gorospe, M.; Sen, R.; et al. Discovery proteomics in aging human skeletal muscle finds change in spliceosome, immunity, proteostasis and mitochondria. eLife 2019, 8, e49874. [Google Scholar] [CrossRef]
- Sugihara, H.; Teramoto, N.; Nakamura, K.; Shiga, T.; Shirakawa, T.; Matsuo, M.; Ogasawara, M.; Nishino, I.; Matsuwaki, T.; Nishihara, M.; et al. Cellular senescence-mediated exacerbation of Duchenne muscular dystrophy. Sci. Rep. 2020, 10, 16385. [Google Scholar] [CrossRef] [PubMed]
- Brehm, M.A.; Kempen, J.C.; van der Kooi, A.J.; de Groot, I.J.; van den Bergen, J.C.; Verschuuren, J.J.; Niks, E.H.; Harlaar, J. Age-related longitudinal changes in metabolic energy expenditure during walking in boys with Duchenne muscular dystrophy. PLoS ONE 2014, 9, e115200. [Google Scholar] [CrossRef] [PubMed]
- Fernando, R.; Drescher, C.; Nowotny, K.; Grune, T.; Castro, J.P. Impaired proteostasis during skeletal muscle aging. Free Radic. Biol. Med. 2019, 132, 58–66. [Google Scholar] [CrossRef]
- Cui, Z.; Hwang, S.M.; Gomes, A.V. Identification of the immunoproteasome as a novel regulator of skeletal muscle differentiation. Mol. Cell. Biol. 2014, 34, 96–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, A.V.; Zong, C.; Edmondson, R.D.; Berhane, B.T.; Wang, G.W.; Le, S.; Young, G.; Zhang, J.; Vondriska, T.M.; Whitelegge, J.P.; et al. The murine cardiac 26S proteasome: An organelle awaiting exploration. Ann. N. Y. Acad. Sci. 2005, 1047, 197–207. [Google Scholar] [CrossRef]
- Basler, M.; Kirk, C.J.; Groettrup, M. The immunoproteasome in antigen processing and other immunological functions. Curr. Opin. Immunol. 2013, 25, 74–80. [Google Scholar] [CrossRef] [Green Version]
- Ferrington, D.A.; Gregerson, D.S. Immunoproteasomes: Structure, function, and antigen presentation. Prog. Mol. Biol. Transl. Sci. 2012, 109, 75–112. [Google Scholar] [CrossRef]
- Liu, H.M.; Ferrington, D.A.; Baumann, C.W.; Thompson, L.V. Denervation-Induced Activation of the Standard Proteasome and Immunoproteasome. PLoS ONE 2016, 11, e0166831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farini, A.; Sitzia, C.; Cassani, B.; Cassinelli, L.; Rigoni, R.; Colleoni, F.; Fusco, N.; Gatti, S.; Bella, P.; Villa, C.; et al. Therapeutic Potential of Immunoproteasome Inhibition in Duchenne Muscular Dystrophy. Mol. Ther. J. Am. Soc. Gene Ther. 2016, 24, 1898–1912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farini, A.; Villa, C.; Di Silvestre, D.; Bella, P.; Tripodi, L.; Rossi, R.; Sitzia, C.; Gatti, S.; Mauri, P.; Torrente, Y. PTX3 Predicts Myocardial Damage and Fibrosis in Duchenne Muscular Dystrophy. Front. Physiol. 2020, 11, 403. [Google Scholar] [CrossRef]
- Adorisio, R.; Mencarelli, E.; Cantarutti, N.; Calvieri, C.; Amato, L.; Cicenia, M.; Silvetti, M.; D’Amico, A.; Grandinetti, M.; Drago, F.; et al. Duchenne Dilated Cardiomyopathy: Cardiac Management from Prevention to Advanced Cardiovascular Therapies. J. Clin. Med. 2020, 9, 3186. [Google Scholar] [CrossRef] [PubMed]
- Farini, A.; Gowran, A.; Bella, P.; Sitzia, C.; Scopece, A.; Castiglioni, E.; Rovina, D.; Nigro, P.; Villa, C.; Fortunato, F.; et al. Fibrosis Rescue Improves Cardiac Function in Dystrophin-Deficient Mice and Duchenne Patient-Specific Cardiomyocytes by Immunoproteasome Modulation. Am. J. Pathol. 2019, 189, 339–353. [Google Scholar] [CrossRef] [Green Version]
- Bansal, D.; Miyake, K.; Vogel, S.S.; Groh, S.; Chen, C.C.; Williamson, R.; McNeil, P.L.; Campbell, K.P. Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature 2003, 423, 168–172. [Google Scholar] [CrossRef]
- Bejaoui, K.; Hirabayashi, K.; Hentati, F.; Haines, J.L.; Ben Hamida, C.; Belal, S.; Miller, R.G.; McKenna-Yasek, D.; Weissenbach, J.; Rowland, L.P.; et al. Linkage of Miyoshi myopathy (distal autosomal recessive muscular dystrophy) locus to chromosome 2p12-14. Neurology 1995, 45, 768–772. [Google Scholar] [CrossRef]
- Nguyen, A.; Duquette, N.; Mamarbachi, M.; Thorin, E. Epigenetic Regulatory Effect of Exercise on Glutathione Peroxidase 1 Expression in the Skeletal Muscle of Severely Dyslipidemic Mice. PLoS ONE 2016, 11, e0151526. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Shin, H.S.; Shireman, P.K.; Vasilaki, A.; Van Remmen, H.; Csete, M.E. Glutathione-peroxidase-1 null muscle progenitor cells are globally defective. Free Radic. Biol. Med. 2006, 41, 1174–1184. [Google Scholar] [CrossRef]
- Reinecke, F.; Smeitink, J.A.; van der Westhuizen, F.H. OXPHOS gene expression and control in mitochondrial disorders. Biochim. Biophys. Acta 2009, 1792, 1113–1121. [Google Scholar] [CrossRef]
- Brown-Borg, H.M.; Johnson, W.T.; Rakoczy, S.G. Expression of oxidative phosphorylation components in mitochondria of long-living Ames dwarf mice. Age 2012, 34, 43–57. [Google Scholar] [CrossRef] [Green Version]
- Gallot, Y.S.; Straughn, A.R.; Bohnert, K.R.; Xiong, G.; Hindi, S.M.; Kumar, A. MyD88 is required for satellite cell-mediated myofiber regeneration in dystrophin-deficient mdx mice. Hum. Mol. Genet. 2018, 27, 3449–3463. [Google Scholar] [CrossRef]
- Bodine, S.C.; Stitt, T.N.; Gonzalez, M.; Kline, W.O.; Stover, G.L.; Bauerlein, R.; Zlotchenko, E.; Scrimgeour, A.; Lawrence, J.C.; Glass, D.J.; et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 2001, 3, 1014–1019. [Google Scholar] [CrossRef]
- Yoon, M.S. mTOR as a Key Regulator in Maintaining Skeletal Muscle Mass. Front. Physiol. 2017, 8, 788. [Google Scholar] [CrossRef] [Green Version]
- Whitley, K.C.; Hamstra, S.I.; Baranowski, R.W.; Watson, C.J.F.; MacPherson, R.E.K.; MacNeil, A.J.; Roy, B.D.; Vandenboom, R.; Fajardo, V.A. GSK3 inhibition with low dose lithium supplementation augments murine muscle fatigue resistance and specific force production. Physiol. Rep. 2020, 8, e14517. [Google Scholar] [CrossRef]
- Pansters, N.A.; Schols, A.M.; Verhees, K.J.; de Theije, C.C.; Snepvangers, F.J.; Kelders, M.C.; Ubags, N.D.; Haegens, A.; Langen, R.C. Muscle-specific GSK-3beta ablation accelerates regeneration of disuse-atrophied skeletal muscle. Biochim. Biophys. Acta 2015, 1852, 490–506. [Google Scholar] [CrossRef] [Green Version]
- Ardanaz, N.; Yang, X.P.; Cifuentes, M.E.; Haurani, M.J.; Jackson, K.W.; Liao, T.D.; Carretero, O.A.; Pagano, P.J. Lack of glutathione peroxidase 1 accelerates cardiac-specific hypertrophy and dysfunction in angiotensin II hypertension. Hypertension 2010, 55, 116–123. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Dennis, J.E.; Muzic, R.F.; Lundberg, M.; Caplan, A.I. The dynamic in vivo distribution of bone marrow-derived mesenchymal stem cells after infusion. Cells Tissues Organs 2001, 169, 12–20. [Google Scholar] [CrossRef]
- Li, M.; Georgakopoulos, D.; Lu, G.; Hester, L.; Kass, D.A.; Hasday, J.; Wang, Y. p38 MAP kinase mediates inflammatory cytokine induction in cardiomyocytes and extracellular matrix remodeling in heart. Circulation 2005, 111, 2494–2502. [Google Scholar] [CrossRef] [Green Version]
- Mareedu, S.; Million, E.D.; Duan, D.; Babu, G.J. Abnormal Calcium Handling in Duchenne Muscular Dystrophy: Mechanisms and Potential Therapies. Front. Physiol. 2021, 12, 647010. [Google Scholar] [CrossRef]
- Rybalka, E.; Timpani, C.A.; Stathis, C.G.; Hayes, A.; Cooke, M.B. Metabogenic and Nutriceutical Approaches to Address Energy Dysregulation and Skeletal Muscle Wasting in Duchenne Muscular Dystrophy. Nutrients 2015, 7, 9734–9767. [Google Scholar] [CrossRef] [Green Version]
- Duan, H.; Bai, H. Is Mitochondrial Oxidative Stress the Key Contributor to Diaphragm Atrophy and Dysfunction in Critically Ill Patients? Crit. Care Res. Pract. 2020, 2020, 8672939. [Google Scholar] [CrossRef] [Green Version]
- Rando, T.A. Oxidative stress and the pathogenesis of muscular dystrophies. Am. J. Phys. Med. Rehabil. 2002, 81, S175–S186. [Google Scholar] [CrossRef]
- Whitehead, N.P.; Yeung, E.W.; Allen, D.G. Muscle damage in mdx (dystrophic) mice: Role of calcium and reactive oxygen species. Clin. Exp. Pharmacol. Physiol. 2006, 33, 657–662. [Google Scholar] [CrossRef]
- Hauser, E.; Hoger, H.; Bittner, R.; Widhalm, K.; Herkner, K.; Lubec, G. Oxyradical damage and mitochondrial enzyme activities in the mdx mouse. Neuropediatrics 1995, 26, 260–262. [Google Scholar] [CrossRef] [PubMed]
- Haycock, J.W.; MacNeil, S.; Jones, P.; Harris, J.B.; Mantle, D. Oxidative damage to muscle protein in Duchenne muscular dystrophy. Neuroreport 1996, 8, 357–361. [Google Scholar] [CrossRef]
- Zhang, H.M.; Fu, J.; Hamilton, R.; Diaz, V.; Zhang, Y. The mammalian target of rapamycin modulates the immunoproteasome system in the heart. J. Mol. Cell. Cardiol. 2015, 86, 158–167. [Google Scholar] [CrossRef]
- Yun, Y.S.; Kim, K.H.; Tschida, B.; Sachs, Z.; Noble-Orcutt, K.E.; Moriarity, B.S.; Ai, T.; Ding, R.; Williams, J.; Chen, L.; et al. mTORC1 Coordinates Protein Synthesis and Immunoproteasome Formation via PRAS40 to Prevent Accumulation of Protein Stress. Mol. Cell 2016, 61, 625–639. [Google Scholar] [CrossRef] [Green Version]
- Pickering, A.M.; Davies, K.J. Differential roles of proteasome and immunoproteasome regulators Pa28alphabeta, Pa28gamma and Pa200 in the degradation of oxidized proteins. Arch. Biochem. Biophys. 2012, 523, 181–190. [Google Scholar] [CrossRef] [Green Version]
- Pickering, A.M.; Koop, A.L.; Teoh, C.Y.; Ermak, G.; Grune, T.; Davies, K.J. The immunoproteasome, the 20S proteasome and the PA28alphabeta proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem. J. 2010, 432, 585–594. [Google Scholar] [CrossRef]
- Chen, C.N.; Graber, T.G.; Bratten, W.M.; Ferrington, D.A.; Thompson, L.V. Immunoproteasome in animal models of Duchenne muscular dystrophy. J. Muscle Res. Cell Motil. 2014, 35, 191–201. [Google Scholar] [CrossRef] [Green Version]
- Jaisson, S.; Gillery, P. Impaired proteostasis: Role in the pathogenesis of diabetes mellitus. Diabetologia 2014, 57, 1517–1527. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, E.; Wiggs, M.; Greathouse, K.L.; Morgan, G.; Gordon, P.M. Impaired proteostasis in obese skeletal muscle relates to altered immunoproteasome activity. Appl. Physiol. Nutr. Metab. 2022, 47, 555–564. [Google Scholar] [CrossRef]
- Johnston-Carey, H.K.; Pomatto, L.C.; Davies, K.J. The Immunoproteasome in oxidative stress, aging, and disease. Crit. Rev. Biochem. Mol. Biol. 2015, 51, 268–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pickering, A.M.; Lehr, M.; Miller, R.A. Lifespan of mice and primates correlates with immunoproteasome expression. J. Clin. Investig. 2015, 125, 2059–2068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deyhle, M.R.; Hyldahl, R.D. The Role of T Lymphocytes in Skeletal Muscle Repair From Traumatic and Contraction-Induced Injury. Front. Physiol. 2018, 9, 768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfister, G.; Weiskopf, D.; Lazuardi, L.; Kovaiou, R.D.; Cioca, D.P.; Keller, M.; Lorbeg, B.; Parson, W.; Grubeck-Loebenstein, B. Naive T cells in the elderly: Are they still there? Ann. N. Y. Acad. Sci. 2006, 1067, 152–157. [Google Scholar] [CrossRef]
- VanderVeen, B.N.; Murphy, E.A.; Carson, J.A. The Impact of Immune Cells on the Skeletal Muscle Microenvironment during Cancer Cachexia. Front. Physiol. 2020, 11, 1037. [Google Scholar] [CrossRef]
- Kang, C.; Li Ji, L. Role of PGC-1alpha signaling in skeletal muscle health and disease. Ann. N. Y. Acad. Sci. 2012, 1271, 110–117. [Google Scholar] [CrossRef]
- Mirzoev, T.M.; Sharlo, K.A.; Shenkman, B.S. The Role of GSK-3beta in the Regulation of Protein Turnover, Myosin Phenotype, and Oxidative Capacity in Skeletal Muscle under Disuse Conditions. Int. J. Mol. Sci. 2021, 22, 5081. [Google Scholar] [CrossRef]
- Yang, S.; Loro, E.; Wada, S.; Kim, B.; Tseng, W.J.; Li, K.; Khurana, T.S.; Arany, Z. Functional effects of muscle PGC-1alpha in aged animals. Skelet. Muscle 2020, 10, 14. [Google Scholar] [CrossRef]
- Opitz, E.; Koch, A.; Klingel, K.; Schmidt, F.; Prokop, S.; Rahnefeld, A.; Sauter, M.; Heppner, F.L.; Volker, U.; Kandolf, R.; et al. Impairment of immunoproteasome function by beta5i/LMP7 subunit deficiency results in severe enterovirus myocarditis. PLoS Pathog. 2011, 7, e1002233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paeschke, A.; Possehl, A.; Klingel, K.; Voss, M.; Voss, K.; Kespohl, M.; Sauter, M.; Overkleeft, H.S.; Althof, N.; Garlanda, C.; et al. The immunoproteasome controls the availability of the cardioprotective pattern recognition molecule Pentraxin3. Eur. J. Immunol. 2016, 46, 619–633. [Google Scholar] [CrossRef] [Green Version]
- Launay, N.; Ruiz, M.; Fourcade, S.; Schluter, A.; Guilera, C.; Ferrer, I.; Knecht, E.; Pujol, A. Oxidative stress regulates the ubiquitin-proteasome system and immunoproteasome functioning in a mouse model of X-adrenoleukodystrophy. Brain J. Neurol. 2013, 136, 891–904. [Google Scholar] [CrossRef] [Green Version]
- Azzu, V.; Brand, M.D. Degradation of an intramitochondrial protein by the cytosolic proteasome. J. Cell Sci. 2010, 123, 578–585. [Google Scholar] [CrossRef] [Green Version]
- Radke, S.; Chander, H.; Schafer, P.; Meiss, G.; Kruger, R.; Schulz, J.B.; Germain, D. Mitochondrial protein quality control by the proteasome involves ubiquitination and the protease Omi. J. Biol. Chem. 2008, 283, 12681–12685. [Google Scholar] [CrossRef] [Green Version]
- Shang, F.; Taylor, A. Ubiquitin-proteasome pathway and cellular responses to oxidative stress. Free Radic. Biol. Med. 2011, 51, 5–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basler, M.; Groettrup, M. On the Role of the Immunoproteasome in Protein Homeostasis. Cells 2021, 10, 3216. [Google Scholar] [CrossRef]
- Burzyn, D.; Kuswanto, W.; Kolodin, D.; Shadrach, J.L.; Cerletti, M.; Jang, Y.; Sefik, E.; Tan, T.G.; Wagers, A.J.; Benoist, C.; et al. A special population of regulatory T cells potentiates muscle repair. Cell 2013, 155, 1282–1295. [Google Scholar] [CrossRef] [Green Version]
- Farini, A.; Sitzia, C.; Villa, C.; Cassani, B.; Tripodi, L.; Legato, M.; Belicchi, M.; Bella, P.; Lonati, C.; Gatti, S.; et al. Defective dystrophic thymus determines degenerative changes in skeletal muscle. Nat. Commun. 2021, 12, 2099. [Google Scholar] [CrossRef] [PubMed]
- Balsa, E.; Marco, R.; Perales-Clemente, E.; Szklarczyk, R.; Calvo, E.; Landazuri, M.O.; Enriquez, J.A. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab. 2012, 16, 378–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.L.; Xu, Y.; Ju, J.Q.; Pan, Z.N.; Liu, J.C.; Sun, S.C. Increased Environment-Related Metabolism and Genetic Expression in the In Vitro Matured Mouse Oocytes by Transcriptome Analysis. Front. Cell Dev. Biol. 2021, 9, 642010. [Google Scholar] [CrossRef]
- Bresolin, N.; Zeviani, M.; Bonilla, E.; Miller, R.H.; Leech, R.W.; Shanske, S.; Nakagawa, M.; DiMauro, S. Fatal infantile cytochrome c oxidase deficiency: Decrease of immunologically detectable enzyme in muscle. Neurology 1985, 35, 802–812. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tripodi, L.; Molinaro, D.; Fortunato, F.; Mella, C.; Cassani, B.; Torrente, Y.; Farini, A. Immunoproteasome Inhibition Ameliorates Aged Dystrophic Mouse Muscle Environment. Int. J. Mol. Sci. 2022, 23, 14657. https://doi.org/10.3390/ijms232314657
Tripodi L, Molinaro D, Fortunato F, Mella C, Cassani B, Torrente Y, Farini A. Immunoproteasome Inhibition Ameliorates Aged Dystrophic Mouse Muscle Environment. International Journal of Molecular Sciences. 2022; 23(23):14657. https://doi.org/10.3390/ijms232314657
Chicago/Turabian StyleTripodi, Luana, Davide Molinaro, Francesco Fortunato, Carolina Mella, Barbara Cassani, Yvan Torrente, and Andrea Farini. 2022. "Immunoproteasome Inhibition Ameliorates Aged Dystrophic Mouse Muscle Environment" International Journal of Molecular Sciences 23, no. 23: 14657. https://doi.org/10.3390/ijms232314657
APA StyleTripodi, L., Molinaro, D., Fortunato, F., Mella, C., Cassani, B., Torrente, Y., & Farini, A. (2022). Immunoproteasome Inhibition Ameliorates Aged Dystrophic Mouse Muscle Environment. International Journal of Molecular Sciences, 23(23), 14657. https://doi.org/10.3390/ijms232314657







