Technologies for Viable Circulating Tumor Cell Isolation
Abstract
1. Introduction
2. Circulating Tumor Cells: Features and Clinical Significance
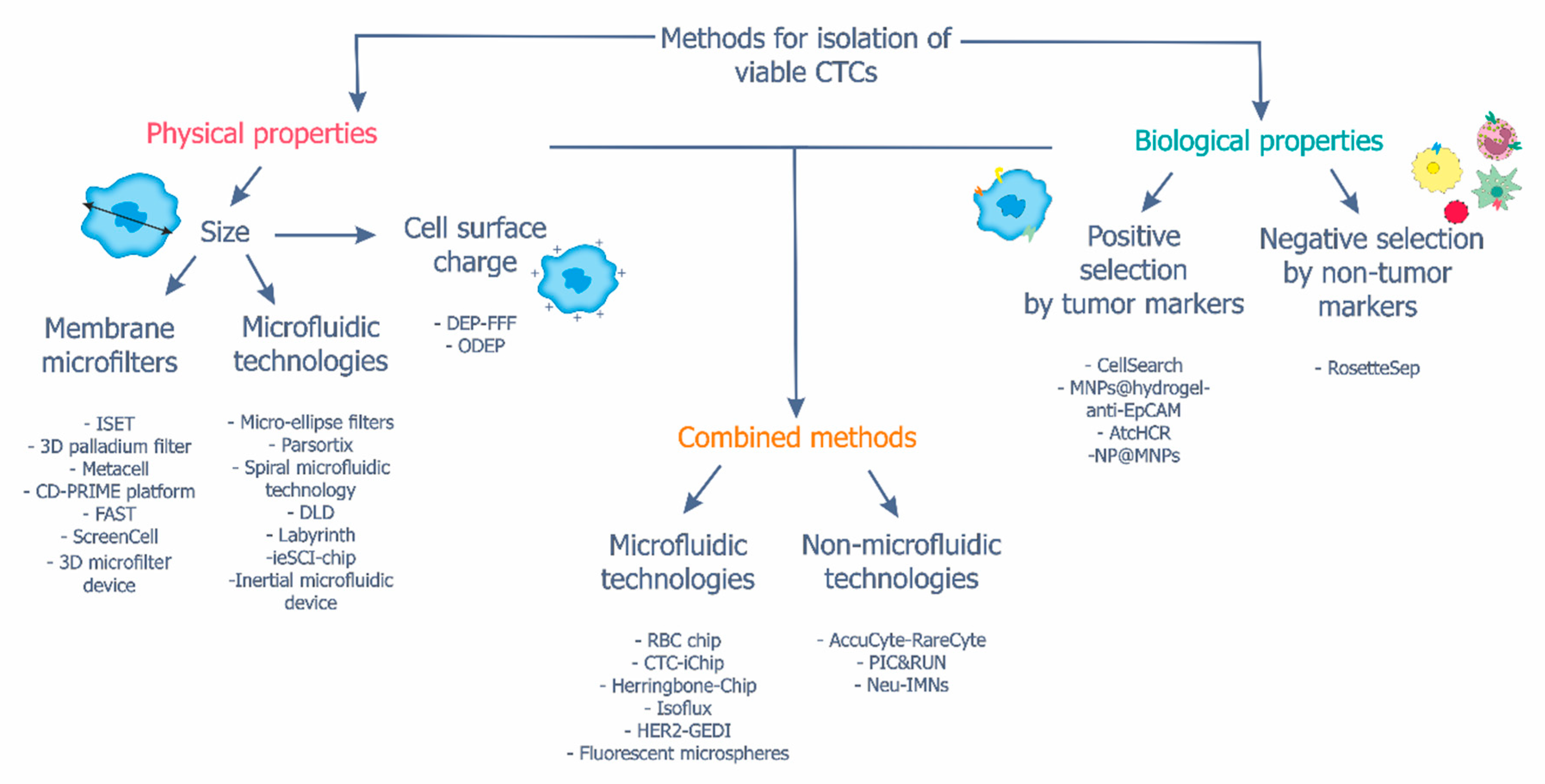
3. Methods for the Isolation of Viable CTCs
3.1. Isolation of CTCs Based on Cell Physical Properties
| Methods | Cancer Type | Further Applications | Advantages (+)/ Disadvantages (−) | References | |
|---|---|---|---|---|---|
| Membrane microfilters | ISET | Non-small cell lung, colorectal cancers, melanoma | Whole-genome sequencing | + 80–90% sensitivity + Isolation of CTC clusters − High loss of small cells | [60,61,86,87] |
| 3D palladium filter | Breast cancer | FISH, KRAS mutation analysis | + 85% cell purity – High loss of small cells | [62] | |
| MetaCell | Colorectal, lung cancers | CTC culturing, gene expression analysis | + Depletion of more than 95% of leukocytes + Isolation of CTC clusters – High loss of small cells | [18,65,88] | |
| CD-PRIME | Pancreatic cancer | NA | + 76% sensitivity – Contamination with leukocytes – High loss of small cells | [66] | |
| FAST | Colorectal, breast, stomach, lung cancers | KRAS mutation analysis | + 95.9 ± 3.1% sensitivity – High loss of small cells | [19,67] | |
| ScreenCell | Laryngeal, pancreatic cancers | FISH, ddPCR | + Fast isolation (<30 min) + Isolation of CTC clusters – High loss of small cells | [89,90] | |
| 3D microfilter device | NA | NA | + 86.5 ± 5.3% capture efficiency – High loss of small cells | [69] | |
| Microfluidic technologies | Micro-ellipse filter | Breast, colorectal, non-small cell lung cancers | Immunofluorescence analysis | + 90% capture efficiency – Contamination with leukocytes – Clogging of filters | [70,91] |
| Parsortix | Breastcancer | Mouse xenograft models, transcriptome or genome analysis, FISH | + 66–96% capture efficiency + Isolating CTC clusters – Contamination with blood cells – High loss of small cells | [71,72,73] | |
| Spiral microfluidic technology | Glioblastoma | FISH | + Fast isolation (15 min) + Isolation of CTC clusters + 90% recovery rate + 1.7 mL min processing rate – High loss of small cells | [76,77] | |
| DLD | Lung cancer | Transcriptome analysis | + 96% capture efficiency + Depletion of more than 99% of leukocytes + 98% viability + 1 mL min processing rate – High loss of small cells – Contamination with blood cells | [79,80] | |
| Labyrinth | Non-small cell lung, liver cancers | FISH | + Isolation of CTC clusters + Depletion of more than 95% of leukocytes – High loss of small cells | [81,92] | |
| ieSCI-chip | Breastcancer | Protein analysis at single-cell resolution | + 89.92 ± 3.37% capture efficiency + Fast isolation (6 min) + 1.4 mL min processing rate | [83] | |
| Inertial microfluidic device | Prostate, laryngeal, thyroid, floor of the mouth, non-small cell lungcancers | NA | + 89%±3.8% capture efficiency + Fast isolation (20 min) – Contamination with blood cells | [84,85] | |
| Dielectric permittivity | DEP-FFF | NA | Immunofluorescence, FISH, and gene mutation analysis | + 70–75% capture efficiency – High loss of small cells | [93,94] |
| ODEP | Head and neck cancer | NA | + 81.6–86.1% cell purity + Isolation of CTC clusters – High loss of small cells | [95,96] | |
3.2. Isolation of CTCs Based on Cell Biological Properties
3.3. Combined Methods
| Methods | Cancer Type | Further Applications | Advantages (+)/Disadvantages (–) | References | |
|---|---|---|---|---|---|
| Positive selection | MNPs@hydrogel-anti-EpCAM | NA | NA | + 96 % viability + Fast isolation (25 min) – Isolation of only EpCAM-positive CTCs | [100] |
| atcHCR | NA | NA | + Isolating CTC clusters – Isolation of only EpCAM-positive CTCs | [101] | |
| NP@MNPs | NA | scRNAseq | + Capture of mesenchymal CTCs + 85% capture efficiency – Isolation of only N-cadherin-positive CTCs | [102] | |
| Negative selection | RosetteSep | Liver, breast cancers | scRNAseq, CTC culturing | + Isolation of CTC clusters + Fast isolation (40 min) + CTC marker-free isolation – High number of untargeted cells | [68,105,106] |
| Methods | Cancer Type | Further Applications | Advantages (+)/Disadvantages (–) | References | |
|---|---|---|---|---|---|
| Based on cell biological properties and microfluidic approaches | RBC-chip | Colorectal cancer | ddPCR | + 96.5% sensitivity and specificity + 96.1% viability – Isolation of only EpCAM-positive CTCs | [104] |
| CTC-iChip | Breast, prostate, lung cancers, melanoma, glioma | CTC culturing | + 89.2 ± 5.7% capture efficiency + Capable of enrichment of CTCs with either positive or negative selection – Multistep nature of the protocol – Loss of CTCs associated with leukocytes | [107,108,109] | |
| HER2-GEDI | Breast, gastric cancers | CTC culturing | + No any blood processing is required + Utilization of only 1 mL of blood – Isolation of only HER2-positive CTCs | [110] | |
| Isoflux | Gastroesophageal cancer | KRAS mutation analysis | + 87% capture efficiency – Isolation of only EpCAM-positive CTCs | [111,112] | |
| Herringbone-Chip | Lung cancer | CTC culturing | + One step method + 95% capture efficiency + Isolation of CTC clusters – Isolation of only EpCAM- and EGFR-positive CTCs | [113] | |
| Fluorescent microspheres | Colorectal, breast, non-small cell lung cancers | NA | + 90% capture efficiency | [91] | |
| Based on cell biological properties and non-microfluidic approaches | AccuCyte- RareCyte/PIC & RUN | Prostate, breast, lung cancers | scRNAseq, CTC culturing | + 90% capture efficiency + 91.6% viability – The presence of false-positive CTCs | [114,115] |
| Neu-IMNs | Breast cancer | PCR, Sanger sequencing, CTC culturing | + 96.82% capture efficiency + 90.68% purity – Contamination with leukocytes | [116] | |
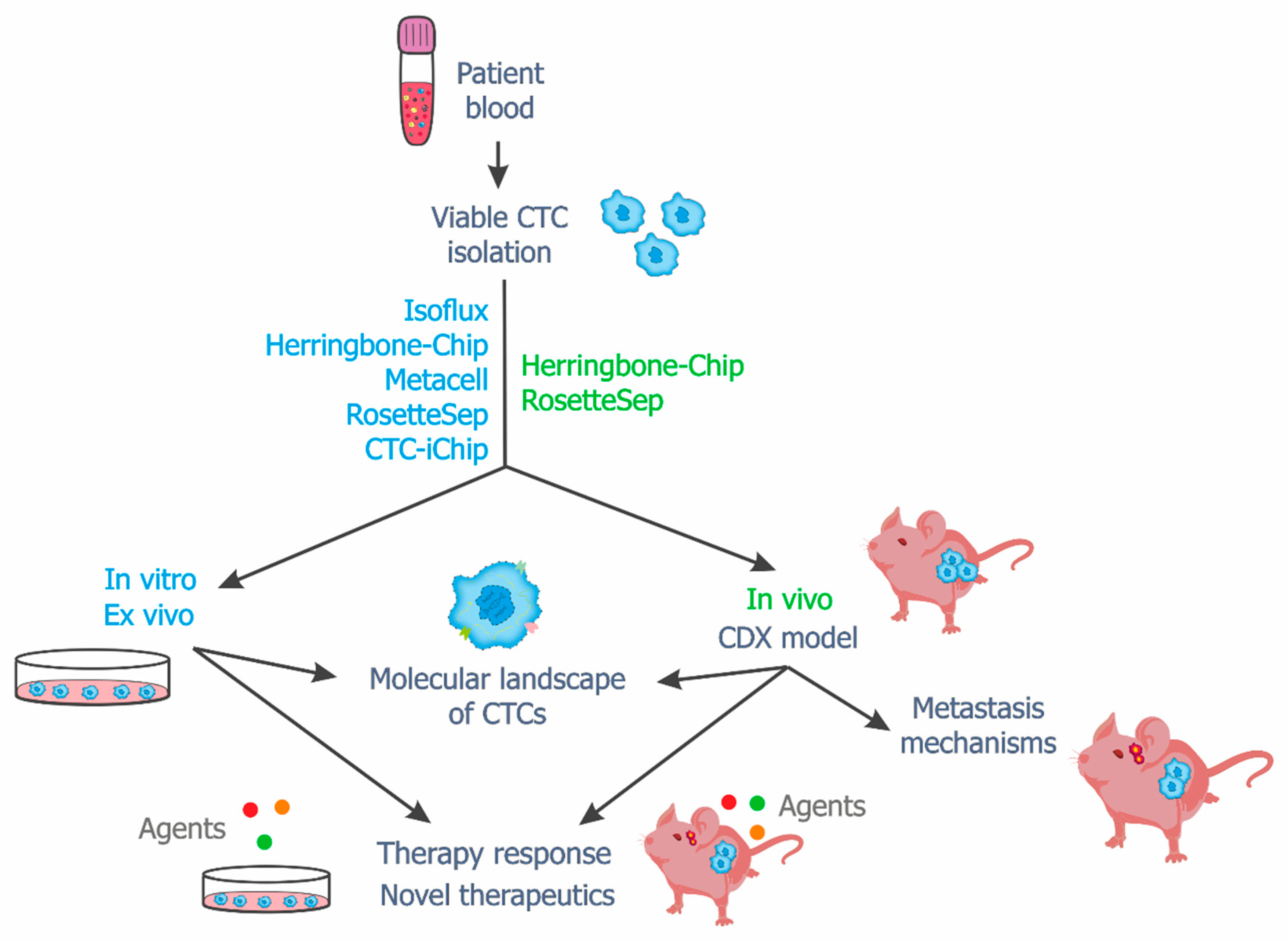
4. Viable CTCs in In Vitro, Ex Vivo and In Vivo Studies
5. Challenges and Trends
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Akhtar, M.; Haider, A.; Rashid, S.; Al-Nabet, A. Paget’s “Seed and Soil” Theory of Cancer Metastasis: An Idea Whose Time has Come. Adv. Anat. Pathol. 2019, 26, 69–74. [Google Scholar] [CrossRef]
- Khoo, B.L.; Chaudhuri, P.K.; Lim, C.T.; Warkiani, M.E. Advancing Techniques and Insights in Circulating Tumor Cell (CTC) Research. In Ex Vivo Engineering of the Tumor Microenvironment; Aref, A.R., Barbie, D., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 71–94. [Google Scholar]
- Agnoletto, C.; Corrà, F.; Minotti, L.; Baldassari, F.; Crudele, F.; Cook, W.J.J.; Di Leva, G.; d’Adamo, A.P.; Gasparini, P.; Volinia, S. Heterogeneity in Circulating Tumor Cells: The Relevance of the Stem-Cell Subset. Cancers 2019, 11, 483. [Google Scholar] [CrossRef]
- Saxena, K.; Subbalakshmi, A.R.; Jolly, M.K. Phenotypic heterogeneity in circulating tumor cells and its prognostic value in metastasis and overall survival. EBioMedicine 2019, 46, 4–5. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Dawson, S.J. Circulating tumor cells and circulating tumor DNA for precision medicine: Dream or reality? Ann. Oncol. 2014, 25, 2304–2313. [Google Scholar] [CrossRef]
- Habli, Z.; AlChamaa, W.; Saab, R.; Kadara, H.; Khraiche, M.L. Circulating Tumor Cell Detection Technologies and Clinical Utility: Challenges and Opportunities. Cancers 2020, 12, 1930. [Google Scholar] [CrossRef]
- Zhang, Z.; Shiratsuchi, H.; Palanisamy, N.; Nagrath, S.; Ramnath, N. Expanded Circulating Tumor Cells from a Patient with ALK-Positive Lung Cancer Present with EML4-ALK Rearrangement Along with Resistance Mutation and Enable Drug Sensitivity Testing: A Case Study. J. Thorac. Oncol. 2017, 12, 397–402. [Google Scholar] [CrossRef]
- Jordan, N.V.; Bardia, A.; Wittner, B.S.; Benes, C.; Ligorio, M.; Zheng, Y.; Yu, M.; Sundaresan, T.K.; Licausi, J.A.; Desai, R.; et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature 2016, 537, 102–106. [Google Scholar] [CrossRef]
- Miyamoto, D.T.; Lee, R.J.; Stott, S.L.; Ting, D.T.; Wittner, B.S.; Ulman, M.; Smas, M.E.; Lord, J.B.; Brannigan, B.W.; Trautwein, J.; et al. Androgen receptor signaling in circulating tumor cells as a marker of hormonally responsive prostate cancer. Cancer Discov. 2012, 2, 995–1003. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Bartkowiak, K.; Pantel, K. Functional studies on circulating and disseminated tumor cells in carcinoma patients. Mol. Oncol. 2016, 10, 443–449. [Google Scholar] [CrossRef]
- Ferreira, M.M.; Ramani, V.C.; Jeffrey, S.S. Circulating tumor cell technologies. Mol. Oncol. 2016, 10, 374–394. [Google Scholar] [CrossRef]
- Grigoriadis, N.; Kyritsis, K.; Akrivou, M.; Lefki-Pavlina, N.; Vizirianakis, I. A Novel Combined Methodology for Isolation and Detection of Circulating Tumor Cells based on Flow Cytometry and Cellular Filtration Technologies. Int. J. Cancer Clin. Res. 2020, 7, 132. [Google Scholar]
- Andree, K.C.; van Dalum, G.; Terstappen, L.W. Challenges in circulating tumor cell detection by the CellSearch system. Mol. Oncol. 2016, 10, 395–407. [Google Scholar] [CrossRef]
- Riethdorf, S.; Fritsche, H.; Müller, V.; Rau, T.; Schindlbeck, C.; Rack, B.; Janni, W.; Coith, C.; Beck, K.; Jänicke, F.; et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: A validation study of the CellSearch system. Clin. Cancer Res. 2007, 13, 920–928. [Google Scholar] [CrossRef]
- Resel Folkersma, L.; Olivier Gómez, C.; San José Manso, L.; Veganzones de Castro, S.; Galante Romo, I.; Vidaurreta Lázaro, M.; de la Orden, G.V.; Arroyo Fernández, M.; Díaz Rubio, E.; Silmi Moyano, A.; et al. Immunomagnetic quantification of circulating tumoral cells in patients with prostate cancer: Clinical and pathological correlation. Arch. Esp. Urol. 2010, 63, 23–31. [Google Scholar]
- Wang, L.; Balasubramanian, P.; Chen, A.P.; Kummar, S.; Evrard, Y.A.; Kinders, R.J. Promise and limits of the CellSearch platform for evaluating pharmacodynamics in circulating tumor cells. Semin. Oncol. 2016, 43, 464–475. [Google Scholar] [CrossRef]
- Kaur, P.; Campo, D.; Porras, T.B.; Ring, A.; Lu, J.; Chairez, Y.; Su, Y.; Kang, I.; Lang, J.E. A Pilot Study for the Feasibility of Exome-Sequencing in Circulating Tumor Cells Versus Single Metastatic Biopsies in Breast Cancer. Int. J. Mol. Sci. 2020, 21, 4826. [Google Scholar] [CrossRef]
- Cegan, M.; Kolostova, K.; Matkowski, R.; Broul, M.; Schraml, J.; Fiutowski, M.; Bobek, V. In vitro culturing of viable circulating tumor cells of urinary bladder cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 7164–7171. [Google Scholar]
- Kim, T.-H.; Lim, M.; Park, J.; Oh, J.M.; Kim, H.; Jeong, H.; Lee, S.J.; Park, H.C.; Jung, S.; Kim, B.C.; et al. FAST: Size-Selective, Clog-Free Isolation of Rare Cancer Cells from Whole Blood at a Liquid–Liquid Interface. Anal. Chem. 2017, 89, 1155–1162. [Google Scholar] [CrossRef]
- Lin, D.; Shen, L.; Luo, M.; Zhang, K.; Li, J.; Yang, Q.; Zhu, F.; Zhou, D.; Zheng, S.; Chen, Y.; et al. Circulating tumor cells: Biology and clinical significance. Signal Transduct. Target. Ther. 2021, 6, 404. [Google Scholar] [CrossRef]
- Harouaka, R.A.; Nisic, M.; Zheng, S.-Y. Circulating Tumor Cell Enrichment Based on Physical Properties. J. Lab. Autom. 2013, 18, 455–468. [Google Scholar] [CrossRef]
- Menyailo, M.E.; Tretyakova, M.S.; Denisov, E.V. Heterogeneity of Circulating Tumor Cells in Breast Cancer: Identifying Metastatic Seeds. Int. J. Mol. Sci. 2020, 21, 1696. [Google Scholar] [CrossRef]
- Hao, S.-J.; Wan, Y.; Xia, Y.-Q.; Zou, X.; Zheng, S.-Y. Size-based separation methods of circulating tumor cells. Adv. Drug Deliv. Rev. 2018, 125, 3–20. [Google Scholar] [CrossRef]
- Zhang, H.; Lin, X.; Huang, Y.; Wang, M.; Cen, C.; Tang, S.; Dique, M.R.; Cai, L.; Luis, M.A.; Smollar, J.; et al. Detection Methods and Clinical Applications of Circulating Tumor Cells in Breast Cancer. Front. Oncol. 2021, 11, 652253. [Google Scholar] [CrossRef]
- Shim, S.; Gascoyne, P.; Noshari, J.; Stemke Hale, K. Dynamic physical properties of dissociated tumor cells revealed by dielectrophoretic field-flow fractionation. Integr. Biol. 2011, 3, 850–862. [Google Scholar] [CrossRef]
- Shaw Bagnall, J.; Byun, S.; Begum, S.; Miyamoto, D.T.; Hecht, V.C.; Maheswaran, S.; Stott, S.L.; Toner, M.; Hynes, R.O.; Manalis, S.R. Deformability of Tumor Cells versus Blood Cells. Sci. Rep. 2015, 5, 18542. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Challenges in circulating tumour cell research. Nat. Rev. Cancer 2014, 14, 623–631. [Google Scholar] [CrossRef]
- Tretyakova, M.S.; Subbalakshmi, A.R.; Menyailo, M.E.; Jolly, M.K.; Denisov, E.V. Tumor Hybrid Cells: Nature and Biological Significance. Front. Cell Dev. Biol. 2022, 10, 814714. [Google Scholar] [CrossRef]
- Gires, O.; Pan, M.; Schinke, H.; Canis, M.; Baeuerle, P.A. Expression and function of epithelial cell adhesion molecule EpCAM: Where are we after 40 years? Cancer Metastasis Rev. 2020, 39, 969–987. [Google Scholar] [CrossRef]
- Jolly, M.K.; Murphy, R.J.; Bhatia, S.; Whitfield, H.J.; Redfern, A.; Davis, M.J.; Thompson, E.W. Measuring and Modelling the Epithelial-Mesenchymal Hybrid State in Cancer: Clinical Implications. Cells Tissues Organs 2022, 211, 110–133. [Google Scholar] [CrossRef]
- Jie, X.-X.; Zhang, X.-Y.; Xu, C.-J. Epithelial-to-mesenchymal transition, circulating tumor cells and cancer metastasis: Mechanisms and clinical applications. Oncotarget 2017, 8, 81558–81571. [Google Scholar] [CrossRef]
- Theodoropoulos, P.A.; Polioudaki, H.; Agelaki, S.; Kallergi, G.; Saridaki, Z.; Mavroudis, D.; Georgoulias, V. Circulating tumor cells with a putative stem cell phenotype in peripheral blood of patients with breast cancer. Cancer Lett. 2010, 288, 99–106. [Google Scholar] [CrossRef]
- Barriere, G.; Fici, P.; Gallerani, G.; Fabbri, F.; Zoli, W.; Rigaud, M. Circulating tumor cells and epithelial, mesenchymal and stemness markers: Characterization of cell subpopulations. Ann. Transl. Med. 2014, 2, 109. [Google Scholar] [CrossRef]
- Savelieva, O.E.; Tashireva, L.A.; Kaigorodova, E.V.; Buzenkova, A.V.; Mukhamedzhanov, R.K.; Grigoryeva, E.S.; Zavyalova, M.V.; Tarabanovskaya, N.A.; Cherdyntseva, N.V.; Perelmuter, V.M. Heterogeneity of Stemlike Circulating Tumor Cells in Invasive Breast Cancer. Int. J. Mol. Sci. 2020, 21, 2780. [Google Scholar] [CrossRef]
- Garrido-Navas, C.; de Miguel-Perez, D.; Exposito-Hernandez, J.; Bayarri, C.; Amezcua, V.; Ortigosa, A.; Valdivia, J.; Guerrero, R.; Garcia Puche, J.L.; Lorente, J.A.; et al. Cooperative and Escaping Mechanisms between Circulating Tumor Cells and Blood Constituents. Cells 2019, 8, 1382. [Google Scholar] [CrossRef]
- Hurtado, P.; Martínez-Pena, I.; Piñeiro, R. Dangerous Liaisons: Circulating Tumor Cells (CTCs) and Cancer-Associated Fibroblasts (CAFs). Cancers 2020, 12, 2861. [Google Scholar] [CrossRef]
- Bork, U.; Rahbari, N.N.; Schölch, S.; Reissfelder, C.; Kahlert, C.; Büchler, M.W.; Weitz, J.; Koch, M. Circulating tumour cells and outcome in non-metastatic colorectal cancer: A prospective study. Br. J. Cancer 2015, 112, 1306–1313. [Google Scholar] [CrossRef]
- van Dalum, G.; Stam, G.J.; Scholten, L.F.; Mastboom, W.J.; Vermes, I.; Tibbe, A.G.; De Groot, M.R.; Terstappen, L.W. Importance of circulating tumor cells in newly diagnosed colorectal cancer. Int. J. Oncol. 2015, 46, 1361–1368. [Google Scholar] [CrossRef]
- Mego, M.; De Giorgi, U.; Dawood, S.; Wang, X.; Valero, V.; Andreopoulou, E.; Handy, B.; Ueno, N.T.; Reuben, J.M.; Cristofanilli, M. Characterization of metastatic breast cancer patients with nondetectable circulating tumor cells. Int. J. Cancer 2011, 129, 417–423. [Google Scholar] [CrossRef]
- Jansson, S.; Bendahl, P.O.; Larsson, A.M.; Aaltonen, K.E.; Rydén, L. Prognostic impact of circulating tumor cell apoptosis and clusters in serial blood samples from patients with metastatic breast cancer in a prospective observational cohort. BMC Cancer 2016, 16, 433. [Google Scholar] [CrossRef]
- Murlidhar, V.; Reddy, R.M.; Fouladdel, S.; Zhao, L.; Ishikawa, M.K.; Grabauskiene, S.; Zhang, Z.; Lin, J.; Chang, A.C.; Carrott, P.; et al. Poor Prognosis Indicated by Venous Circulating Tumor Cell Clusters in Early-Stage Lung Cancers. Cancer Res. 2017, 77, 5194–5206. [Google Scholar] [CrossRef]
- Bidard, F.C.; Peeters, D.J.; Fehm, T.; Nolé, F.; Gisbert-Criado, R.; Mavroudis, D.; Grisanti, S.; Generali, D.; Garcia-Saenz, J.A.; Stebbing, J.; et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: A pooled analysis of individual patient data. Lancet Oncol. 2014, 15, 406–414. [Google Scholar] [CrossRef] [PubMed]
- de Bono, J.S.; Scher, H.I.; Montgomery, R.B.; Parker, C.; Miller, M.C.; Tissing, H.; Doyle, G.V.; Terstappen, L.W.; Pienta, K.J.; Raghavan, D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2008, 14, 6302–6309. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.G.; Sloane, R.; Priest, L.; Lancashire, L.; Hou, J.M.; Greystoke, A.; Ward, T.H.; Ferraldeschi, R.; Hughes, A.; Clack, G.; et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J. Clin. Oncol. 2011, 29, 1556–1563. [Google Scholar] [CrossRef]
- Cohen, S.J.; Punt, C.J.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef]
- Bocci, F.; Gearhart-Serna, L.; Boareto, M.; Ribeiro, M.; Ben-Jacob, E.; Devi, G.R.; Levine, H.; Onuchic, J.N.; Jolly, M.K. Toward understanding cancer stem cell heterogeneity in the tumor microenvironment. Proc. Natl. Acad. Sci. USA 2019, 116, 148–157. [Google Scholar] [CrossRef]
- Strati, A.; Nikolaou, M.; Georgoulias, V.; Lianidou, E.S. Prognostic Significance of TWIST1, CD24, CD44, and ALDH1 Transcript Quantification in EpCAM-Positive Circulating Tumor Cells from Early Stage Breast Cancer Patients. Cells 2019, 8, 652. [Google Scholar] [CrossRef] [PubMed]
- Su, P.; Lai, W.; Liu, L.; Zeng, Y.; Xu, H.; Lan, Q.; Chu, Z.; Chu, Z. Mesenchymal and Phosphatase of Regenerating Liver-3 Status in Circulating Tumor Cells May Serve as a Crucial Prognostic Marker for Assessing Relapse or Metastasis in Postoperative Patients With Colorectal Cancer. Clin. Transl. Gastroenterol. 2020, 11, e00265. [Google Scholar] [CrossRef]
- Wang, X.Q.; Liu, B.; Li, B.Y.; Wang, T.; Chen, D.Q. Effect of CTCs and INHBA level on the effect and prognosis of different treatment methods for patients with early breast cancer. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 12735–12740. [Google Scholar] [CrossRef]
- Magbanua, M.J.M.; Savenkov, O.; Asmus, E.J.; Ballman, K.V.; Scott, J.H.; Park, J.W.; Dickler, M.; Partridge, A.; Carey, L.A.; Winer, E.P.; et al. Clinical Significance of Circulating Tumor Cells in Hormone Receptor-positive Metastatic Breast Cancer Patients who Received Letrozole with or Without Bevacizumab. Clin. Cancer Res. 2020, 26, 4911–4920. [Google Scholar] [CrossRef]
- Wei, T.; Zhang, X.; Zhang, Q.; Yang, J.; Chen, Q.; Wang, J.; Li, X.; Chen, J.; Ma, T.; Li, G.; et al. Vimentin-positive circulating tumor cells as a biomarker for diagnosis and treatment monitoring in patients with pancreatic cancer. Cancer Lett. 2019, 452, 237–243. [Google Scholar] [CrossRef]
- Hüsemann, Y.; Geigl, J.B.; Schubert, F.; Musiani, P.; Meyer, M.; Burghart, E.; Forni, G.; Eils, R.; Fehm, T.; Riethmüller, G.; et al. Systemic spread is an early step in breast cancer. Cancer Cell 2008, 13, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, H.; Obradović, M.M.S.; Hoffmann, M.; Harper, K.L.; Sosa, M.S.; Werner-Klein, M.; Nanduri, L.K.; Werno, C.; Ehrl, C.; Maneck, M.; et al. Early dissemination seeds metastasis in breast cancer. Nature 2016, 540, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, J.; Kokuryo, T.; Yokoyama, Y.; Ebata, T.; Ochiai, Y.; Nagino, M. Premalignant pancreatic cells seed stealth metastasis in distant organs in mice. Oncogene 2021, 40, 2273–2284. [Google Scholar] [CrossRef] [PubMed]
- Rothé, F.; Venet, D.; Peeters, D.; Rouas, G.; Rediti, M.; Smeets, D.; Dupont, F.; Campbell, P.; Lambrechts, D.; Dirix, L.; et al. Interrogating breast cancer heterogeneity using single and pooled circulating tumor cell analysis. NPJ Breast Cancer 2022, 8, 79. [Google Scholar] [CrossRef]
- Sharma, S.; Zhuang, R.; Long, M.; Pavlovic, M.; Kang, Y.; Ilyas, A.; Asghar, W. Circulating tumor cell isolation, culture, and downstream molecular analysis. Biotechnol. Adv. 2018, 36, 1063–1078. [Google Scholar] [CrossRef]
- Bankó, P.; Lee, S.Y.; Nagygyörgy, V.; Zrínyi, M.; Chae, C.H.; Cho, D.H.; Telekes, A. Technologies for circulating tumor cell separation from whole blood. J. Hematol. Oncol. 2019, 12, 48. [Google Scholar] [CrossRef]
- Gabriel, M.T.; Calleja, L.R.; Chalopin, A.; Ory, B.; Heymann, D. Circulating Tumor Cells: A Review of Non–EpCAM-Based Approaches for Cell Enrichment and Isolation. Clin. Chem. 2016, 62, 571–581. [Google Scholar] [CrossRef]
- Hvichia, G.E.; Parveen, Z.; Wagner, C.; Janning, M.; Quidde, J.; Stein, A.; Müller, V.; Loges, S.; Neves, R.P.; Stoecklein, N.H.; et al. A novel microfluidic platform for size and deformability based separation and the subsequent molecular characterization of viable circulating tumor cells. Int. J. Cancer 2016, 138, 2894–2904. [Google Scholar] [CrossRef]
- Laget, S.; Broncy, L.; Hormigos, K.; Dhingra, D.M.; BenMohamed, F.; Capiod, T.; Osteras, M.; Farinelli, L.; Jackson, S.; Paterlini-Bréchot, P. Technical Insights into Highly Sensitive Isolation and Molecular Characterization of Fixed and Live Circulating Tumor Cells for Early Detection of Tumor Invasion. PLoS ONE 2017, 12, e0169427. [Google Scholar] [CrossRef]
- Tamminga, M.; Andree, K.C.; Hiltermann, T.J.N.; Jayat, M.; Schuuring, E.; van den Bos, H.; Spierings, D.C.J.; Lansdorp, P.M.; Timens, W.; Terstappen, L.W.M.M.; et al. Detection of Circulating Tumor Cells in the Diagnostic Leukapheresis Product of Non-Small-Cell Lung Cancer Patients Comparing CellSearch® and ISET. Cancers 2020, 12, 896. [Google Scholar] [CrossRef]
- Yusa, A.; Toneri, M.; Masuda, T.; Ito, S.; Yamamoto, S.; Okochi, M.; Kondo, N.; Iwata, H.; Yatabe, Y.; Ichinosawa, Y.; et al. Development of a New Rapid Isolation Device for Circulating Tumor Cells (CTCs) Using 3D Palladium Filter and Its Application for Genetic Analysis. PLoS ONE 2014, 9, e88821. [Google Scholar] [CrossRef] [PubMed]
- Kolostova, K.; Pinkas, M.; Jakabova, A.; Pospisilova, E.; Svobodova, P.; Spicka, J.; Cegan, M.; Matkowski, R.; Bobek, V. Molecular characterization of circulating tumor cells in ovarian cancer. Am. J. Cancer Res. 2016, 6, 973–980. [Google Scholar] [PubMed]
- Bobek, V.; Gurlich, R.; Eliasova, P.; Kolostova, K. Circulating tumor cells in pancreatic cancer patients: Enrichment and cultivation. World J. Gastroenterol. 2014, 20, 17163–17170. [Google Scholar] [CrossRef] [PubMed]
- Vasantharajan, S.S.; Barnett, E.; Gray, E.S.; McCall, J.L.; Rodger, E.J.; Eccles, M.R.; Munro, F.; Pattison, S.; Chatterjee, A. Assessment of a Size-Based Method for Enriching Circulating Tumour Cells in Colorectal Cancer. Cancers 2022, 14, 3446. [Google Scholar] [CrossRef]
- Park, Y.; Jun, H.R.; Choi, H.W.; Hwang, D.W.; Lee, J.H.; Song, K.B.; Lee, W.; Kwon, J.; Ha, S.H.; Jun, E.; et al. Circulating tumour cells as an indicator of early and systemic recurrence after surgical resection in pancreatic ductal adenocarcinoma. Sci. Rep. 2021, 11, 1644. [Google Scholar] [CrossRef]
- Baek, D.H.; Kim, G.H.; Song, G.A.; Han, I.S.; Park, E.Y.; Kim, H.S.; Jo, H.J.; Ko, S.H.; Park, D.Y.; Cho, Y.K. Clinical Potential of Circulating Tumor Cells in Colorectal Cancer: A Prospective Study. Clin. Transl. Gastroenterol. 2019, 10, e00055. [Google Scholar] [CrossRef]
- Drucker, A.; Teh, E.M.; Kostyleva, R.; Rayson, D.; Douglas, S.; Pinto, D.M. Comparative performance of different methods for circulating tumor cell enrichment in metastatic breast cancer patients. PLoS ONE 2020, 15, e0237308. [Google Scholar] [CrossRef]
- Zheng, S.; Lin, H.K.; Lu, B.; Williams, A.; Datar, R.; Cote, R.J.; Tai, Y.C. 3D microfilter device for viable circulating tumor cell (CTC) enrichment from blood. Biomed. Microdevices 2011, 13, 203–213. [Google Scholar] [CrossRef]
- Chen, H.; Cao, B.; Sun, B.; Cao, Y.; Yang, K.; Lin, Y.-S. Highly-sensitive capture of circulating tumor cells using micro-ellipse filters. Sci. Rep. 2017, 7, 610. [Google Scholar] [CrossRef]
- Lampignano, R.; Yang, L.; Neumann, M.H.D.; Franken, A.; Fehm, T.; Niederacher, D.; Neubauer, H. A Novel Workflow to Enrich and Isolate Patient-Matched EpCAM(high) and EpCAM(low/negative) CTCs Enables the Comparative Characterization of the PIK3CA Status in Metastatic Breast Cancer. Int. J. Mol. Sci. 2017, 18, 1885. [Google Scholar] [CrossRef]
- Miller, M.C.; Robinson, P.S.; Wagner, C.; O’Shannessy, D.J. The Parsortix™ Cell Separation System—A versatile liquid biopsy platform. Cytom. Part A 2018, 93, 1234–1239. [Google Scholar] [CrossRef]
- Kitz, J.; Lowes, L.E.; Goodale, D.; Allan, A.L. Circulating Tumor Cell Analysis in Preclinical Mouse Models of Metastasis. Diagnostics 2018, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Jesenko, T.; Modic, Z.; Kuhar, C.G.; Cemazar, M.; Matkovic, U.; Miceska, S.; Varl, J.; Kuhar, A.; Kloboves-Prevodnik, V. Morphological features of breast cancer circulating tumor cells in blood after physical and biological type of isolation. Radiol. Oncol. 2021, 55, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Philippron, A.; Depypere, L.; Oeyen, S.; De Laere, B.; Vandeputte, C.; Nafteux, P.; De Preter, K.; Pattyn, P. Evaluation of a marker independent isolation method for circulating tumor cells in esophageal adenocarcinoma. PLoS ONE 2021, 16, e0251052. [Google Scholar] [CrossRef]
- Müller Bark, J.; Kulasinghe, A.; Hartel, G.; Leo, P.; Warkiani, M.E.; Jeffree, R.L.; Chua, B.; Day, B.W.; Punyadeera, C. Isolation of Circulating Tumour Cells in Patients With Glioblastoma Using Spiral Microfluidic Technology—A Pilot Study. Front. Oncol. 2021, 11, 681130. [Google Scholar] [CrossRef] [PubMed]
- Warkiani, M.E.; Khoo, B.L.; Wu, L.; Tay, A.K.P.; Bhagat, A.A.S.; Han, J.; Lim, C.T. Ultra-fast, label-free isolation of circulating tumor cells from blood using spiral microfluidics. Nat. Protoc. 2016, 11, 134–148. [Google Scholar] [CrossRef]
- Aceto, N.; Bardia, A.; Miyamoto, D.T.; Donaldson, M.C.; Wittner, B.S.; Spencer, J.A.; Yu, M.; Pely, A.; Engstrom, A.; Zhu, H. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 2014, 158, 1110–1122. [Google Scholar] [CrossRef]
- Tottori, N.; Hatsuzawa, T.; Nisisako, T. Separation of main and satellite droplets in a deterministic lateral displacement microfluidic device. RSC Adv. 2017, 7, 35516–35524. [Google Scholar] [CrossRef]
- Liu, Z.; Huang, Y.; Liang, W.; Bai, J.; Feng, H.; Fang, Z.; Tian, G.; Zhu, Y.; Zhang, H.; Wang, Y.; et al. Cascaded filter deterministic lateral displacement microchips for isolation and molecular analysis of circulating tumor cells and fusion cells. Lab Chip 2021, 21, 2881–2891. [Google Scholar] [CrossRef]
- Wan, S.; Kim, T.H.; Smith, K.J.; Delaney, R.; Park, G.S.; Guo, H.; Lin, E.; Plegue, T.; Kuo, N.; Steffes, J.; et al. New Labyrinth Microfluidic Device Detects Circulating Tumor Cells Expressing Cancer Stem Cell Marker and Circulating Tumor Microemboli in Hepatocellular Carcinoma. Sci. Rep. 2019, 9, 18575. [Google Scholar] [CrossRef]
- Rivera-Báez, L.; Lohse, I.; Lin, E.; Raghavan, S.; Owen, S.; Harouaka, R.; Herman, K.; Mehta, G.; Lawrence, T.S.; Morgan, M.A.; et al. Expansion of Circulating Tumor Cells from Patients with Locally Advanced Pancreatic Cancer Enable Patient Derived Xenografts and Functional Studies for Personalized Medicine. Cancers 2020, 12, 1011. [Google Scholar] [CrossRef] [PubMed]
- Abdulla, A.; Zhang, T.; Li, S.; Guo, W.; Warden, A.R.; Xin, Y.; Maboyi, N.; Lou, J.; Xie, H.; Ding, X. Integrated microfluidic single-cell immunoblotting chip enables high-throughput isolation, enrichment and direct protein analysis of circulating tumor cells. Microsyst. Nanoeng. 2022, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Rzhevskiy, A.S.; Kapitannikova, A.Y.; Vasilescu, S.A.; Karashaeva, T.A.; Razavi Bazaz, S.; Taratkin, M.S.; Enikeev, D.V.; Lekarev, V.Y.; Shpot, E.V.; Butnaru, D.V.; et al. Isolation of Circulating Tumor Cells from Seminal Fluid of Patients with Prostate Cancer Using Inertial Microfluidics. Cancers 2022, 14, 3364. [Google Scholar] [CrossRef]
- Volovetskiy, A.B.; Malinina, P.A.; Kapitannikova, A.Y.; Smetanina, S.V.; Kruglova, I.A.; Maslennikova, A.V. Isolation of Circulating Tumor Cells from Peripheral Blood Samples of Cancer Patients Using Microfluidic Technology. Sovrem. Tekhnologii Meditsine 2021, 12, 62–68. [Google Scholar] [CrossRef] [PubMed]
- De Giorgi, V.; Pinzani, P.; Salvianti, F.; Grazzini, M.; Orlando, C.; Lotti, T.; Pazzagli, M.; Massi, D. Circulating Benign Nevus Cells Detected by ISET Technique: Warning for Melanoma Molecular Diagnosis. Arch. Dermatol. 2010, 146, 1120–1124. [Google Scholar] [CrossRef][Green Version]
- Chen, F.; Wang, S.; Fang, Y.; Zheng, L.; Zhi, X.; Cheng, B.; Chen, Y.; Zhang, C.; Shi, D.; Song, H.; et al. Feasibility of a novel one-stop ISET device to capture CTCs and its clinical application. Oncotarget 2017, 8, 3029–3041. [Google Scholar] [CrossRef] [PubMed]
- Kolostova, K.; Zhang, Y.; Hoffman, R.M.; Bobek, V. In vitro culture and characterization of human lung cancer circulating tumor cells isolated by size exclusion from an orthotopic nude-mouse model expressing fluorescent protein. J. Fluoresc. 2014, 24, 1531–1536. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, M.I.; Ralli, M.; Nicolazzo, C.; Gradilone, A.; Carletti, R.; Di Gioia, C.; De Vincentiis, M.; Greco, A. Detection of circulating tumor cells in patients with laryngeal cancer using ScreenCell: Comparative pre- and post-operative analysis and association with prognosis. Oncol. Lett. 2020, 19, 4183–4188. [Google Scholar] [CrossRef]
- Amantini, C.; Morelli, M.B.; Nabissi, M.; Piva, F.; Marinelli, O.; Maggi, F.; Bianchi, F.; Bittoni, A.; Berardi, R.; Giampieri, R.; et al. Expression Profiling of Circulating Tumor Cells in Pancreatic Ductal Adenocarcinoma Patients: Biomarkers Predicting Overall Survival. Front. Oncol. 2019, 9, 874. [Google Scholar] [CrossRef]
- Chen, H.; Peng, G.; Li, Y.; Wang, S.; Lin, C. Clinical immunofluorescence comparison analysis of microfluidic chips for highly efficient detection of CTCs with colorectal cancer and other cancers. Microfluid. Nanofluidics 2022, 26, 62. [Google Scholar] [CrossRef]
- Zeinali, M.; Lee, M.; Nadhan, A.; Mathur, A.; Hedman, C.; Lin, E.; Harouaka, R.; Wicha, M.S.; Zhao, L.; Palanisamy, N.; et al. High-Throughput Label-Free Isolation of Heterogeneous Circulating Tumor Cells and CTC Clusters from Non-Small-Cell Lung Cancer Patients. Cancers 2020, 12, 127. [Google Scholar] [CrossRef] [PubMed]
- Shim, S.; Stemke-Hale, K.; Tsimberidou, A.M.; Noshari, J.; Anderson, T.E.; Gascoyne, P.R. Antibody-independent isolation of circulating tumor cells by continuous-flow dielectrophoresis. Biomicrofluidics 2013, 7, 11807. [Google Scholar] [CrossRef] [PubMed]
- Gascoyne, P.R.C.; Noshari, J.; Anderson, T.J.; Becker, F.F. Isolation of rare cells from cell mixtures by dielectrophoresis. Electrophoresis 2009, 30, 1388–1398. [Google Scholar] [CrossRef]
- Chu, P.-Y.; Hsieh, C.-H.; Wu, M.-H. The Combination of Immunomagnetic Bead-Based Cell Isolation and Optically Induced Dielectrophoresis (ODEP)-Based Microfluidic Device for the Negative Selection-Based Isolation of Circulating Tumor Cells (CTCs). Front. Bioeng. Biotechnol. 2020, 8, 921. [Google Scholar] [CrossRef]
- Chiu, T.-K.; Chao, A.C.; Chou, W.-P.; Liao, C.-J.; Wang, H.-M.; Chang, J.-H.; Chen, P.-H.; Wu, M.-H. Optically-induced-dielectrophoresis (ODEP)-based cell manipulation in a microfluidic system for high-purity isolation of integral circulating tumor cell (CTC) clusters based on their size characteristics. Sens. Actuators B Chem. 2018, 258, 1161–1173. [Google Scholar] [CrossRef]
- Waheed, W.; Sharaf, O.Z.; Alazzam, A.; Abu-Nada, E. Dielectrophoresis-field flow fractionation for separation of particles: A critical review. J. Chromatogr. A 2021, 1637, 461799. [Google Scholar] [CrossRef]
- Königsberg, R.; Obermayr, E.; Bises, G.; Pfeiler, G.; Gneist, M.; Wrba, F.; de Santis, M.; Zeillinger, R.; Hudec, M.; Dittrich, C. Detection of EpCAM positive and negative circulating tumor cells in metastatic breast cancer patients. Acta Oncol. 2011, 50, 700–710. [Google Scholar] [CrossRef]
- Mikolajczyk, S.D.; Millar, L.S.; Tsinberg, P.; Coutts, S.M.; Zomorrodi, M.; Pham, T.; Bischoff, F.Z.; Pircher, T.J. Detection of EpCAM-Negative and Cytokeratin-Negative Circulating Tumor Cells in Peripheral Blood. J. Oncol. 2011, 2011, 252361. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, Z.; Sun, N.; Cao, Y.; Cai, X.; Yuan, F.; Zou, H.; Xing, C.; Pei, R. Antifouling hydrogel-coated magnetic nanoparticles for selective isolation and recovery of circulating tumor cells. J. Mater. Chem. B 2021, 9, 677–682. [Google Scholar] [CrossRef]
- Song, P.; Ye, D.; Zuo, X.; Li, J.; Wang, J.; Liu, H.; Hwang, M.T.; Chao, J.; Su, S.; Wang, L.; et al. DNA Hydrogel with Aptamer-Toehold-Based Recognition, Cloaking, and Decloaking of Circulating Tumor Cells for Live Cell Analysis. Nano Lett. 2017, 17, 5193–5198. [Google Scholar] [CrossRef]
- Jia, F.; Wang, Y.; Fang, Z.; Dong, J.; Shi, F.; Zhang, W.; Wang, Z.; Hu, Z. Novel peptide-based magnetic nanoparticle for mesenchymal circulating tumor cells detection. Anal. Chem. 2021, 93, 5670–5675. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabières, C.; Pantel, K. Technologies for detection of circulating tumor cells: Facts and vision. Lab Chip 2014, 14, 57–62. [Google Scholar] [CrossRef]
- Shen, H.; Su, R.; Peng, J.; Zhu, L.; Deng, K.; Niu, Q.; Song, Y.; Yang, L.; Wu, L.; Zhu, Z.; et al. Antibody-engineered red blood cell interface for high-performance capture and release of circulating tumor cells. Bioact. Mater. 2022, 11, 32–40. [Google Scholar] [CrossRef] [PubMed]
- D’Avola, D.; Villacorta-Martin, C.; Martins-Filho, S.N.; Craig, A.; Labgaa, I.; von Felden, J.; Kimaada, A.; Bonaccorso, A.; Tabrizian, P.; Hartmann, B.M.; et al. High-density single cell mRNA sequencing to characterize circulating tumor cells in hepatocellular carcinoma. Sci. Rep. 2018, 8, 11570. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Ule, N.; González-Conde, M.; Abuín, C.; Cueva, J.F.; Palacios, P.; López-López, R.; Costa, C.; Dávila-Ibáñez, A.B. Short-Term Ex Vivo Culture of CTCs from Advance Breast Cancer Patients: Clinical Implications. Cancers 2021, 13, 2668. [Google Scholar] [CrossRef] [PubMed]
- Fachin, F.; Spuhler, P.; Martel-Foley, J.M.; Edd, J.F.; Barber, T.A.; Walsh, J.; Karabacak, M.; Pai, V.; Yu, M.; Smith, K.; et al. Monolithic Chip for High-throughput Blood Cell Depletion to Sort Rare Circulating Tu103mor Cells. Sci. Rep. 2017, 7, 10936. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Dubash, T.D.; Edd, J.F.; Jewett, M.K.; Garre, S.G.; Karabacak, N.M.; Rabe, D.C.; Mutlu, B.R.; Walsh, J.R.; Kapur, R.; et al. Ultrahigh-throughput magnetic sorting of large blood volumes for epitope-agnostic isolation of circulating tumor cells. Proc. Natl. Acad. Sci. USA 2020, 117, 16839–16847. [Google Scholar] [CrossRef]
- Zhang, H.; Yuan, F.; Qi, Y.; Liu, B.; Chen, Q. Circulating Tumor Cells for Glioma. Front. Oncol. 2021, 11, 607150. [Google Scholar] [CrossRef]
- Galletti, G.; Sung, M.S.; Vahdat, L.T.; Shah, M.A.; Santana, S.M.; Altavilla, G.; Kirby, B.J.; Giannakakou, P. Isolation of breast cancer and gastric cancer circulating tumor cells by use of an anti HER2-based microfluidic device. Lab Chip 2014, 14, 147–156. [Google Scholar] [CrossRef]
- Ruiz-Rodríguez, A.J.; Molina-Vallejo, M.P.; Aznar-Peralta, I.; González Puga, C.; Cañas García, I.; González, E.; Lorente, J.A.; Serrano, M.J.; Garrido-Navas, M.C. Deep Phenotypic Characterisation of CTCs by Combination of Microfluidic Isolation (IsoFlux) and Imaging Flow Cytometry (ImageStream). Cancers 2021, 13, 6386. [Google Scholar] [CrossRef]
- Brungs, D.; Lynch, D.; Luk, A.W.; Minaei, E.; Ranson, M.; Aghmesheh, M.; Vine, K.L.; Carolan, M.; Jaber, M.; de Souza, P.; et al. Cryopreservation for delayed circulating tumor cell isolation is a valid strategy for prognostic association of circulating tumor cells in gastroesophageal cancer. World J. Gastroenterol. 2018, 24, 810–818. [Google Scholar] [CrossRef] [PubMed]
- Que, Z.; Luo, B.; Zhou, Z.; Dong, C.; Jiang, Y.; Wang, L.; Shi, Q.; Tian, J. Establishment and characterization of a patient-derived circulating lung tumor cell line in vitro and in vivo. Cancer Cell Int. 2019, 19, 21. [Google Scholar] [CrossRef] [PubMed]
- Kaldjian, E.P.; Ramirez, A.B.; Sun, Y.; Campton, D.E.; Werbin, J.L.; Varshavskaya, P.; Quarre, S.; George, T.; Madan, A.; Blau, C.A.; et al. The RareCyte® platform for next-generation analysis of circulating tumor cells. Cytom. Part A 2018, 93, 1220–1225. [Google Scholar] [CrossRef] [PubMed]
- Kamal, M.; Saremi, S.; Klotz, R.; Iriondo, O.; Amzaleg, Y.; Chairez, Y.; Tulpule, V.; Lang, J.E.; Kang, I.; Yu, M. PIC&RUN: An integrated assay for the detection and retrieval of single viable circulating tumor cells. Sci. Rep. 2019, 9, 17470. [Google Scholar] [CrossRef]
- Wu, X.; Lin, Z.; Zhao, C.; Liu, L.; Zhang, K.; Lai, J.; Meng, Q.-F.; Yao, G.; Huang, Q.; Zhao, X.-Z.; et al. Neutrophil membrane-coated immunomagnetic nanoparticles for efficient isolation and analysis of circulating tumor cells. Biosens. Bioelectron. 2022, 213, 114425. [Google Scholar] [CrossRef]
- Harb, W.; Fan, A.; Tran, T.; Danila, D.C.; Keys, D.; Schwartz, M.; Ionescu-Zanetti, C. Mutational Analysis of Circulating Tumor Cells Using a Novel Microfluidic Collection Device and qPCR Assay. Transl. Oncol. 2013, 6, 528-IN521. [Google Scholar] [CrossRef]
- Chen, B.; Zheng, J.; Gao, K.; Hu, X.; Guo, S.-S.; Zhao, X.-Z.; Liao, F.; Yang, Y.; Liu, W. Noninvasive Optical Isolation and Identification of Circulating Tumor Cells Engineered by Fluorescent Microspheres. ACS Appl. Bio Mater. 2022, 5, 2768–2776. [Google Scholar] [CrossRef]
- Yu, M.; Bardia, A.; Aceto, N.; Bersani, F.; Madden, M.W.; Donaldson, M.C.; Desai, R.; Zhu, H.; Comaills, V.; Zheng, Z.; et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 2014, 345, 216–220. [Google Scholar] [CrossRef]
- Zhang, L.; Ridgway, L.D.; Wetzel, M.D.; Ngo, J.; Yin, W.; Kumar, D.; Goodman, J.C.; Groves, M.D.; Marchetti, D. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci. Transl. Med. 2013, 5, 180ra148. [Google Scholar] [CrossRef]
- Brungs, D.; Minaei, E.; Piper, A.-K.; Perry, J.; Splitt, A.; Carolan, M.; Ryan, S.; Wu, X.J.; Corde, S.; Tehei, M.; et al. Establishment of novel long-term cultures from EpCAM positive and negative circulating tumour cells from patients with metastatic gastroesophageal cancer. Sci. Rep. 2020, 10, 539. [Google Scholar] [CrossRef]
- Bobek, V.; Matkowski, R.; Gürlich, R.; Grabowski, K.; Szelachowska, J.; Lischke, R.; Schützner, J.; Harustiak, T.; Pazdro, A.; Rzechonek, A.; et al. Cultivation of circulating tumor cells in esophageal cancer. Folia Histochem. Cytobiol. 2014, 52, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Soler, A.; Cayrefourcq, L.; Mazard, T.; Babayan, A.; Lamy, P.-J.; Assou, S.; Assenat, E.; Pantel, K.; Alix-Panabières, C. Autologous cell lines from circulating colon cancer cells captured from sequential liquid biopsies as model to study therapy-driven tumor changes. Sci. Rep. 2018, 8, 15931. [Google Scholar] [CrossRef]
- Cayrefourcq, L.; Thomas, F.; Mazard, T.; Assenat, E.; Assou, S.; Alix-Panabières, C. Selective treatment pressure in colon cancer drives the molecular profile of resistant circulating tumor cell clones. Mol. Cancer 2021, 20, 30. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Cayrefourcq, L.; Mazard, T.; Maudelonde, T.; Assenat, E.; Assou, S. Molecular Portrait of Metastasis-Competent Circulating Tumor Cells in Colon Cancer Reveals the Crucial Role of Genes Regulating Energy Metabolism and DNA Repair. Clin. Chem. 2017, 63, 700–713. [Google Scholar] [CrossRef] [PubMed]
- Drapkin, B.J.; George, J.; Christensen, C.L.; Mino-Kenudson, M.; Dries, R.; Sundaresan, T.; Phat, S.; Myers, D.T.; Zhong, J.; Igo, P.; et al. Genomic and Functional Fidelity of Small Cell Lung Cancer Patient-Derived Xenografts. Cancer Discov. 2018, 8, 600–615. [Google Scholar] [CrossRef] [PubMed]
- Rupp, B.; Ball, H.; Wuchu, F.; Nagrath, D.; Nagrath, S. Circulating tumor cells in precision medicine: Challenges and opportunities. Trends Pharmacol. Sci. 2022, 43, 378–391. [Google Scholar] [CrossRef]
- Pereira-Veiga, T.; Abreu, M.; Robledo, D.; Matias-Guiu, X.; Santacana, M.; Sánchez, L.; Cueva, J.; Palacios, P.; Abdulkader, I.; López-López, R.; et al. CTCs-derived xenograft development in a triple negative breast cancer case. Int. J. Cancer 2019, 144, 2254–2265. [Google Scholar] [CrossRef]
- Stewart, C.A.; Gay, C.M.; Xi, Y.; Sivajothi, S.; Sivakamasundari, V.; Fujimoto, J.; Bolisetty, M.; Hartsfield, P.M.; Balasubramaniyan, V.; Chalishazar, M.D.; et al. Single-cell analyses reveal increased intratumoral heterogeneity after the onset of therapy resistance in small-cell lung cancer. Nat. Cancer 2020, 1, 423–436. [Google Scholar] [CrossRef]
- Vishnoi, M.; Liu, N.H.; Yin, W.; Boral, D.; Scamardo, A.; Hong, D.; Marchetti, D. The identification of a TNBC liver metastasis gene signature by sequential CTC-xenograft modeling. Mol. Oncol. 2019, 13, 1913–1926. [Google Scholar] [CrossRef]
- Faugeroux, V.; Pailler, E.; Oulhen, M.; Deas, O.; Brulle-Soumare, L.; Hervieu, C.; Marty, V.; Alexandrova, K.; Andree, K.C.; Stoecklein, N.H.; et al. Genetic characterization of a unique neuroendocrine transdifferentiation prostate circulating tumor cell-derived eXplant model. Nat. Commun. 2020, 11, 1884. [Google Scholar] [CrossRef]
- May, A.N.; Crawford, B.D.; Nedelcu, A.M. In Vitro Model-Systems to Understand the Biology and Clinical Significance of Circulating Tumor Cell Clusters. Front. Oncol. 2018, 8, 63. [Google Scholar] [CrossRef] [PubMed]
- Girotti, M.R.; Gremel, G.; Lee, R.; Galvani, E.; Rothwell, D.; Viros, A.; Mandal, A.K.; Lim, K.H.; Saturno, G.; Furney, S.J.; et al. Application of Sequencing, Liquid Biopsies, and Patient-Derived Xenografts for Personalized Medicine in Melanoma. Cancer Discov. 2016, 6, 286–299. [Google Scholar] [CrossRef] [PubMed]
- Menyailo, M.E.; Zainullina, V.R.; Tashireva, L.A.; Zolotareva, S.Y.; Gerashchenko, T.S.; Alifanov, V.V.; Savelieva, O.E.; Grigoryeva, E.S.; Tarabanovskaya, N.A.; Popova, N.O.; et al. Heterogeneity of circulating epithelial cells in breast cancer at single-cell resolution: Identifying tumor and hybrid cells. bioRxiv 2022. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.T.; Zhang, W.; Wang, G.Z.; Zhou, B.; Yang, G.W.; Qu, X.D.; Liu, R.; Qian, S.; Zhu, L.; Liu, L.X.; et al. Circulating tumor cells in the central and peripheral venous compartment—Assessing hematogenous dissemination after transarterial chemoembolization of hepatocellular carcinoma. OncoTargets Ther. 2014, 7, 1311–1318. [Google Scholar] [CrossRef] [PubMed]
- Gray, E.S. Arterial or Venous: Where Are the Circulating Tumor Cells? EBioMedicine 2015, 2, 1596–1597. [Google Scholar] [CrossRef]
- Terai, M.; Mu, Z.; Eschelman, D.J.; Gonsalves, C.F.; Kageyama, K.; Chervoneva, I.; Orloff, M.; Weight, R.; Mastrangelo, M.J.; Cristofanilli, M.; et al. Arterial Blood, Rather Than Venous Blood, is a Better Source for Circulating Melanoma Cells. EBioMedicine 2015, 2, 1821–1826. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tretyakova, M.S.; Menyailo, M.E.; Schegoleva, A.A.; Bokova, U.A.; Larionova, I.V.; Denisov, E.V. Technologies for Viable Circulating Tumor Cell Isolation. Int. J. Mol. Sci. 2022, 23, 15979. https://doi.org/10.3390/ijms232415979
Tretyakova MS, Menyailo ME, Schegoleva AA, Bokova UA, Larionova IV, Denisov EV. Technologies for Viable Circulating Tumor Cell Isolation. International Journal of Molecular Sciences. 2022; 23(24):15979. https://doi.org/10.3390/ijms232415979
Chicago/Turabian StyleTretyakova, Maria S., Maxim E. Menyailo, Anastasia A. Schegoleva, Ustinia A. Bokova, Irina V. Larionova, and Evgeny V. Denisov. 2022. "Technologies for Viable Circulating Tumor Cell Isolation" International Journal of Molecular Sciences 23, no. 24: 15979. https://doi.org/10.3390/ijms232415979
APA StyleTretyakova, M. S., Menyailo, M. E., Schegoleva, A. A., Bokova, U. A., Larionova, I. V., & Denisov, E. V. (2022). Technologies for Viable Circulating Tumor Cell Isolation. International Journal of Molecular Sciences, 23(24), 15979. https://doi.org/10.3390/ijms232415979







