Tumor Suppressor miRNA-503 Inhibits Cell Invasion in Head and Neck Cancer through the Wnt Signaling Pathway via the WNT3A/MMP Molecular Axis
Abstract
1. Introduction
2. Results
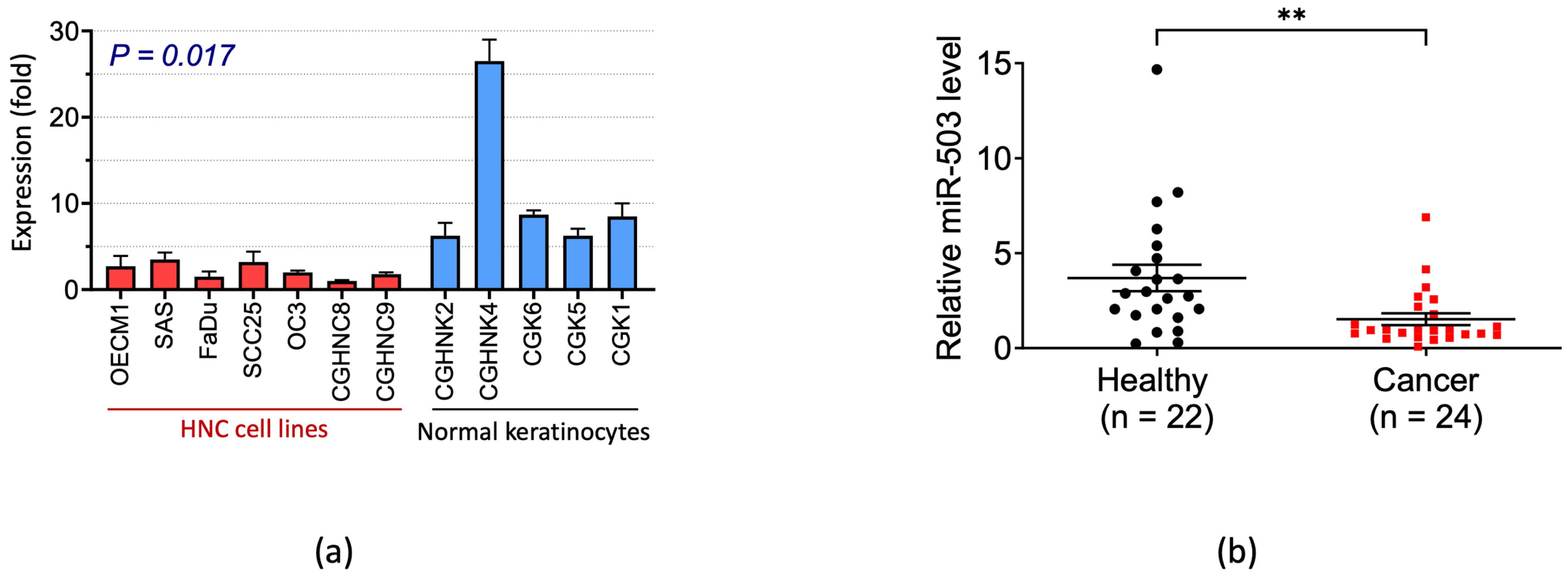
2.1. miRNA-503 Is Downexpressed in Patients with HNC as Well as HNC Cell Lines
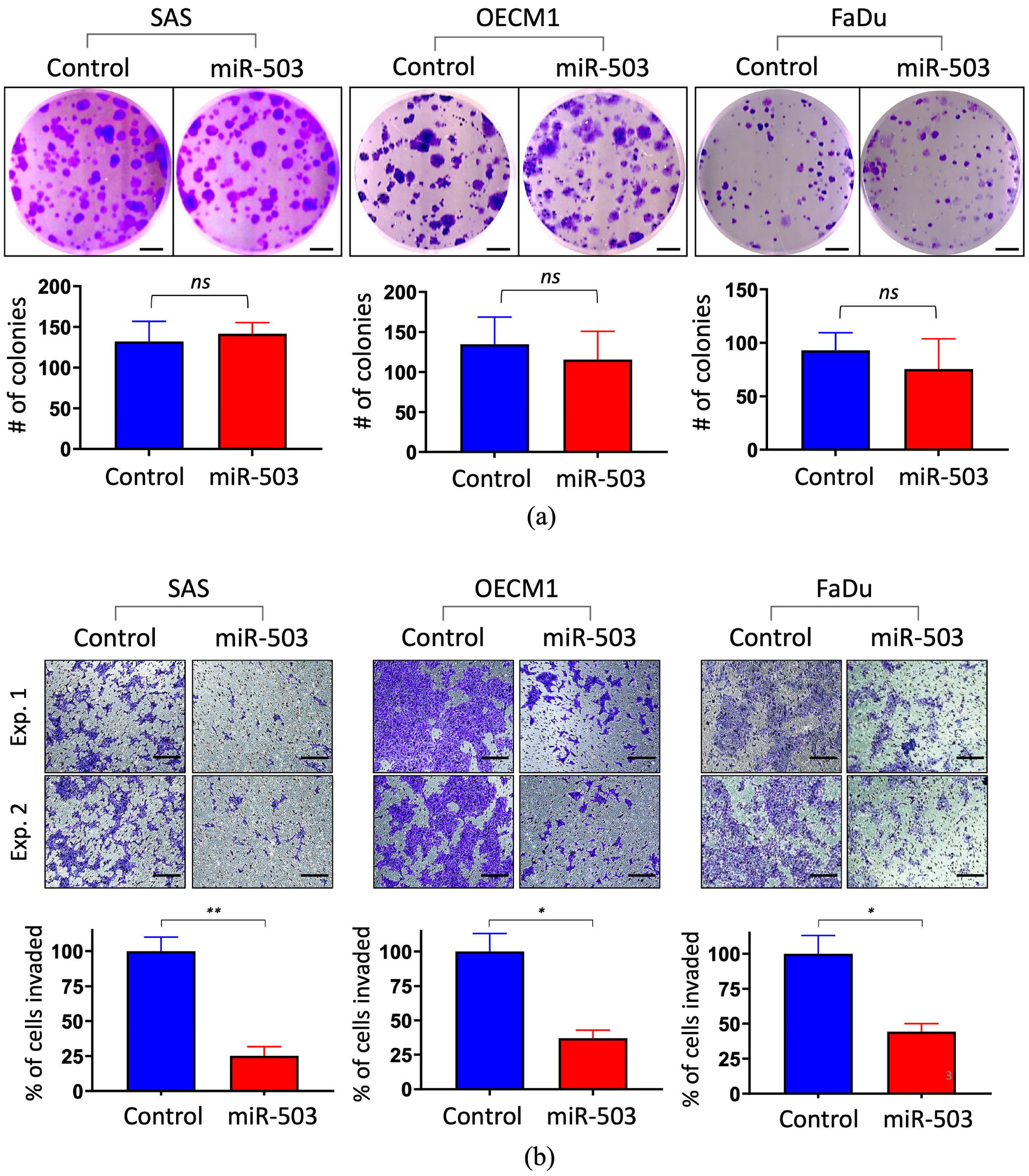
2.2. miRNA-503 Has a Minimal Effect on Cell Growth but Suppresses Cell Invasion in HNC Cells
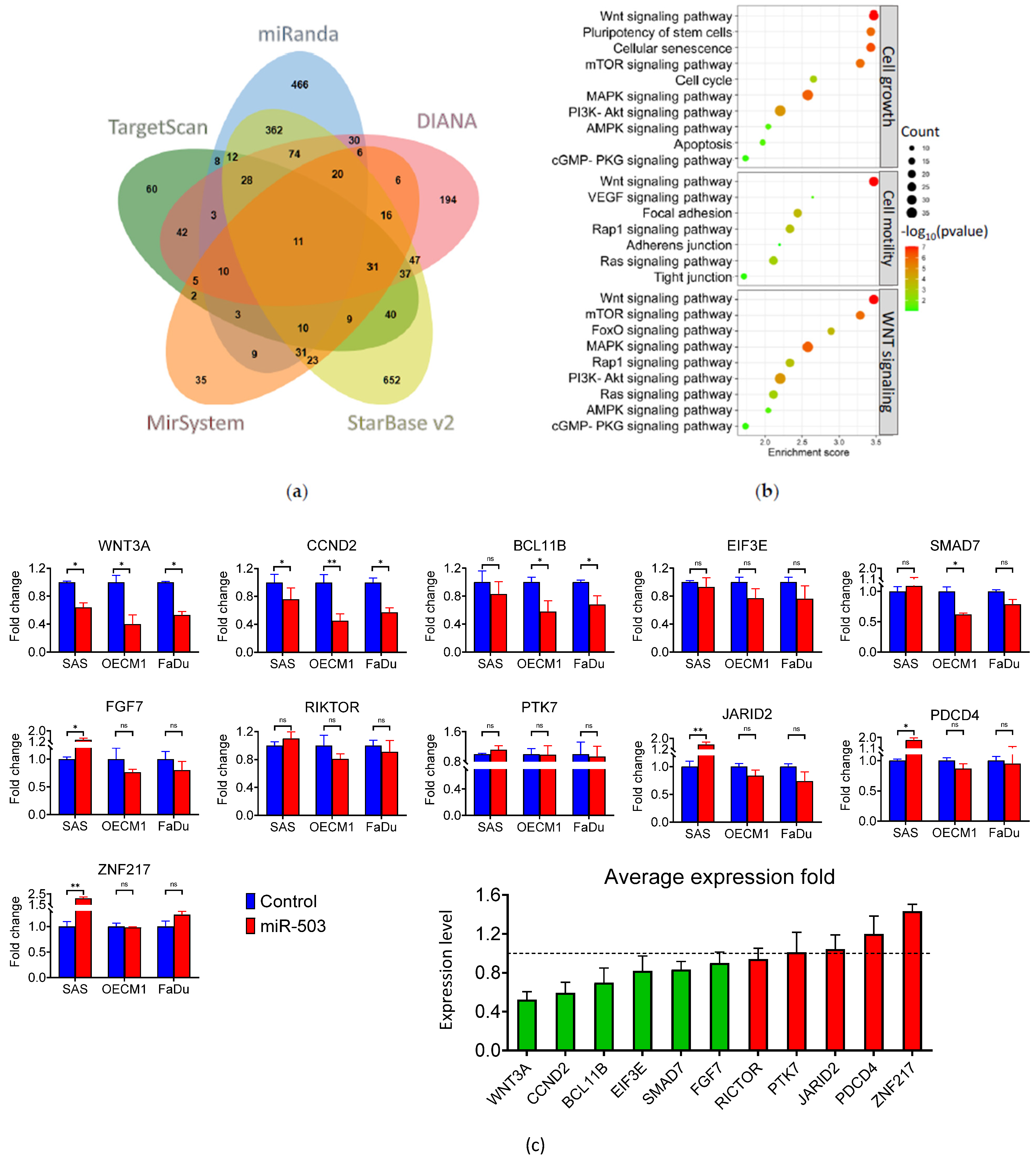
2.3. WNT3A Is Directly Inhibited by miRNA-503 in HNC
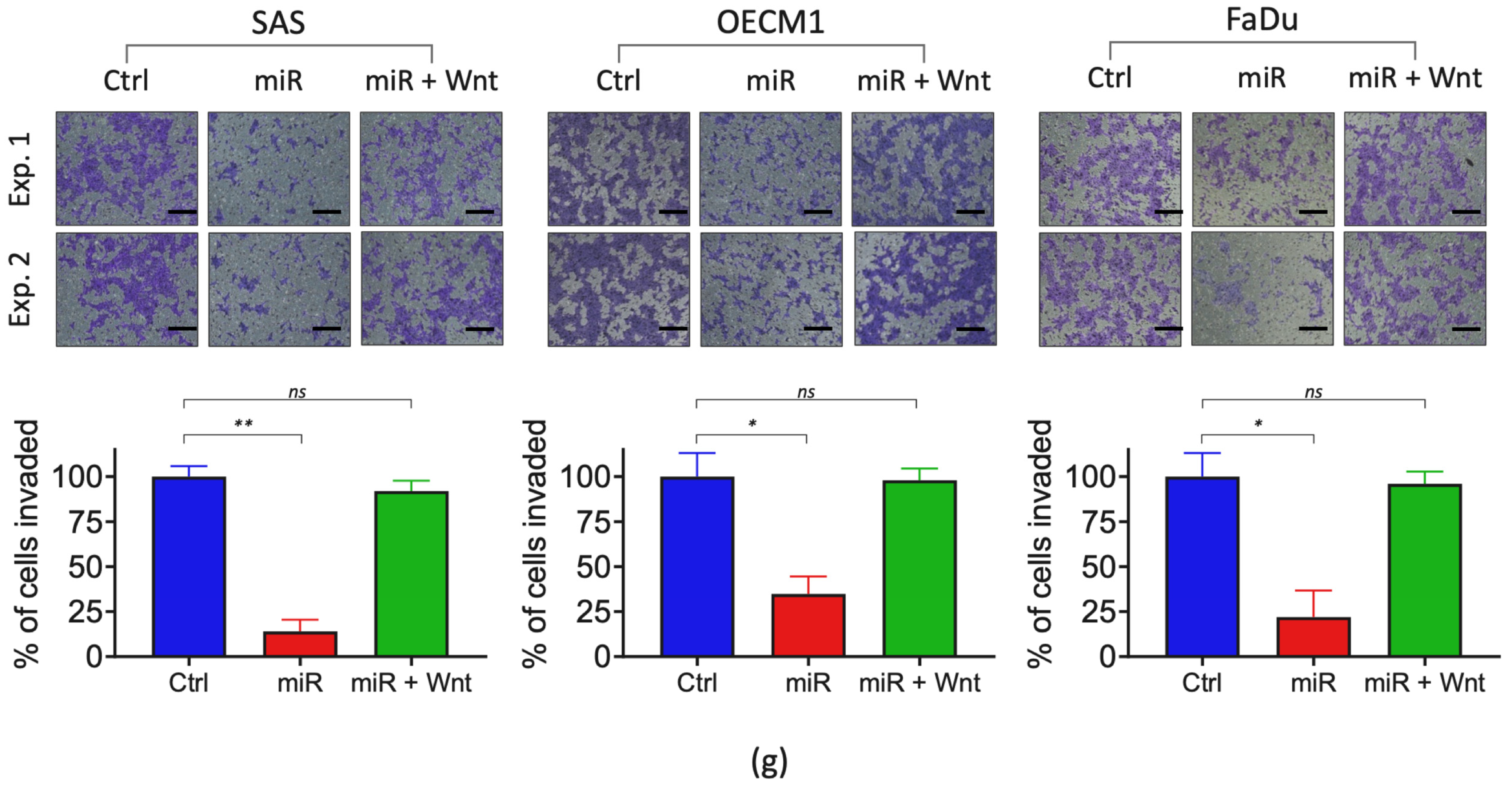
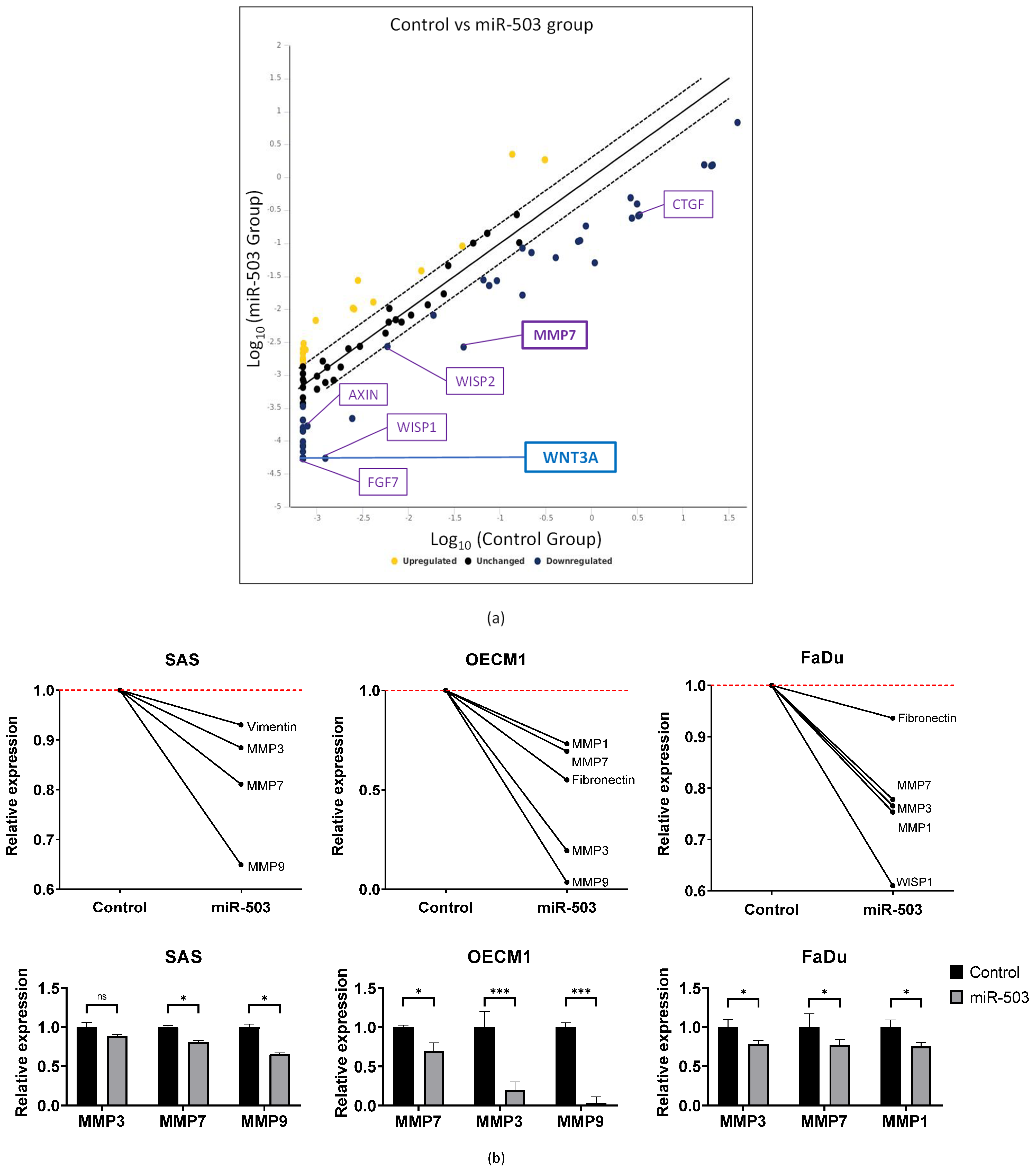
2.4. miRNA-503 Modulates Multiple Invasion-Associated Genes, including MMPs, through the WNT3A Downstream Signaling Pathway
3. Discussion
4. Materials and Methods
4.1. Cells and Cell Cultures
4.2. miRNA-503 Mimic Construction and Transfection
4.3. RT-qPCR Analysis for miRNA-503 and Target Gene Expression
4.4. Cell Proliferation Assay for Cell Growth Analysis
4.5. Colony Formation Assay for Cell Growth Analysis
4.6. Wound-Healing Migration Assay
4.7. Matrigel Transwell Invasion Assay
4.8. Western Blot Analysis of WNT3A Protein Expression
4.9. Bioinformatic Algorithm for the Target Gene Prediction of miRNA-503
4.10. Dual-Luciferase Reporter Assay to Confirm WNT3A Binding with miRNA-503
4.11. KEGG Enrichment Analysis to Examine the miRNA-503 Regulation Pathways
4.12. PCR Array Profiling of the WNT Signaling Target Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, B.; Zhang, S.; Yue, K.; Wang, X.D. The recurrence and survival of oral squamous cell carcinoma: A report of 275 cases. Chin. J. Cancer 2013, 32, 614–618. [Google Scholar] [CrossRef] [PubMed]
- DiMeo, T.A.; Anderson, K.; Phadke, P.; Feng, C.; Perou, C.M.; Naber, S.; Kuperwasser, C. A novel lung metastasis signature links wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. Cancer Res. 2009, 69, 5364–5373. [Google Scholar] [CrossRef]
- Heroiu Cataloiu, A.D.; Danciu, C.E.; Popescu, C.R. Multiple cancers of the head and neck. Maedica 2013, 8, 80–85. [Google Scholar] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Argiris, A.; Karamouzis, M.V.; Raben, D.; Ferris, R.L. Head and neck cancer. Lancet 2008, 371, 1695–1709. [Google Scholar] [CrossRef]
- Howlader, N.; Noone, A.M.; Krapcho, M.; Miller, D.; Bishop, K.; Altekruse, S.F.; Kosary, C.L.; Yu, M.; Ruhl, J.; Tatalovich, Z.; et al. SEER Cancer Statistics Review, 1975–2013. National Cancer Institute: Bethesda, MD, USA, 2016. Available online: https://seer.cancer.gov/archive/csr/1975_2013/ (accessed on 24 April 2018).
- Mayeux, R. Biomarkers: Potential uses and limitations. NeuroRx 2004, 1, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, K.; Dhankhar, R. Updated overview of current biomarkers in head and neck carcinoma. World J. Methodol. 2016, 6, 77–86. [Google Scholar] [CrossRef]
- Ueda, M.; Shimada, T.; Goto, Y.; Tei, K.; Nakai, S.; Hisa, Y.; Kannagi, R. Expression of CC-chemokine receptor 7 (CCR7) and CXC-chemokine receptor 4 (CXCR4) in head and neck squamous cell carcinoma. Auris Nasus Larynx 2010, 37, 488–495. [Google Scholar] [CrossRef]
- Barak, V.; Goike, H.; Panaretakis, K.W.; Einarsson, R. Clinical utility of cytokeratins as tumor markers. Clin. Biochem. 2004, 37, 529–540. [Google Scholar] [CrossRef]
- Lee, K.D.; Lee, H.S.; Jeon, C.H. Body fluid biomarkers for early detection of head and neck squamous cell carcinomas. Anticancer Res. 2011, 31, 1161–1167. [Google Scholar]
- Yen, C.Y.; Chen, C.H.; Chang, C.H.; Tseng, H.F.; Liu, S.Y.; Chuang, L.Y.; Wen, C.H.; Chang, H.W. Matrix metalloproteinases (MMP) 1 and MMP10 but not MMP12 are potential oral cancer markers. Biomark. Biochem. Indic. Expo. Response Susceptibility Chem. 2009, 14, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids—The mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef]
- Komatsu, S.; Imamura, T.; Kiuchi, J.; Takashima, Y.; Kamiya, H.; Ohashi, T.; Konishi, H.; Shiozaki, A.; Kubota, T.; Okamoto, K.; et al. Depletion of tumor suppressor miRNA-148a in plasma relates to tumor progression and poor outcomes in gastric cancer. Am. J. Cancer Res. 2021, 11, 6133–6146. [Google Scholar]
- Huang, Z.; Huang, D.; Ni, S.; Peng, Z.; Sheng, W.; Du, X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int. J. Cancer 2010, 127, 118–126. [Google Scholar] [CrossRef]
- Lin, P.Y.; Yu, S.L.; Yang, P.C. MicroRNA in lung cancer. Br. J. Cancer 2010, 103, 1144–1148. [Google Scholar] [CrossRef]
- Le Quesne, J.; Caldas, C. Micro-RNAs and breast cancer. Mol. Oncol. 2010, 4, 230–241. [Google Scholar] [CrossRef]
- Catania, A.; Maira, F.; Skarmoutsou, E.; D'Amico, F.; Abounader, R.; Mazzarino, M.C. Insight into the role of microRNAs in brain tumors (review). Int. J. Oncol. 2012, 40, 605–624. [Google Scholar] [CrossRef]
- Kulkarni, V.; Uttamani, J.R.; Naqvi, A.R.; Nares, S. microRNAs: Emerging players in oral cancers and inflammatory disorders. Tumor Biol. 2017, 39, 1010428317698379. [Google Scholar] [CrossRef]
- Baba, O.; Hasegawa, S.; Nagai, H.; Uchida, F.; Yamatoji, M.; Kanno, N.I.; Yamagata, K.; Sakai, S.; Yanagawa, T.; Bukawa, H. MicroRNA-155-5p is associated with oral squamous cell carcinoma metastasis and poor prognosis. J. Oral Pathol. Med. 2016, 45, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.C.; Chang, J.T.; Liao, C.T.; Kang, C.J.; Huang, S.F.; Chen, I.H.; Huang, C.C.; Huang, Y.C.; Chen, W.H.; Tsai, C.Y.; et al. OncomiR-196 promotes an invasive phenotype in oral cancer through the NME4-JNK-TIMP1-MMP signaling pathway. Mol. Cancer 2014, 13, 218. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.F.; Wei, Y.Y.; Yang, C.C.; Liu, C.J.; Yeh, L.Y.; Chou, C.H.; Chang, K.W.; Lin, S.C. miR-125b suppresses oral oncogenicity by targeting the anti-oxidative gene PRXL2A. Redox Biol. 2019, 22, 101140. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, P.L.; Liao, Y.W.; Hsieh, C.W.; Chen, P.N.; Yu, C.C. Soy isoflavone genistein impedes cancer stemness and mesenchymal transition in head and neck cancer through activating miR-34a/RTCB axis. Nutrients 2020, 12, 1924. [Google Scholar] [CrossRef]
- Xu, K.; Lin, J.; Zandi, R.; Roth, J.A.; Ji, L. MicroRNA-mediated target mRNA cleavage and 3′-uridylation in human cells. Sci. Rep. 2016, 6, 30242. [Google Scholar] [CrossRef]
- Valinezhad Orang, A.; Safaralizadeh, R.; Kazemzadeh-Bavili, M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int. J. Genom. 2014, 2014, 970607. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O'Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Lu, Y.C.; Chen, Y.J.; Wang, H.M.; Tsai, C.Y.; Chen, W.H.; Huang, Y.C.; Fan, K.H.; Tsai, C.N.; Huang, S.F.; Kang, C.J.; et al. Oncogenic function and early detection potential of miRNA-10b in oral cancer as identified by microRNA profiling. Cancer Prev. Res. 2012, 5, 665–674. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, W.; Yang, A.G.; Zhang, R. The microRNA-424/503 cluster: A master regulator of tumorigenesis and tumor progression with paradoxical roles in cancer. Cancer Lett. 2020, 494, 58–72. [Google Scholar] [CrossRef]
- Zhou, R.S.; Zhang, E.X.; Sun, Q.F.; Ye, Z.J.; Liu, J.W.; Zhou, D.H.; Tang, Y. Integrated analysis of lncRNA-miRNA-mRNA ceRNA network in squamous cell carcinoma of tongue. BMC Cancer 2019, 19, 779. [Google Scholar] [CrossRef] [PubMed]
- Lv, T.; Liu, Y.; Li, Z.; Huang, R.; Zhang, Z.; Li, J. miR-503 is down-regulated in osteosarcoma and suppressed MG63 proliferation and invasion by targeting VEGFA/Rictor. Cancer Biomark. 2018, 23, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Chen, Y.; Du, E.; Yang, K.; Zhang, Z.; Qi, S.; Xu, Y. GATA3-driven expression of miR-503 inhibits prostate cancer progression by repressing ZNF217 expression. Cell. Signal. 2016, 28, 1216–1224. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, L.; Xing, S.; Pan, Y.; Shi, Y.; Zhang, L.; Shen, Q. miR-503-3p induces apoptosis of lung cancer cells by regulating p21 and CDK4 expression. Cancer Biomark. 2017, 20, 597–608. [Google Scholar] [CrossRef]
- Xiao, F.; Zhang, W.; Chen, L.; Chen, F.; Xie, H.; Xing, C.; Yu, X.; Ding, S.; Chen, K.; Guo, H.; et al. MicroRNA-503 inhibits the G1/S transition by downregulating cyclin D3 and E2F3 in hepatocellular carcinoma. J. Transl. Med. 2013, 11, 195. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Liu, Y.-M.; Li, L.-C.; Wang, L.-L.; Wu, X.-L. microRNA-503 inhibits gastric cancer cell growth and epithelial-to-mesenchymal transition. Oncol. Lett. 2014, 7, 1233–1238. [Google Scholar] [CrossRef]
- Li, J.; Jin, C.; Sun, L.; Wang, B.; Hua, P.; Zhang, Y. HDAC2 enhances esophageal squamous cell carcinoma development through down-regulating microRNA-503-5p and promoting CXCL10. Clin. Epigenet. 2021, 13, 96. [Google Scholar] [CrossRef]
- Noguchi, T.; Toiyama, Y.; Kitajima, T.; Imaoka, H.; Hiro, J.; Saigusa, S.; Tanaka, K.; Inoue, Y.; Mohri, Y.; Toden, S.; et al. miRNA-503 promotes tumor progression and is associated with early recurrence and poor prognosis in human colorectal cancer. Oncology 2016, 90, 221–231. [Google Scholar] [CrossRef]
- Ide, S.; Toiyama, Y.; Shimura, T.; Kawamura, M.; Yasuda, H.; Saigusa, S.; Ohi, M.; Tanaka, K.; Mohri, Y.; Kusunoki, M. MicroRNA-503 promotes tumor progression and acts as a novel biomarker for prognosis in oesophageal cancer. Anticancer Res. 2015, 35, 1447–1451. [Google Scholar]
- Kolokythas, A.; Zhou, Y.; Schwartz, J.L.; Adami, G.R. Similar squamous cell carcinoma epithelium microrna expression in never smokers and ever smokers. PLoS ONE 2015, 10, e0141695. [Google Scholar] [CrossRef]
- Ma, R.; Jiang, T.; Kang, X. Circulating microRNAs in cancer: Origin, function and application. J. Exp. Clin. Cancer Res. 2012, 31, 38. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Qu, W.; Zhong, Z. Down-regulation of miR-503 expression predicate advanced mythological features and poor prognosis in patients with NSCLC. Int. J. Clin. Exp. Pathol. 2015, 8, 5609–5613. [Google Scholar] [PubMed]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Xiao, Y.; Tian, Q.; He, J.; Huang, M.; Yang, C.; Gong, L. MiR-503 inhibits hepatocellular carcinoma cell growth via inhibition of insulin-like growth factor 1 receptor. OncoTargets Ther. 2016, 9, 3535–3544. [Google Scholar] [CrossRef]
- Gupta, G.; Chellappan, D.K.; de Jesus Andreoli Pinto, T.; Hansbro, P.M.; Bebawy, M.; Dua, K. Tumor suppressor role of miR-503. Panminerva Med. 2018, 60, 17–24. [Google Scholar] [CrossRef]
- Zhao, Z.; Fan, X.; Jiang, L.; Xu, Z.; Xue, L.; Zhan, Q.; Song, Y. miR-503-3p promotes epithelial-mesenchymal transition in breast cancer by directly targeting SMAD2 and E-cadherin. J. Genet. Genom. 2017, 44, 75–84. [Google Scholar] [CrossRef]
- Cheng, Y.; Liu, W. MicroRNA-503 serves an oncogenic role in retinoblastoma progression by directly targeting PTPN12. Exp. Ther. Med. 2019, 18, 2285–2292. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.; Rheinheimer, S.; Meder, B.; Stähler, C.; Meese, E.; et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016, 44, 3865–3877. [Google Scholar] [CrossRef]
- Fu, Y.; Meng, Y.; Gu, X.; Tian, S.; Hou, X.; Ji, M. miR-503 expression is downregulated in cervical cancer and suppresses tumor growth by targeting AKT2. J. Cell. Biochem. 2019, 120, 8177–8184. [Google Scholar] [CrossRef]
- Li, W.; Li, J.; Mu, H.; Guo, M.; Deng, H. MiR-503 suppresses cell proliferation and invasion of gastric cancer by targeting HMGA2 and inactivating WNT signaling pathway. Cancer Cell Int. 2019, 19, 164. [Google Scholar] [CrossRef]
- Sun, S.L.; Shu, Y.G.; Tao, M.Y. miR-503 inhibits proliferation, migration, and angiogenesis of glioma by acting on VEGFA through targeting lRIG2. Cancer Manag. Res. 2019, 11, 10599–10608. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.P.; Li, Z.R. MiR-503-5p regulates cell epithelial-to-mesenchymal transition, metastasis and prognosis of hepatocellular carcinoma through inhibiting WEE1. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 2028–2037. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Sun, C.; Zhang, Y.; Han, N.; Sun, S. miR-503-5p inhibits colon cancer tumorigenesis, angiogenesis, and lymphangiogenesis by directly downregulating VEGF-A. Gene Ther. 2022, 29, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Ribatti, D.; Tamma, R.; Annese, T. Epithelial-Mesenchymal Transition in Cancer: A Historical Overview. Transl. Oncol. 2020, 13, 100773. [Google Scholar] [CrossRef] [PubMed]
- Popova, N.V.; Jücker, M. The functional role of extracellular matrix proteins in cancer. Cancers 2022, 14, 238. [Google Scholar] [CrossRef]
- Crowe, D.L.; Shuler, C.F. Regulation of tumor cell invasion by extracellular matrix. Histol. Histopathol. 1999, 14, 665–671. [Google Scholar] [CrossRef]
- Gobin, E.; Bagwell, K.; Wagner, J.; Mysona, D.; Sandirasegarane, S.; Smith, N.; Bai, S.; Sharma, A.; Schleifer, R.; She, J.X. A pan-cancer perspective of matrix metalloproteases (MMP) gene expression profile and their diagnostic/prognostic potential. BMC Cancer 2019, 19, 581. [Google Scholar] [CrossRef]
- Jia, S.; Zhai, H.; Zhao, M. MicroRNAs regulate immune system via multiple targets. Discov. Med. 2014, 18, 237–247. [Google Scholar]
- Ali Syeda, Z.; Langden, S.S.S.; Munkhzul, C.; Lee, M.; Song, S.J. Regulatory Mechanism of MicroRNA Expression in Cancer. Int. J. Mol. Sci. 2020, 21, 1723. [Google Scholar] [CrossRef]
- Kabekkodu, S.P.; Shukla, V.; Varghese, V.K.; D'Souza, J.; Chakrabarty, S.; Satyamoorthy, K. Clustered miRNAs and their role in biological functions and diseases. Biol. Rev. Camb. Philos. Soc. 2018, 93, 1955–1986. [Google Scholar] [CrossRef]
- Riffo-Campos, Á.L.; Riquelme, I.; Brebi-Mieville, P. Tools for Sequence-Based miRNA Target Prediction: What to Choose? Int. J. Mol. Sci. 2016, 17, 1987. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.-P.; Lee, C.-Y.; Tsai, M.-H.; Chiu, Y.-C.; Hsiao, C.K.; Lai, L.-C.; Chuang, E.Y. miRSystem: An integrated system for characterizing enriched functions and pathways of microRNA targets. PLoS ONE 2012, 7, e42390. [Google Scholar] [CrossRef] [PubMed]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-H.; Liu, S.; Zhou, H.; Qu, L.-H.; Yang, J.-H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2013, 42, D92–D97. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.H.; Wang, S.; Zhang, Y.; Huang, T.; Cai, Y.D. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS ONE 2017, 12, e0184129. [Google Scholar] [CrossRef]
- Li, P.; Burke, S.; Wang, J.; Chen, X.; Ortiz, M.; Lee, S.C.; Lu, D.; Campos, L.; Goulding, D.; Ng, B.L.; et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science 2010, 329, 85–89. [Google Scholar] [CrossRef]
- Helsten, T.; Kato, S.; Schwaederle, M.; Tomson, B.N.; Buys, T.P.; Elkin, S.K.; Carter, J.L.; Kurzrock, R. Cell-Cycle Gene Alterations in 4,864 Tumors Analyzed by Next-Generation Sequencing: Implications for Targeted Therapeutics. Mol. Cancer Ther. 2016, 15, 1682–1690. [Google Scholar] [CrossRef]
- Nusse, R.; Clevers, H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Liu, L.; Zheng, M.; Ye, Z.; Chen, R.; Lan, X. MiR-503 Contributes to Glucocorticoid Sensitivity in Acute Lymphoblastic Leukaemia via Targeting WNT3A. Folia Biol. 2021, 67, 199–207. [Google Scholar]
- Nusse, R. Wnt signaling in disease and in development. Cell Res. 2005, 15, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Logan, C.Y.; Nusse, R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004, 20, 781–810. [Google Scholar] [CrossRef]
- Wu, B.; Crampton, S.P.; Hughes, C.C.W. Wnt signaling induces MMP expression and regulates T cell transmigration. Immunity 2007, 26, 227–239. [Google Scholar] [CrossRef]
- Sedgwick, A.E.; D’Souza-Schorey, C. Wnt Signaling in Cell Motility and Invasion: Drawing Parallels between Development and Cancer. Cancers 2016, 8, 80. [Google Scholar] [CrossRef]
- You, Y.; Que, K.; Zhou, Y.; Zhang, Z.; Zhao, X.; Gong, J.; Liu, Z. MicroRNA-766-3p Inhibits Tumour Progression by Targeting Wnt3a in Hepatocellular Carcinoma. Mol. Cells 2018, 41, 830–841. [Google Scholar] [CrossRef]
- Lin, Z.; Liu, J. lncRNA DQ786243 promotes hepatocellular carcinoma cell invasion and proliferation by regulating the miR-15b-5p/Wnt3A axis. Mol. Med. Rep. 2021, 23, 318. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, Y.; Han, M.; Lu, H.; Chen, X.; Liu, S.; Yuan, X.; Han, K.; Liang, P.; Cheng, J. P7TP3 inhibits tumor development, migration, invasion and adhesion of liver cancer through the Wnt/β-catenin signaling pathway. Cancer Sci. 2020, 111, 994–1007. [Google Scholar] [CrossRef]
- Lu, F.; Ye, Y.; Zhang, H.; He, X.; Sun, X.; Yao, C.; Mao, H.; He, X.; Qian, C.; Wang, B.; et al. miR-497/Wnt3a/c-jun feedback loop regulates growth and epithelial-to-mesenchymal transition phenotype in glioma cells. Int. J. Biol. Macromol. 2018, 120, 985–991. [Google Scholar] [CrossRef]
- Liu, G.; Wang, P.; Zhang, H. MiR-6838-5p suppresses cell metastasis and the EMT process in triple-negative breast cancer by targeting WNT3A to inhibit the Wnt pathway. J. Gene Med. 2019, 21, e3129. [Google Scholar] [CrossRef] [PubMed]
- Oguma, J.; Ozawa, S.; Kazuno, A.; Nitta, M.; Ninomiya, Y.; Kajiwara, H. Wnt3a expression is associated with poor prognosis of esophageal squamous cell carcinoma. Oncol. Lett. 2018, 15, 3100–3108. [Google Scholar] [CrossRef]
- Li, B.; Wang, S.; Wang, S. MiR-195 suppresses colon cancer proliferation and metastasis by targeting WNT3A. Mol. Genet. Genom. 2018, 293, 1245–1253. [Google Scholar] [CrossRef] [PubMed]
- Koni, M.; Pinnarò, V.; Brizzi, M.F. The Wnt signalling pathway: A tailored target in cancer. Int. J. Mol. Sci. 2020, 21, 7697. [Google Scholar] [CrossRef]
- Alamoud, K.A.; Kukuruzinska, M.A. Emerging insights into Wnt/β-catenin signaling in head and neck cancer. J. Dent. Res. 2018, 97, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Huang, L.; Lu, Y.G.; Zheng, D.L. Roles of the Wnt signaling pathway in head and neck squamous cell carcinoma. Front. Mol. Biosci. 2020, 7, 590912. [Google Scholar] [CrossRef] [PubMed]
- Javed, Z.; Muhammad Farooq, H.; Ullah, M.; Zaheer Iqbal, M.; Raza, Q.; Sadia, H.; Pezzani, R.; Salehi, B.; Sharifi-Rad, J.; Cho, W.C. Wnt signaling: A potential therapeutic target in head and neck squamous cell carcinoma. Asian Pac. J. Cancer Prev. 2019, 20, 995–1003. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zhang, H.W.; Cao, W.H.; Mao, Y.; Cheng, R.C. Exploration of the Potential Biomarkers of Papillary Thyroid Cancer (PTC) Based on RT(2) Profiler PCR Arrays and Bioinformatics Analysis. Cancer Manag. Res. 2020, 12, 9235–9246. [Google Scholar] [CrossRef]
- Shen, Y.W.; Zhou, Y.D.; Chen, H.Z.; Luan, X.; Zhang, W.D. Targeting CTGF in cancer: An emerging therapeutic opportunity. Trends Cancer 2021, 7, 511–524. [Google Scholar] [CrossRef] [PubMed]
- Emon, B.; Bauer, J.; Jain, Y.; Jung, B.; Saif, T. Biophysics of tumor microenvironment and cancer metastasis—A mini review. Comput. Struct. Biotechnol. J. 2018, 16, 279–287. [Google Scholar] [CrossRef]
- Pukrop, T.; Klemm, F.; Hagemann, T.; Gradl, D.; Schulz, M.; Siemes, S.; Trümper, L.; Binder, C. Wnt 5a signaling is critical for macrophage-induced invasion of breast cancer cell lines. Proc. Natl. Acad. Sci. USA 2006, 103, 5454–5459. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Lee, B.; Jiang, Y. Cell-ECM interactions in tumor invasion. Adv. Exp. Med. Biol. 2016, 936, 73–91. [Google Scholar] [CrossRef]
- Lu, P.; Takai, K.; Weaver, V.M.; Werb, Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb. Perspect. Biol. 2011, 3, a005058. [Google Scholar] [CrossRef] [PubMed]
- Bassiouni, W.; Ali, M.A.M.; Schulz, R. Multifunctional intracellular matrix metalloproteinases: Implications in disease. FEBS J. 2021, 288, 7162–7182. [Google Scholar] [CrossRef]
- Pittayapruek, P.; Meephansan, J.; Prapapan, O.; Komine, M.; Ohtsuki, M. Role of matrix metalloproteinases in photoaging and photocarcinogenesis. Int. J. Mol. Sci. 2016, 17, 868. [Google Scholar] [CrossRef]
- Jabłońska-Trypuć, A.; Matejczyk, M.; Rosochacki, S. Matrix metalloproteinases (MMPs), the main extracellular matrix (ECM) enzymes in collagen degradation, as a target for anticancer drugs. J. Enzyme Inhib. Med. Chem. 2016, 31, 177–183. [Google Scholar] [CrossRef]
- Tang, S.-J.; You, G.-R.; Chang, J.T.; Cheng, A.-J. Systematic analysis and identification of dysregulated panel lncrnas contributing to poor prognosis in head-neck cancer. Front. Oncol. 2021, 11, 731752. [Google Scholar] [CrossRef] [PubMed]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. Jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, S.-J.; Fan, K.-H.; You, G.-R.; Huang, S.-F.; Kang, C.-J.; Huang, Y.-F.; Huang, Y.-C.; Chang, J.T.-C.; Cheng, A.-J. Tumor Suppressor miRNA-503 Inhibits Cell Invasion in Head and Neck Cancer through the Wnt Signaling Pathway via the WNT3A/MMP Molecular Axis. Int. J. Mol. Sci. 2022, 23, 15900. https://doi.org/10.3390/ijms232415900
Tang S-J, Fan K-H, You G-R, Huang S-F, Kang C-J, Huang Y-F, Huang Y-C, Chang JT-C, Cheng A-J. Tumor Suppressor miRNA-503 Inhibits Cell Invasion in Head and Neck Cancer through the Wnt Signaling Pathway via the WNT3A/MMP Molecular Axis. International Journal of Molecular Sciences. 2022; 23(24):15900. https://doi.org/10.3390/ijms232415900
Chicago/Turabian StyleTang, Shang-Ju, Kang-Hsing Fan, Guo-Rung You, Shiang-Fu Huang, Chung-Jan Kang, Yi-Fang Huang, Yu-Chen Huang, Joseph Tung-Chieh Chang, and Ann-Joy Cheng. 2022. "Tumor Suppressor miRNA-503 Inhibits Cell Invasion in Head and Neck Cancer through the Wnt Signaling Pathway via the WNT3A/MMP Molecular Axis" International Journal of Molecular Sciences 23, no. 24: 15900. https://doi.org/10.3390/ijms232415900
APA StyleTang, S.-J., Fan, K.-H., You, G.-R., Huang, S.-F., Kang, C.-J., Huang, Y.-F., Huang, Y.-C., Chang, J. T.-C., & Cheng, A.-J. (2022). Tumor Suppressor miRNA-503 Inhibits Cell Invasion in Head and Neck Cancer through the Wnt Signaling Pathway via the WNT3A/MMP Molecular Axis. International Journal of Molecular Sciences, 23(24), 15900. https://doi.org/10.3390/ijms232415900





