MiR-223 Exclusively Impairs In Vitro Tumor Growth through IGF1R Modulation in Rhabdomyosarcoma of Adolescents and Young Adults
Abstract
1. Introduction
2. Results
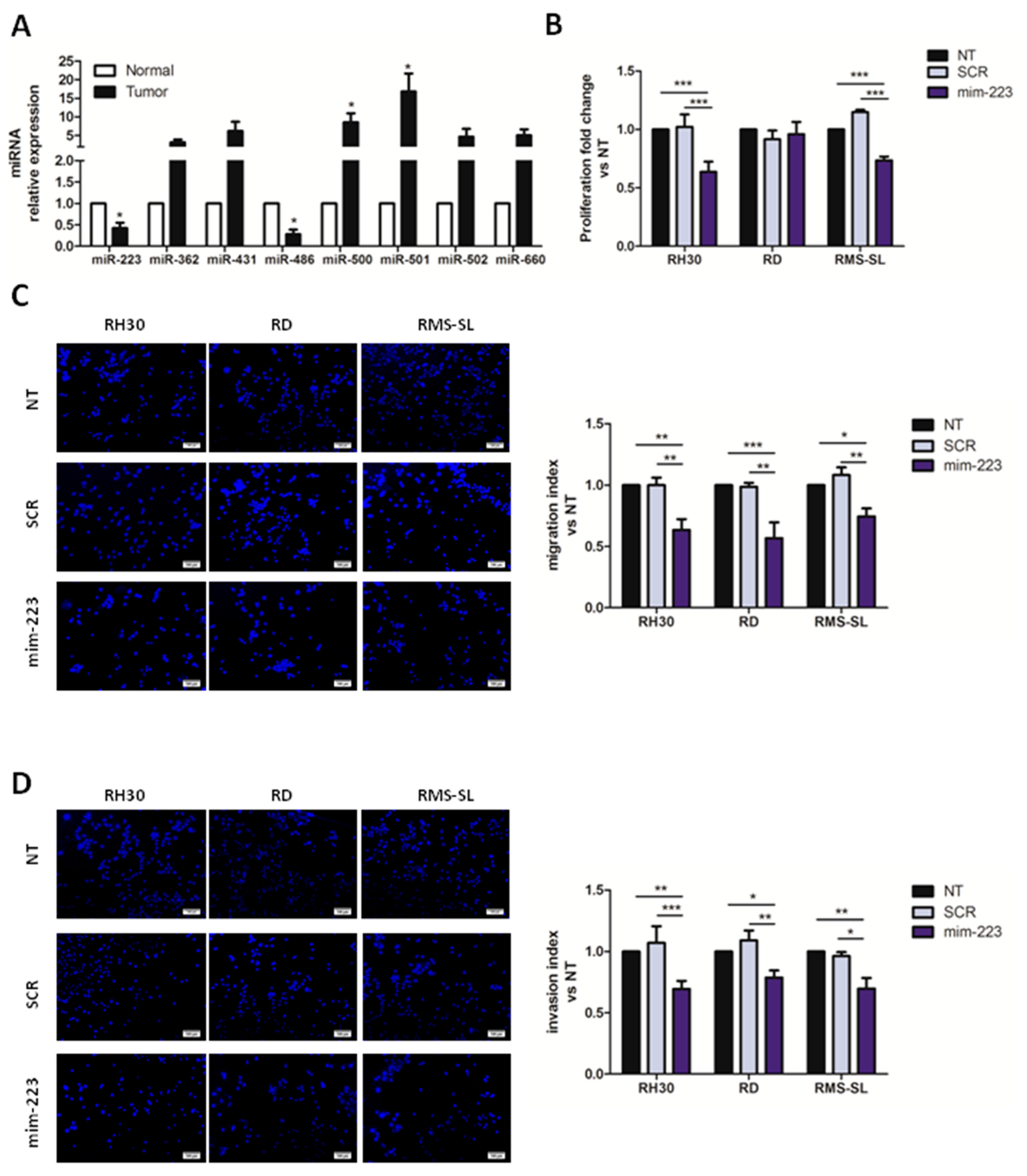
2.1. MiR-223 Affects Proliferation and Aggressiveness of AYA-RMS Cell Lines
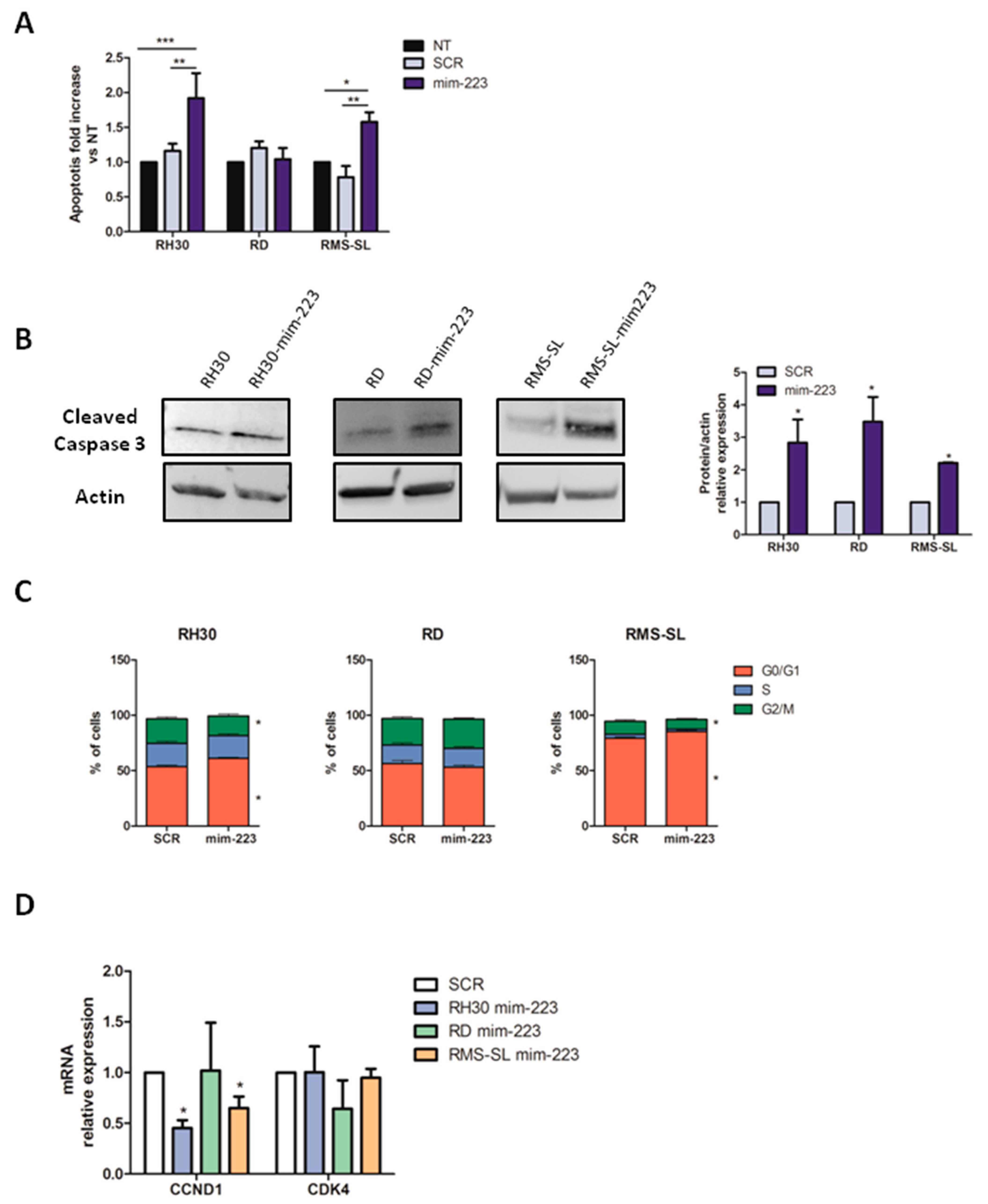
2.2. MiR-223 Induces Apoptosis and Determines Cell Cycle Arrest
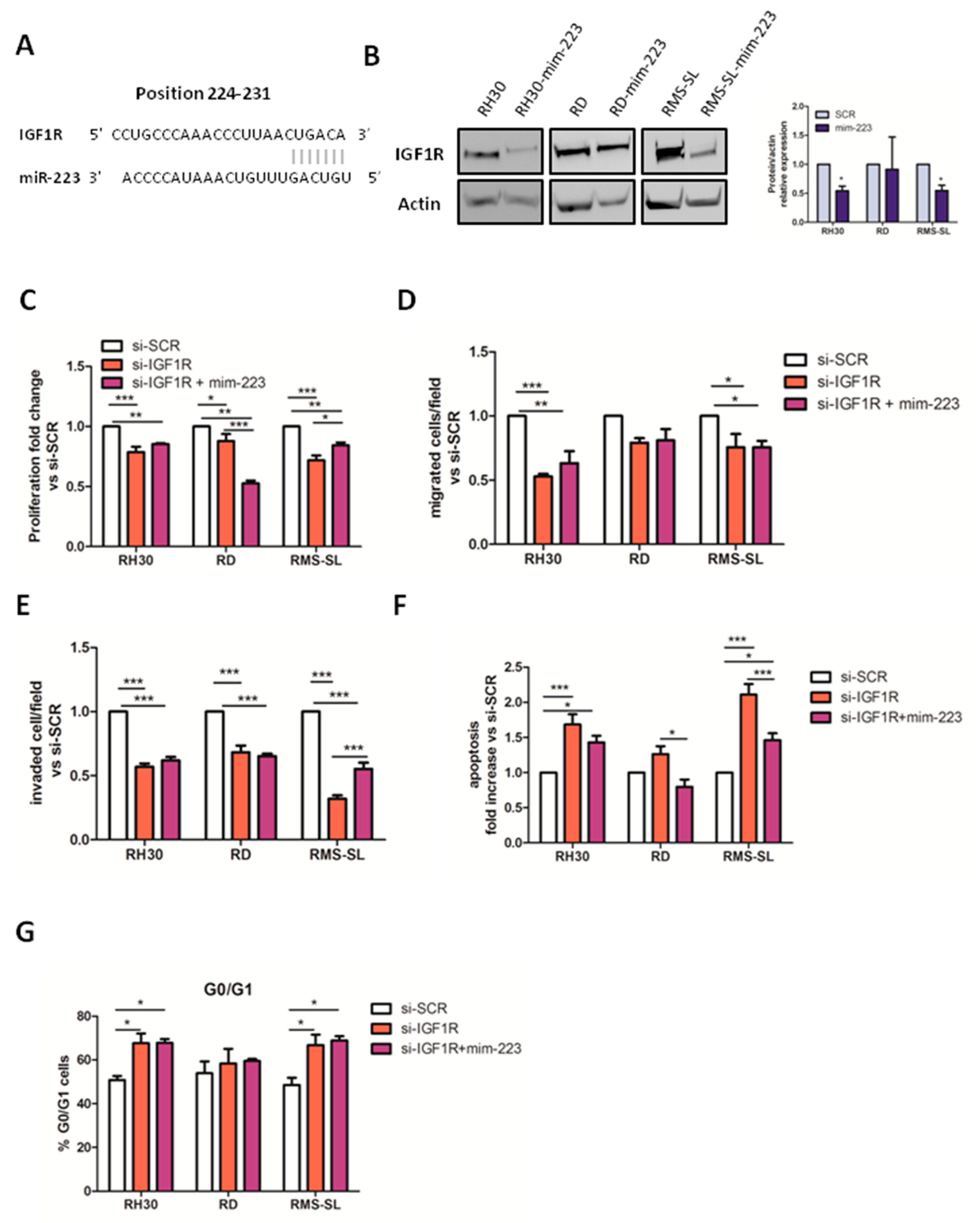
2.3. IGF1R Is a Direct Target of miR-223 in RMS Cells
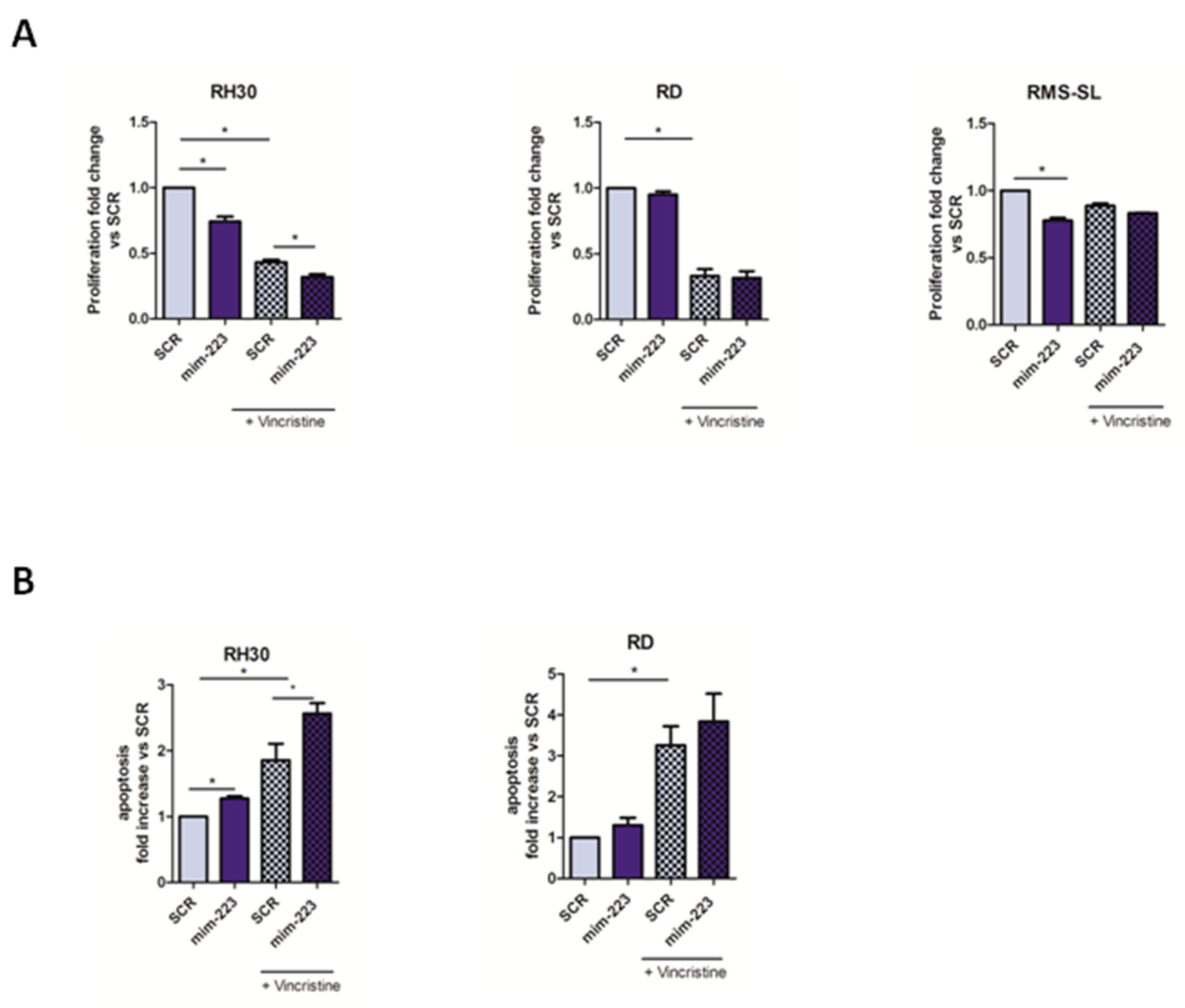
2.4. MiR-223 Improved Chemotherapy Efficacy in RMS Cells
3. Discussion
4. Materials and Methods
4.1. RMS Clinical Specimen and Cell Lines
4.2. RNA Extraction from FFPE and miRNAs Analysis
4.3. MiRNA and siRNA Transfection
4.4. Luciferase-Based Proliferation Assays
4.5. Migration and Invasion Assays
4.6. Early and Late Apoptosis Assessment
4.7. Cell Cycle Analysis
4.8. Western Blot Analysis
4.9. Quantitative Real Time PCR
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferrari, A.; Sultan, I.; Huang, T.T.; Rodriguez-Galindo, C.; Shehadeh, A.; Meazza, C.; Ness, K.K.; Casanova, M.; Spunt, S.L. Soft tissue sarcoma across the age spectrum: A population-based study from the Surveillance Epidemiology and End Results database. Pediatr. Blood Cancer 2011, 57, 943–949. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, A.; Gasparini, P.; Gill, J.; Gorlick, R. Challenges of Clinical Management of Adolescent and Young Adults with Bone and Soft Tissue Sarcoma. Cancer J. 2018, 24, 301–306. [Google Scholar] [CrossRef]
- Ferrari, A.; Bleyer, A.; Patel, S.; Chiaravalli, S.; Gasparini, P.; Casanova, M. The challenge of the management of adolescents and young adults with soft tissue sarcomas. Pediatr. Blood Cancer 2018, 65, e27013. [Google Scholar] [CrossRef] [PubMed]
- Trama, A.; Botta, L.; Foschi, R.; Ferrari, A.; Stiller, C.; Desandes, E.; Maule, M.M.; Merletti, F.; Gatta, G.; EUROCARE-5 Working Group. Survival of European adolescents and young adults diagnosed with cancer in 2000-07: Population-based data from EUROCARE-5. Lancet Oncol. 2016, 17, 896–906. [Google Scholar] [CrossRef]
- Ferrari, A.; Bernasconi, A.; Bergamaschi, L.; Botta, L.; Andreano, A.; Castaing, M.; Rugge, M.; Bisogno, G.; Falcini, F.; Sacerdote, C.; et al. Impact of Rhabdomyosarcoma Treatment Modalities by Age in a Population-Based Setting. J. Adolesc. Young Adult Oncol. 2021, 10, 309–315. [Google Scholar] [CrossRef]
- Sultan, I.; Qaddoumi, I.; Yaser, S.; Rodriguez-Galindo, C.; Ferrari, A. A Comparing adult and pediatric rhabdomyosarcoma in the surveillance, epidemiology and end results program, 1973 to 2005: An analysis of 2600 patients. J. Clin. Oncol. 2009, 27, 3391–3397. [Google Scholar] [CrossRef]
- Gasparini, P.; Fortunato, O.; De, C.L.; Casanova, M.; Ianno, M.F.; Carenzo, A.; Centonze, G.; Milione, M.; Collini, P.; Boeri, M.; et al. Age-Related Alterations in Immune Contexture Are Associated with Aggressiveness in Rhabdomyosarcoma. Cancers 2019, 11, 1380. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Siomi, H.; Siomi, M.C. Posttranscriptional regulation of microRNA biogenesis in animals. Mol. Cell 2010, 38, 323–332. [Google Scholar] [CrossRef]
- Liu, X.; Fortin, K.; Mourelatos, Z. MicroRNAs: Biogenesis and molecular functions. Brain Pathol. 2008, 18, 113–121. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E.; Liu, N.; Olson, E.N. MicroRNAs flex their muscles. Trends Genet 2008, 24, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Cardinali, B.; Castellani, L.; Fasanaro, P.; Basso, A.; Alema, S.; Martelli, F.; Falcone, G. Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PLoS ONE 2009, 4, e7607. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, F.; Fahs, A.; Ghayad, S.E.; Saab, R. Signaling pathways in Rhabdomyosarcoma invasion and metastasis. Cancer Metastasis Rev. 2020, 39, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Gasparini, P.; Ferrari, A.; Casanova, M.; Limido, F.; Massimino, M.; Sozzi, G.; Fortunato, O. MiRNAs as Players in Rhabdomyosarcoma Development. Int. J. Mol. Sci. 2019, 20, 5818. [Google Scholar] [CrossRef]
- Wu, L.; Li, H.; Jia, C.Y.; Cheng, W.; Yu, M.; Peng, M.; Zhu, Y.; Zhao, Q.; Dong, Y.W.; Shao, K.; et al. MicroRNA-223 regulates FOXO1 expression and cell proliferation. FEBS Lett. 2012, 586, 1038–1043. [Google Scholar] [CrossRef]
- Aziz, F.; Chakraborty, A.; Khan, I.; Monts, J. Relevance of miR-223 as Potential Diagnostic and Prognostic Markers in Cancer. Biology 2022, 11, 249. [Google Scholar] [CrossRef]
- Rota, R.; Ciarapica, R.; Giordano, A.; Miele, L.; Locatelli, F. MicroRNAs in rhabdomyosarcoma: Pathogenetic implications and translational potentiality. Mol. Cancer 2011, 10, 120. [Google Scholar] [CrossRef]
- Megiorni, F.; Cialfi, S.; McDowell, H.P.; Felsani, A.; Camero, S.; Guffanti, A.; Pizer, B.; Clerico, A.; De Grazia, A.; Pizzuti, A. Deep Sequencing the microRNA profile in rhabdomyosarcoma reveals down-regulation of miR-378 family members. BMC Cancer 2014, 14, 880. [Google Scholar] [CrossRef]
- Taulli, R.; Bersani, F.; Foglizzo, V.; Linari, A.; Vigna, E.; Ladanyi, M.; Tuschl, T.; Ponzetto, C. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J. Clin. Investig 2009, 119, 2366–2378. [Google Scholar] [CrossRef]
- Luo, P.; Wang, Q.; Ye, Y.; Zhang, J.; Lu, D.; Cheng, L.; Zhou, H.; Xie, M.; Wang, B. MiR-223-3p functions as a tumor suppressor in lung squamous cell carcinoma by miR-223-3p-mutant p53 regulatory feedback loop. J. Exp. Clin. Cancer Res. 2019, 38, 74. [Google Scholar] [CrossRef] [PubMed]
- Du, T.; Wang, D.; Wan, X.; Xu, J.; Xiao, Q.; Liu, B. Regulatory effect of microRNA-223-3p on breast cancer cell processes via the Hippo/Yap signaling pathway. Oncol. Lett. 2021, 22, 516. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yao, Q.; Hou, Y.; Xu, M.; Liu, S.; Yang, L.; Zhang, L.; Xu, H. MiR-223/Ect2/p21 signaling regulates osteosarcoma cell cycle progression and proliferation. Biomed. Pharmacother. 2013, 67, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Ji, Q.; Xu, X.; Song, Q.; Xu, Y.; Tai, Y.; Goodman, S.B.; Bi, W.; Xu, M.; Jiao, S.; Maloney, W.J.; et al. miR-223-3p Inhibits Human Osteosarcoma Metastasis and Progression by Directly Targeting CDH6. Mol. Ther. 2018, 26, 1299–1312. [Google Scholar] [CrossRef] [PubMed]
- Pappo, A.S.; Vassal, G.; Crowley, J.J.; Bolejack, V.; Hogendoorn, P.C.; Chugh, R.; Ladanyi, M.; Grippo, J.F.; Dall, G.; Staddone, A.P.; et al. A phase 2 trial of R1507, a monoclonal antibody to the insulin-like growth factor-1 receptor (IGF-1R), in patients with recurrent or refractory rhabdomyosarcoma, osteosarcoma, synovial sarcoma, and other soft tissue sarcomas: Results of a Sarcoma Alliance for Research through Collaboration study. Cancer 2014, 120, 2448–2456. [Google Scholar] [PubMed]
- Zhu, B.; Davie, J.K. New insights into signalling-pathway alterations in rhabdomyosarcoma. Br. J. Cancer 2015, 112, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Haneklaus, M.; Gerlic, M.; O’Neill, L.A.; Masters, S.L. miR-223: Infection, inflammation and cancer. J. Intern. Med. 2013, 274, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Aziz, F. The emerging role of miR-223 as novel potential diagnostic and therapeutic target for inflammatory disorders. Cell. Immunol. 2016, 303, 1–6. [Google Scholar] [CrossRef]
- Fortunato, O.; Boeri, M.; Verri, C.; Conte, D.; Mensah, M.; Suatoni, P.; Pastorino, U.; Sozzi, G. Assessment of circulating microRNAs in plasma of lung cancer patients. Molecules 2014, 19, 3038–3054. [Google Scholar] [CrossRef]
| RMS (n = 9) | |
|---|---|
| Gender | |
| Male | 6 (66.7%) |
| Female | 3 (33.3%) |
| Age (years) | 19.3 + 10.3 |
| PEDS (0–14) | 2 |
| AYA (>15) | 7 |
| Hystology | |
| ARMS | 3 (33.3%) |
| FOXO1/PAX3 | 1 |
| Fusion negative | 2 |
| ERMS | 6 (55.6%) |
| Fusion negative | 6 |
| IRS grade | |
| I | 3 (33.3%) |
| III | 6 (66.7%) |
| Site of onset | |
| Testicle | 4 |
| Prostate | 1 |
| Limbs | 3 |
| Trunk | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Casanova, M.; Pontis, F.; Ghidotti, P.; Petraroia, I.; Venturini, L.V.; Bergamaschi, L.; Chiaravalli, S.; De Cecco, L.; Massimino, M.; Sozzi, G.; et al. MiR-223 Exclusively Impairs In Vitro Tumor Growth through IGF1R Modulation in Rhabdomyosarcoma of Adolescents and Young Adults. Int. J. Mol. Sci. 2022, 23, 13989. https://doi.org/10.3390/ijms232213989
Casanova M, Pontis F, Ghidotti P, Petraroia I, Venturini LV, Bergamaschi L, Chiaravalli S, De Cecco L, Massimino M, Sozzi G, et al. MiR-223 Exclusively Impairs In Vitro Tumor Growth through IGF1R Modulation in Rhabdomyosarcoma of Adolescents and Young Adults. International Journal of Molecular Sciences. 2022; 23(22):13989. https://doi.org/10.3390/ijms232213989
Chicago/Turabian StyleCasanova, Michela, Francesca Pontis, Patrizia Ghidotti, Ilaria Petraroia, Lara Veronica Venturini, Luca Bergamaschi, Stefano Chiaravalli, Loris De Cecco, Maura Massimino, Gabriella Sozzi, and et al. 2022. "MiR-223 Exclusively Impairs In Vitro Tumor Growth through IGF1R Modulation in Rhabdomyosarcoma of Adolescents and Young Adults" International Journal of Molecular Sciences 23, no. 22: 13989. https://doi.org/10.3390/ijms232213989
APA StyleCasanova, M., Pontis, F., Ghidotti, P., Petraroia, I., Venturini, L. V., Bergamaschi, L., Chiaravalli, S., De Cecco, L., Massimino, M., Sozzi, G., Ferrari, A., Fortunato, O., & Gasparini, P. (2022). MiR-223 Exclusively Impairs In Vitro Tumor Growth through IGF1R Modulation in Rhabdomyosarcoma of Adolescents and Young Adults. International Journal of Molecular Sciences, 23(22), 13989. https://doi.org/10.3390/ijms232213989








