Abstract
Our understanding of the regulatory processes of reepithelialization during wound healing is incomplete. In an attempt to map the genes involved in epidermal regeneration and differentiation, we measured gene expression in formalin-fixed, paraffin-embedded standardized epidermal wounds induced by the suction-blister technique with associated nonwounded skin using NanoString technology. The transcripts of 139 selected genes involved in clotting, immune response to tissue injury, signaling pathways, cell adhesion and proliferation, extracellular matrix remodeling, zinc transport and keratinocyte differentiation were evaluated. We identified 22 upregulated differentially expressed genes (DEGs) in descending order of fold change (MMP1, MMP3, IL6, CXCL8, SERPINE1, IL1B, PTGS2, HBEGF, CXCL5, CXCL2, TIMP1, CYR61, CXCL1, MMP12, MMP9, HGF, CTGF, ITGB3, MT2A, FGF7, COL4A1 and PLAUR). The expression of the most upregulated gene, MMP1, correlated strongly with MMP3 followed by IL6 and IL1B. rhIL-1β, but not rhIL-6, exposure of cultured normal human epidermal keratinocytes and normal human dermal fibroblasts increased both MMP1 mRNA and MMP-1 protein levels, as well as TIMP1 mRNA levels. The increased TIMP1 in wounds was validated by immunohistochemistry. The six downregulated DEGs (COL7A1, MMP28, SLC39A2, FLG1, KRT10 and FLG2) were associated with epidermal maturation. KLK8 showed the strongest correlation with MKI67 mRNA levels and is a potential biomarker for keratinocyte proliferation. The observed gene expression changes correlate well with the current knowledge of physiological reepithelialization. Thus, the gene expression panel described in this paper could be used in patients with impaired healing to identify possible therapeutic targets.
1. Introduction
Reepithelialization during wound healing is crucial for restoring the skin barrier. Our understanding of the regulatory processes is incomplete for this fundamental process. The factors responsible have not been fully delineated in humans [1,2], although there are gene expression data from partial-thickness excisional and burn wounds [3,4] as well as full-thickness skin wounds in patients with basal cell carcinoma [5].
The suction-blister wound healing model is excellent for studies of the reepithelialization process, and this model has been used to evaluate systemic and topical factors and interventions [1,6,7,8,9,10,11,12,13,14,15,16,17]. For example, treatment with a general matrix metalloproteinase (MMP) inhibitor has been shown to delay reepithelialization [11]. Many clinical outcomes have been validated, but more knowledge of the underlying molecular mechanisms is needed.
The nCounter® (NanoString Technologies, Seattle, WA, USA) gene expression assay is based on direct digital detection of mRNA molecules using target-specific, color-coded probe pairs. It does not require the conversion of mRNA into cDNA by reverse transcription or the amplification of the resulting cDNA by PCR, limiting analytical bias. Another important feature is that the technology can be applied directly to RNA extracted from formalin-fixed, paraffin-embedded (FFPE) tissues [18].
The aim of this study was to validate a customized gene expression panel using archival FFPE tissues of epidermal wounds induced by the suction blister technique from a randomized, double-blind controlled trial in healthy volunteers [1,14,15].
2. Results
To screen candidate genes involved in normal epidermal wound healing, we designed a gene expression panel composed of 139 different target genes encoding transcription factors, cytokines/chemokines, growth factors, receptors, extracellular matrix (ECM) molecules, proteinases/antiproteinases, zinc importers/exporters, antibacterial peptides, adhesion molecules and epidermal stratification markers (Appendix A, Table A1).
We analyzed FFPE tissues from 4-day-old epidermal wounds (reepithelialized to 30–40% as determined by histology [14]), including adjacent normal skin from 20 nondiabetic participants (age 19–43 years old, 27.2 ± 6.0 years). The 8 women and 12 men were included in the period from 30 March 2014 to 4 May 2014 [1,14,15]. Six participants had skin type I, three had type II, seven had type III and four had skin type IV [19].
2.1. Determination of mRNAs by NanoString
The raw barcode counts of the immobilized labeled mRNA complexes were obtained by automated scanning using inverted fluorescence microscopy. The data for all samples and genes are shown in Supplementary Table S1. The expression of the genes was then normalized to the spiked-in positive controls and six different housekeeping genes using nSolver software. The housekeeping genes were expressed at similar levels for all samples. The normalized values are shown in Supplementary Table S2 and were used for the subsequent analyses.
Skin samples of three participants (24, 27 and 28) were flagged according to the nSolver default algorithm due to low RNA contents; therefore, paired analyses with the wounds were not possible. The comparisons between wounds and skin, expressed as fold changes (FC) with p and q values, for the remaining 17 participants are shown for the 139 genes in Supplementary Table S3.
2.2. Differentially Expressed Genes (DEGs) in Human Epidermal Wounds
We defined a DEG as a gene with a mean 2-FC and p < 0.05 in the wound group compared to the associated nonwounded skin. When using this criterion, the following 28 DEGs (all p < 0.05, q < 0.05) arranged in descending order of FC were: MMP1, MMP3, IL6, CXCL8, SERPINE1, IL1B, PTGS2, HBEGF, CXCL5, CXCL2, TIMP1, CYR61, CXCL1, MMP12, MMP9, HGF, CTGF, ITGB3, MT2A, FGF7, COL4A1 and PLAUR (22 upregulated DEGs) and COL7A1, MMP28, SLC39A2, FLG1, KRT10 and FLG2 (6 downregulated DEGs), as illustrated in Figure 1.
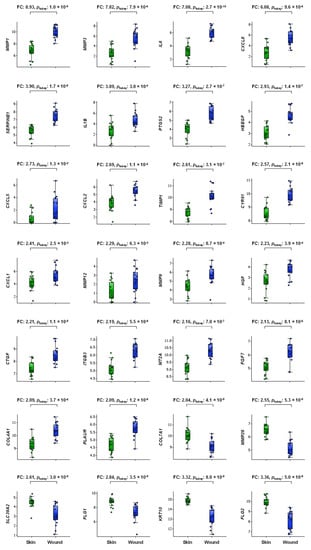
Figure 1.
Box plot of injury-induced DEGs in wound tissue (blue symbols) vs. adjacent normal skin (green symbols) tissue of 4-day-old deroofed suction blisters (n = 17) arranged in descending FC order. Boxes represent the 25th–75th percentiles, whiskers represent the 5th–95th percentiles, and the horizontal dashed lines within the boxes indicate the median values. The y-axis shows log2-transformed expression values. FC, fold change.
2.3. MMP1 mRNA Correlations in Wounds
MMP1 was the most upregulated gene in the wounds compared to the skin. A first step to elucidate the possible regulatory mechanisms of MMP1 was to correlate MMP1 mRNA with the expression of the other upregulated genes. The strongest correlation was found for MMP3 (r = 0.83, p = 4.9 × 10−6) followed by the expression of IL6 (r = 0.70, p < 0.001) and IL1B (r = 0.59, p < 0.01), as shown in Figure 2. IL1B mRNA levels correlated strongly (r = 0.69, p < 0.001) with IL6 mRNA levels in wounds (Supplementary Figure S1).
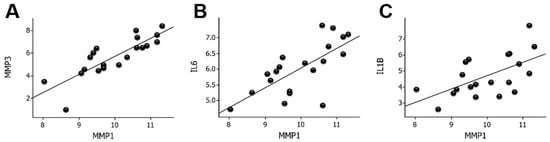
Figure 2.
Correlation between MMP1 mRNA levels and MMP3 (A), IL6 (B), and IL1B (C) mRNA levels in wounds. Log2-transformed expression values are shown on the y- and x-axes. MMP, matrix metalloproteinase; IL, interleukin.
2.4. Effects of Cytokines on MMP1, MMP3 and TIMP1 Gene Expression and Secretion of MMP-1 and TIMP-1 into the Medium of Normal Human Epidermal Keratinocytes (NHEKs) and Normal Human Dermal Fibroblasts (NHDFs)
To study the transcriptional regulation of MMP1, MMP3 and TIMP1 and translation into MMP-1 and TIMP-1, NHEKs and NHDFs were exposed to 30 ng/mL rhIL-6 and 1 ng/mL rhIL-1β separately and together. The IL-6 and IL-1β concentrations were chosen from measurements of 1-day-old suction blister wounds [20,21] and were found to be noncytotoxic, as indicated by the similar LDH activities in the media (Table 1).

Table 1.
LDH activity (mU/mL) in media from treated NHEKs and NHDFs.
Previous studies in NHDFs indicated that IL-6 is anti-proliferative at ≥10 ng/mL while IL-1β (0.1 ng/mL) is proliferative [22]. In our studies of NHDFs using the BrdU incorporation assay [23,24], IL-6 and IL-1β did not significantly influence DNA synthesis at 30 ng/mL and 1 ng/mL, while IL-6 at 100 ng/mL tended to reduce (p = 0.052) BrdU incorporation into NHDFs (Supplementary Figure S2).
2.4.1. MMP1 and TIMP1 mRNA and MMP-1 and TIMP-1 Protein Levels
IL-6 treatment for 24 h increased MMP1 mRNA levels 1.6-fold in NHEKs and 2.5-fold in NHDFs. The corresponding increases for IL-1β were 2.0-fold and 85-fold. Utani et al. [25] reported a 2.5-fold increase in MMP1 mRNA in NHEKs and a 63-fold increase in NHDFs after 8 h of IL-1β (10 ng/mL) treatment. There were no additive effects of combining IL-6 with IL-1β treatment on the MMP1 mRNA levels in NHEKs (p = 0.641) or in NHDFs (p = 0.362). MMP-1 protein levels in control conditioned media of NHEKs were 46.5 ± 6.6 ng/mL and of NHDFs 322 ± 53 ng/mL. IL-6 had no significant effect on MMP-1 protein levels while IL-1β increased MMP-1 protein levels in NHEKs (1.7-fold) and NHDFs (3.3-fold) compared to the controls. No additive effect of IL-6 was observed in either cell type (Figure 3).
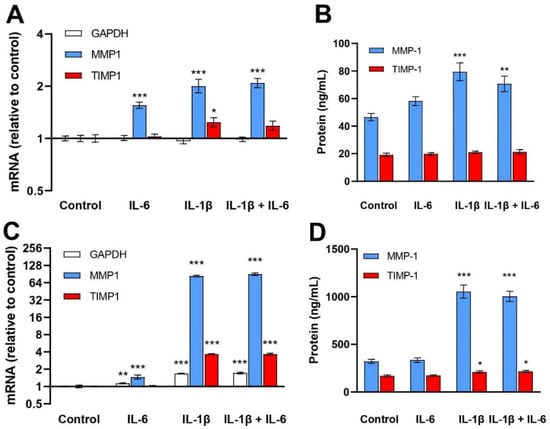
Figure 3.
Effects of IL-6 (30 ng/mL) and IL-1β (1 ng/mL) treatment of NHEKs (A,B) and NHDFs (C,D) on MMP1 and TIMP1 mRNA (A,C) and protein media levels of MMP-1 and TIMP-1 (B,D). mRNA levels were determined by RT–qPCR and normalized to RPLP0, expressed as FC relative to control-treated NHEKs and NHDFs. The geometric mean ± back-transformed SEM of 6 replicates is shown (A,C). MMP-1 and TIMP-1 protein levels were determined by ELISA. The mean ± SEM of 6 replicates is shown (B,D). * p < 0.05, ** p < 0.01, *** p < 0.001 vs. control.
TIMP1 mRNA levels were increased in NHEKs (1.2-fold) and in NHDFs (3.6-fold) with IL-1β, but not with IL-6 exposure. The TIMP-1 protein concentration of the control medium of NHEKs was 19.1 ± 3.1 ng/mL and that of the NHDFs was 170 ± 23 ng/mL. IL-1β increased TIMP-1 protein levels in conditioned media of NHDFs (1.2-fold) but not of NHEKs. IL-6 treatment was ineffective (Figure 3).
2.4.2. MMP3 mRNA Levels
In NHEKs, only the combination of IL-6 and IL-1β upregulated MMP3 mRNA levels. IL-6 had no significant effect on MMP3 mRNA levels in NHDFs, in contrast to the 121-fold stimulation (p < 0.001) by IL-1β of MMP3 mRNA in NHDFs (Supplementary Figure S3).
2.5. TIMP-1 and Collagen I Protein Expression
We applied immunohistochemistry to validate the increased TIMP1 mRNA levels in wounds at the protein level and to elucidate the cellular sources of TIMP-1 in 19 wounds with adjoining skin. TIMP-1 was undetectable in the epidermal compartments of skin and wounds. In the dermal compartments, a few fibroblasts revealed TIMP-1 staining in normal skin, while the expression of the TIMP-1 protein was markedly increased in fibroblasts and in endothelial cells, as well as lymphocytes in wounds, compared to normal skin. TIMP-1 was observed in hair follicle epithelium more often in wounds than in the skin. Acrosyringium of the eccrine sweat gland was positive in one wound. In general, TIMP-1 stained granularly in the cytoplasm of the cells (Figure 4A,B).
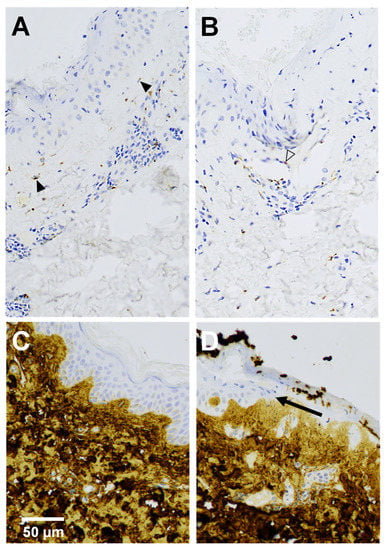
Figure 4.
TIMP-1 (A,B) and collagen α1 (I) (C,D) immunohistochemical analysis of a 4-day-old wound (A,B,D) with adjacent skin (C). (A) Fibroblasts (solid arrowheads) below neoepidermis and (B) endothelial cell (outline arrowhead) of small blood vessel show positive immunoreactivity of TIMP-1 in upper dermis. (D) Arrow indicates the tip of the migrating neoepidermal tongue.
Collagen I strongly stained the dermis in both wounds and skin and was observed below the basement membrane but not in epidermis (Figure 4C,D).
2.6. Proliferation Markers in Wounds
To delineate conceivable biomarkers of cell proliferation, we examined correlations between MKI67 mRNA levels and the entire panel of genes. KLK8 (r = 0.90, p = 7.2 × 10−8) and KRT6A (r = 0.83, p = 5.8 × 10−6) showed strong correlations with MKI67 mRNA levels, as shown in Figure 5.
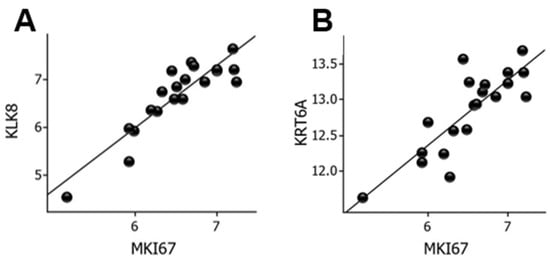
Figure 5.
Correlation between MKI67 and KLK8 mRNA levels (A) and between MKI67 and KRT6A mRNA levels (B) in wounds. Log2-transformed expression values are shown on the y- and x-axes. KLK, kallikrein; KRT, keratin.
3. Discussion
The aim of this study was to design and validate an expression panel of genes involved in reepithelialization during wound healing. Unlike high-throughput next-generation technologies, such as microarray and RNASeq, which produce a comprehensive set of gene expression profiles, we selected genes known to be involved in wound healing from the literature. This focused strategy lowers the false discovery rate (FDR) with accompanying increased power. Of the 22 upregulated DEGs, nine were related to ECM remodeling, six encoded cytokines/chemokines and five were growth factor-transcribing genes, and all of these genes are considered important for reepithelialization.
MMP1 was the most upregulated gene in the wounds. Nuutila et al. [3] reported that MMP1 was the top overexpressed gene in split-thickness skin graft (STSG) donor site wounds using genome-wide transcriptomic methodology. MMP-1 is unambiguously associated with reepithelialization [26] and we recently demonstrated the exclusive presence of the MMP-1 protein in the neoepidermis and in fibroblasts in the dermis beneath the neoepidermis [1].
The regulation of the MMP1 gene involves several signaling pathways [1,27,28]. The strong correlation between IL6 and MMP1 mRNA levels indicated a possible role of IL-6 in the induction of MMP1. The effect of IL-6 treatment of cultured NHEKs and NHDFs was weak in contrast to IL-1β. These somewhat contradictory results could be explained by the indirect effect of IL-1β on IL6 expression [29]. We also found that IL1B mRNA levels correlated strongly with IL6 mRNA levels in the wounds.
It has been suggested that collagen I is the primary inducer of MMP1 via the α2β1 integrin [30]. Collagen I was observed to be juxtaposed to the basement membrane by immunohistochemistry, implying no direct contact of keratinocytes with collagen I, indicating that collagen I-keratinocyte interactions are subordinate in the regulation of MMP1. It should be emphasized that the basement membrane remains essentially intact during reepithelialization [1,28]. The elevated COL4A1 may indicate the requirement for collagen IV by migrating keratinocytes on the basement membrane but also for angiogenesis [28,31].
MMP1 belongs to the MMP family, consisting of 23 human members [2,32]. In another study, MMP1 and MMP3 expression was coordinately induced [33]; the MMP1 and MMP3 genes are closely located on chromosome 11q22.2. MMP3 was also the second most upregulated gene and was increased by IL-1β but not by IL-6. In earlier studies, investigators failed to detect MMP3 in suction blister wounds by in situ hybridization [11,28]. This discrepancy might be attributed to the high sensitivity of our analyses. MMP-3 contributes to the conversion of latent into active MMP-1 [34]. Apart from MMP1 and MMP3, MMP12 and MMP9 were upregulated. MMP-12 appears to play a key role in cytoskeletal rearrangements in migrating keratinocytes [35]. MMP-9 protein increases during suction blister wound healing [7,11].
MMP activity is antagonized by tissue inhibitors of metalloproteinases (TIMPs), and TIMP1 transcripts were increased in wounds compared with adjacent skin. Our immunohistochemical analysis clearly demonstrated increased TIMP-1 protein expression in wounds vs. skin primarily due to TIMP-1-producing fibroblasts, endothelial cells, and lymphocytes. In one study, TIMP1 mRNA was expressed in fibroblasts below the neo-epidermis but absent in keratinocytes [28]. Mechanistically, inhibition of the proteolytic action of MMP-1 slows epidermal tongue movement. Local TIMP-1 overexpression retards keratinocyte migration in wounds [36]. It has also been suggested that TIMP-1 is a beneficial angiogenic factor [37].
Regulation of TIMP1 has rarely been described. IL-6 did not induce the TIMP1 gene in NHDFs, corroborating earlier findings [38]. We found that IL-1β induced TIMP1, which also resulted in increased TIMP-1 secretion from NHDFs [39].
Neither COL1A1 expression nor proliferation of stromal cells were increased in dermis [1]. The lack of a fibroproliferative response explains the fact that these lesions leave no scar.
The precursor plasminogen of the serine proteinase plasmin acts together with MMPs for maximal keratinocyte migration [40,41]. Plasminogen activator inhibitor SERPINE1 and the receptor for urokinase plasminogen activator PLAUR were upregulated, indicating the need for the plasminogen system in coordinating reepithelialization [42,43,44].
Different soluble mediators are involved in the immunoregulatory and inflammatory processes in these lesions [20]. The protein levels of IL-6, IL-8 and IL-1β were dramatically increased 1 day after wound infliction while IL-1α levels remained low [20]. These data are convincingly consistent with our mRNA measurements of IL6, CXCL8 and IL1B. In contrast, CSF2, IFNG, IL1A and TNF were not upregulated in the wounds vs. nonwounded skin. This finding does not exclude the possibility that these cytokines were upregulated earlier. The proinflammatory chemokines CXCL5, CXCL2 and CXCL1 in addition to CXCL8 were upregulated in wounds, presumably as a response to injury [45]. The main receptor CXCR2 is expressed on innate immune cells and on keratinocytes; CXCR2-deficient keratinocytes display impaired migration [46]. Prostaglandin-endoperoxide synthase 2 (PTGS2), also known as COX-2, is induced by trauma in the epidermis [47] and provides proinflammatory prostaglandins.
Five growth factors (HBEGF, CYR61/CCN1, HGF, CTGF/CCN2 and FGF7) were upregulated in the wounds as indicated in previous studies [48,49,50,51,52]. The neoepidermis is regenerated by the combined action of keratinocyte migration and proliferation. Previously, we showed that keratinocyte proliferation was increased to the same magnitude in neoepidermis and adjacent epidermis [1]. This finding might explain the lack of difference in gene expression of the proliferation marker MKI67 between wounds and skin. Collectively, these findings imply that the primary mode of action of the upregulated growth factors is the stimulation of keratinocyte migration independent of their mitogenic effects; these factors also have the ability to enhance the migration of epidermal keratinocytes in vitro [48,49,50,51,53].
The serine proteinase kallikrein-related peptidase 8 (KLK8) showed the strongest correlation with MKI67. KLK8 is one of 15 different kallikreins in the epidermis involved in epidermal homeostasis [54]. Interestingly, delayed wound healing in KLK8-knockout mice was accompanied by decreased Ki-67 immunolabeling of the neoepidermis [55]. Keratin-6A (KRT6A) mRNA levels also correlated strongly with MKI67 gene expression. Keratin-6A protein levels were increased in proliferating vs. differentiating NHEKs in vitro [56], which was the reason why we investigated the usefulness of keratin-6A protein as a biomarker for keratinocyte proliferation in human wounds in a previous study [56]. However, keratin-6A was undetectable possibly because it is an intracellular protein [56].
The increased MT2A mRNA levels in wounds corroborate our previous immunohistochemical results using an anti-MT antibody that reacted not only with MT2A but also with the subisoform MT1A [1]. Because MT1A was not significantly upregulated in the wounds compared to skin, MT2A was most likely the predominant metallothionein isoform detected in the wounds [1].
The downregulated genes were associated with epidermal maturation. Collagen VII is the main component of anchoring fibrils. Nyström et al. [57] observed collagen VII in migrating epidermis, but this finding was in wounds devoid of basement membrane. Leivo et al. [10] found no differences in collagen VII between suction blister floor and normal skin. The role of MMP-28 is unclear; MMP28 mRNA was detected distal to the leading edge in the proliferating epidermal compartment [58]. The zinc importer SLC39A2 (ZIP2) has been suggested to participate in keratinocyte differentiation [59]. K10 is an early epidermal differentiation marker. Filaggrins (FLG1 and FLG2) are essential for the formation of a functional stratum corneum [60,61].
4. Materials and Methods
4.1. Ethical Statements
The study was approved by the Committee on Biomedical Research Ethics for the Capital Region of Denmark (H-6-2014-001) and was registered at ClinicalTrials.gov (NCT02116725) on 15 April 2014, and conducted at the Department of Dermatology, Bispebjerg Hospital, University of Copenhagen, Copenhagen, Denmark [14].
4.2. Participants
Healthy nonsmoking volunteers between 18 and 65 years of age were included after providing written informed consent. Individuals with skin disorders; those who were pregnant, breastfeeding or receiving systemic immunosuppressive treatment; and/or those who were hypersensitive to zinc were excluded [1,14,15].
4.3. Induction of Epidermal (Suction Blister) Wounds, Treatment and Tissue Procedures
Suction blisters (10 mm in diameter) were raised on each buttock in 30 remunerated participants, and the blister roofs were excised. Two of the three treatments (zinc sulfate, placebo, or control) were randomized to the left or right wound by concealed allocation, i.e., each of the 3 treatments was applied to 20 wounds in 20 participants. In the present study, the participants of the control arm of this three-arm randomized, double-blind trial were included [14]. Wounds and adjoining skin were treated once daily with distilled water and covered with a bacteria-proof and moisture-retaining dressing (Mepore Film & Pad, Mölnlycke Health Care, Göteborg, Sweden) [1,15]. On post-wounding day 4, wounds, including uninjured skin, were excised [1]. The biopsies were fixed in 4% phosphate-buffered paraformaldehyde (pH 7.4) overnight at 4 °C and embedded in paraffin.
4.4. Macrodissection and Isolation of RNA from FFPE Wound and Skin Compartments
Tissue sections were cut at 5 µm and dissected into one central wound piece and two adjacent pieces of normal nonwounded skin (Figure 6).
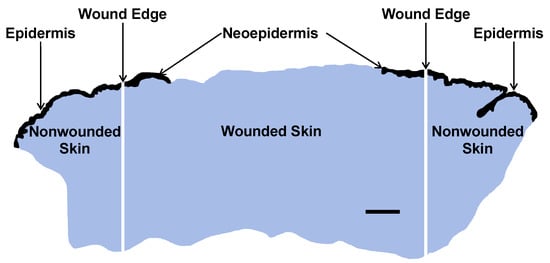
Figure 6.
Macrodissection of FFPE tissue section of a day-4 suction blister wound into central wound (1 piece) including neoepidermis and adjoining nonwounded normal skin including epidermis (2 pieces). Reepithelialization was calculated using the following formula: Neoepidermis (mm)/Wound length (mm) × 100%. Total RNA was extracted, and the extracts were subjected to nCounter mRNA analysis. Epidermal compartments are black and dermal compartments blue. Scale bar, 1 mm.
The tissue pieces were deparaffinized in xylene and absolute ethanol, and the tissue was scratched into reaction vials using a scalpel. Total RNA was extracted with a High Pure FFPE RNA Micro Kit (Roche, Mannheim, Germany). The concentration and purity of RNA in 20 µL elution buffer were determined by NanoDrop (NanoDrop Technologies, Wilmington, DE, USA) spectrophotometry. RNA purity was indicated by the OD260 nm/OD280 nm ratio (≥1.5 was acceptable).
4.5. Design of Gene Panel and nCounter Analyses
The NanoString human Preselected PlexSet Wound Healing Panel was used encompassing 90 target genes related to clotting, immune response to tissue injury, and ECM remodeling, along with relevant signaling pathway genes coordinating wound healing. Forty-nine wound-healing related genes expressed in skin were added to this panel by the authors, as indicated in Appendix A (Table A1). The details of the design of our gene expression panel are found in Supplementary Table S4.
The isolated total RNA (100 ng) was hybridized to the code sets at 65 °C overnight on the nCounter Prep Station. Gene expression was analyzed with an nCounter Digital Analyzer. Fluorescence was determined using a built-in inverted fluorescence microscope using all 550 possible counting areas of the NanoString cartridges.
nSolver software, version 4.0, was used to export, quality check, and normalize the hybridization results. Background subtraction was performed by negative control thresholding using the average of the included 8 probe-set negative controls. Normalization was performed in two steps. First, based on the geometric mean of the spiked-in positive controls, the positive control normalization factor was calculated to adjust for differences in various steps of the process. Default settings between 0.3 and 3 were used. Second, to adjust for differences in analyte abundance and quality across samples, a normalization factor was calculated based on the geometric mean of the included housekeeping genes (ABCF1, GUSB, HPRT1, LDHA, PTEN and RPLP0). The default settings (0.1–10) were applied.
4.6. Immunohistochemical Analysis of TIMP-1 and Collagen I
Sections with a thickness of 4 μm were cut, and slides were deparaffinized and rehydrated. The sections were pretreated and stained using the Omnis automated slide-processing system from Agilent (Glostrup, Denmark). The tissue sections were subjected to heat-induced epitope retrieval pretreatment using EnVision™ FLEX Target Retrieval Solution High pH (GV804, Dako Omnis, Agilent) for TIMP-1 or EnVision™ FLEX Target Retrieval Solution Low pH (GV805, Dako Omnis, Agilent) for collagen I for 30 min, followed by incubation with rabbit monoclonal antibodies against TIMP-1 (clone EPR18352, 1:500, ab211926, Abcam, Cambridge, UK) or collagen α1 (I) (E8F4L, 1:100, #72026, Cell Signaling, Danvers, MA, USA) for 30 min at 32 °C. The reactions were detected using the standard polymer technique EnVision™ FLEX/HRP Detection Reagent (GV800, Dako Omnis, Agilent), and signal intensity was enhanced using the EnVision™ FLEX+ Rabbit LINKER (GV809, Dako Omnis, Agilent) and visualized using EnVision™ Flex DAB+ Chromogen system (GV825, Dako Omnis, Agilent). Finally, the sections were counterstained with hematoxylin and mounted with Pertex. The immunostained sections were evaluated by a senior consultant pathologist (L.M.R.G.).
4.7. Studies in NHEKs and NHDFs
NHEKs (C-12006) were derived from 24- to 57-year-old Caucasian women and purchased from PromoCell (Heidelberg, Germany). NHEKs were cultured in keratinocyte growth medium-2 medium (PromoCell) composed of keratinocyte basal medium (KBM)-2 with penicillin (100 IU/mL), streptomycin (100 μg/mL), and amphotericin-B (50 ng/mL) and supplemented with bovine pituitary extract (30 µg/mL), recombinant human epidermal growth factor (0.125 ng/mL), insulin (5 µg/mL) and transferrin (10 µg/mL), hydrocortisone (0.33 µg/mL), epinephrine (0.39 µg/mL) and CaCl2 (0.06 mM) and on collagen I-coated surfaces. NHDFs (CC-2511) were derived from a 37-year-old Caucasian woman and purchased from Lonza (Basel, Switzerland). NHDFs were cultured in DMEM with GlutaMAX™, glucose (4.5 g/L) and pyruvate (Gibco, Life Technologies, Grand Island, NY, USA) with 10% fetal bovine serum (FBS; Gibco heat-inactivated qualified FBS, 10500064, Thermo Fisher Scientific, Waltham, MA USA), penicillin (100 IU/mL) and streptomycin (100 μg/mL). Cells were incubated in a humidified atmosphere of 5% CO2/air at 37 °C and were passaged using 0.05% trypsin-0.02% EDTA (Biological Industries, Kibbutz Bet-Haemek, Israel) [24].
NHEKs and NHDFs were seeded (1 × 105 in 1 mL/well) in 24-well tissue culture plates (CellStar®, Greiner Bio-One). NHEKs were grown in wells coated with collagen I [14]. Cells were incubated for 72 h. The confluent cell layers were then washed with Dulbecco’s PBS (pH 7.4) and starved for 24 h in serum-free KBM-2/DMEM with 1.8 mM CaCl2 containing 1 mg/mL bovine serum albumin. The cells were then treated with 30 ng/mL rhIL-6 (206-IL, R&D Systems, Minneapolis, MN, USA) and 1 ng/mL rhIL-1β (201-LB, R&D Systems) separately and combined for 24 h [62]. Media were collected, spun (2000× g, 10 min, 4 °C) and the supernatants kept at −80 °C until analysis for LDH activity [14], and MMP-1 (RAB0361, Sigma-Aldrich, St. Louis, MO, USA) and TIMP-1 (ab187394, Abcam) levels by ELISA [1,63].
Total RNA of the treated NHEKs and NHDFs was extracted with 1 mL of TriReagent® (Molecular Research Center, Cincinnati, OH, USA). Bromochloropropane (100 µL) was added to isolate the aqueous phase containing the RNA, which was precipitated using isopropanol. The RNA pellet was then washed in ethanol and subsequently dissolved in 10 μL RNAse-free water. Total RNA concentrations were determined with the Ribo-Green assay (R11490, Life Technologies).
Total RNA (500 ng) was converted into cDNA in 20 μL using OmniScript reverse transcriptase (Qiagen, Valencia, CA, USA) and 1 μM poly-dT (Invitrogen) according to the manufacturer’s protocol (Qiagen). For each target mRNA, 0.5 μL of cDNA was amplified in 25 μL of SYBR Green polymerase chain reaction (PCR) containing 1× QuantiTect SYBR Green Master Mix (Qiagen) and 100 nM of each primer, as shown in Table 2. The amplification was monitored in real time using an MX3005P Real-time PCR machine (Stratagene, La Jolla, CA, USA). Ct values were related to a standard curve made with known concentrations of cloned PCR products or DNA oligonucleotides (Ultramer™ oligos, Integrated DNA Technologies, Leuven, Belgium) with a DNA sequence corresponding to the sequence of the expected PCR product. The specificity of the PCR products was confirmed by melting curve analysis after amplification. RPLP0 mRNA was chosen as an internal control.

Table 2.
Primer sequences for RT–qPCR analyses of treated NHEKs and NHDFs.
4.8. Statistical Analysis
DEGs were identified by the paired t test (participant eliminated as factor, p < 0.05, log2FC ≥ |1.00|). All data were log2-transformed prior to analysis using Qlucore Omics Explorer software, version 3.7 (Qlucore AB, Lund, Sweden). Gene expression correlations with MMP1 and MKI67 were calculated using Pearson’s correlation coefficients. Cell culture data (LDH, BrdU incorporation, RT–qPCR and ELISAs) were analyzed with one-way ANOVA and the Holm-Sidak post hoc method using SigmaPlot software, version 14.0 (Systat, Palo Alto, CA, USA). The level of statistical significance was set to p < 0.05. FDR-adjusted p values, i.e., q values, were calculated [64].
5. Conclusions
The major genes involved in human epidermal wound healing were successfully quantified using a customized panel for the NanoString platform. The obtained wound healing gene expression signature, consisting of 28 DEGs, is a start in identifying possible therapeutic targets to accelerate reepithelialization and epidermal stratification. The overlapping functions of upregulated cytokines/chemokines and growth factors could indicate biological redundancies.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms232415746/s1.
Author Contributions
Conceptualization, L.M.R.G. and M.S.Å.; methodology, J.O.E.; formal analysis, M.B., J.O.E., L.M.R.G., M.S.Å., P.S. and T.L.; writing—original draft preparation, M.S.Å.; supervision, M.B., J.O.E., L.M.R.G., M.S.Å. and P.S.; project administration, L.M.R.G. and M.S.Å.; funding acquisition, L.M.R.G. and M.S.Å. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Aage Bangs Fond and Mölnlycke Health Care.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved 3 March 2014 by the Ethics Committee (H-6-2014-001).
Informed Consent Statement
Written informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
| BrdU | bromodeoxyuridine |
| DEG | differentially expressed gene |
| ECM | extracellular matrix |
| ELISA | enzyme-linked immunosorbent assay |
| FBS | fetal bovine serum |
| FC | fold change |
| FDR | false discovery rate |
| FFPE | formalin-fixed, paraffin-embedded |
| KBM | keratinocyte basal medium |
| LDH | lactate dehydrogenase |
| NHDF | normal human dermal fibroblast |
| NHEK | normal human epidermal keratinocyte |
| PBS | phosphate-buffered saline |
| RT–qPCR | reverse transcription quantitative polymerase chain reaction |
| STSG | split-thickness skin graft |
Appendix A

Table A1.
Gene expression panel of 139 targets (genes included in the nCounter PlexSet Preselected Human Wound Healing Panel arehighlighted in grey).
Table A1.
Gene expression panel of 139 targets (genes included in the nCounter PlexSet Preselected Human Wound Healing Panel arehighlighted in grey).
| Gene Name | GenBank ID | |
|---|---|---|
| ACTC1 | NM_005159.4 | actin, alpha, cardiac muscle 1 |
| ACVRL1 | NM_000020.1 | activin A receptor type II-like 1 |
| ADAM17 | NM_003183.4 | ADAM metallopeptidase domain 17 |
| ADORA2A | NM_000675.3 | adenosine A2a receptor |
| AGER | NM_001136.3 | advanced glycosylation end product-specific receptor |
| ANGPT1 | NM_001146.3 | angiopoietin 1 |
| BMP1 | NM_001199.1 | bone morphogenetic protein 1 |
| CCL2 | NM_002982.3 | chemokine (C-C motif) ligand 2 |
| CCL7 | NM_006273.2 | chemokine (C-C motif) ligand 7 |
| CD36 | NM_000072.3 | thrombospondin receptor |
| CD40LG | NM_000074.2 | CD40 ligand |
| CD59 | NM_000611.4 | complement regulatory protein |
| CDH1 | NM_004360.2 | E-cadherin (epithelial) |
| COL1A1 | NM_000088.3 | collagen type I, alpha 1 chain |
| COL3A1 | NM_000090.3 | collagen type III, alpha 1 chain |
| COL4A1 | NM_001845.4 | collagen, type IV, alpha 1 chain |
| COL4A3 | NM_000091.4 | collagen, type IV, alpha 3 (Goodpasture antigen) |
| COL5A2 | NM_000393.3 | collagen, type V, alpha 2 chain |
| COL5A3 | NM_015719.3 | collagen, type V, alpha 3 chain |
| COL7A1 | NM_000094.2 | collagen type VII, alpha 1 chain |
| COL14A1 | NM_021110.1 | collagen, type XIV, alpha 1 chain |
| COL17A1 | NM_000494.3 | collagen type XVII, alpha 1 chain |
| CSF2 | NM_000758.2 | colony stimulating factor 2 (granulocyte-macrophage) |
| CSF2RA | NM_006140.3 | colony stimulating factor 2 receptor alpha subunit |
| CSF3 | NM_000759.3 | colony stimulating factor 3 (granulocyte) |
| CTGF | NM_001901.2 | connective tissue growth factor |
| CTNNB1 | NM_001098210.1 | catenin (cadherin-associated protein), beta 1, 88 kDa |
| CTSG | NM_001911.2 | cathepsin G |
| CTSK | NM_000396.2 | cathepsin K |
| CTSV | NM_001333.3 | cathepsin V |
| CXCL1 | NM_001511.1 | chemokine (C-X-C motif) ligand 1 |
| CXCL2 | NM_002089.3 | chemokine (C-X-C motif) ligand 2 |
| CXCL5 | NM_002994.3 | chemokine (C-X-C motif) ligand 5 |
| CXCL8 | NM_000584.2 | chemokine (C-X-C motif) ligand 8 |
| CXCL11 | NM_005409.4 | chemokine (C-X-C motif) ligand 11 |
| CYR61 | NM_001554.3 | cysteine-rich angiogenic inducer 61 |
| DEFB4 | NM_004942.2 | beta-defensin 2 |
| EGF | NM_001963.4 | epidermal growth factor |
| EGFR | NM_201282.1 | epidermal growth factor receptor |
| EGR1 | NM_001964.2 | early growth response 1 |
| ENTPD1 | NM_001098175.1 | ectonucleoside triphosphate diphosphohydrolase 1 |
| EREG | NM_001432.2 | epiregulin |
| F3 | NM_001993.3 | coagulation factor III (thromboplastin, tissue factor) |
| F5 | NM_000130.2 | coagulation factor V (proaccelerin, labile factor) |
| F13A1 | NM_000129.3 | coagulation factor XIII, A1 polypeptide |
| FGF2 | NM_002006.4 | fibroblast growth factor 2 (basic) |
| FGF7 | NM_002009.3 | fibroblast growth factor 7 |
| FGF10 | NM_004465.1 | fibroblast growth factor 10 |
| FLG1 | NM_002016.1 | filaggrin |
| FLG2 | NM_001014342.2 | filaggrin family member 2 |
| FN1 | NM_212482.1 | fibronectin |
| GNAQ | NM_002072.2 | guanine nucleotide binding protein (G protein) |
| HBEGF | NM_001945.1 | heparin-binding EGF-like growth factor |
| HGF | NM_000601.4 | hepatocyte growth factor (hepapoietin A, scatter factor) |
| HIF1A | NM_001530.2 | hypoxia inducible factor 1 alpha subunit |
| IFNG | NM_000619.2 | interferon gamma |
| IGF1 | NM_000618.3 | insulin-like growth factor 1 (somatomedin C) |
| IL1A | NM_000575.3 | interleukin 1 alpha |
| IL1B | NM_000576.2 | interleukin 1 beta |
| IL2 | NM_000586.2 | interleukin 2 |
| IL4 | NM_000589.2 | interleukin 4 |
| IL6 | NM_000600.3 | interleukin 6 (interferon, beta 2) |
| IL6ST | NM_002184.2 | interleukin 6 signal transducer (gp130) |
| IL10 | NM_000572.2 | interleukin 10 |
| ITGA1 | NM_181501.1 | integrin, alpha 1 |
| ITGA2 | NM_002203.2 | integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
| ITGA3 | NM_002204.2 | integrin, alpha 3 (antigen CD49C) |
| ITGA4 | NM_000885.4 | integrin, alpha 4 (antigen CD49D) |
| ITGA5 | NM_002205.2 | integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| ITGA6 | NM_000210.1 | integrin, alpha 6 |
| ITGAV | NM_002210.2 | integrin, alpha V |
| ITGB1 | NM_002211.3 | integrin, beta 1 (fibronectin receptor, antigen CD29) |
| ITGB3 | NM_000212.2 | integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) |
| ITGB5 | NM_002213.3 | integrin, beta 5 |
| ITGB6 | NM_000888.3 | integrin, beta 6 |
| IVL | NM_005547.2 | involucrin |
| KLK8 | NM_144507.1 | kallikrein-related peptidase 8 |
| KRT6A | NM_005554.3 | keratin 6A |
| KRT10 | NM_000421.3 | keratin 10 |
| LAMA3 | NM_000227.3 | laminin subunit alpha 3 |
| MAPK1 | NM_138957.2 | mitogen-activated protein kinase 1 |
| MAPK3 | NM_001040056.1 | mitogen-activated protein kinase 3 |
| MIF | NM_002415.1 | macrophage migration inhibitory factor |
| MKI67 | NM_002417.2 | marker of proliferation Ki-67 |
| MMP1 | NM_002421.3 | matrix metallopeptidase-1 (interstitial collagenase) |
| MMP2 | NM_004530.2 | matrix metallopeptidase-2 (gelatinase A) |
| MMP3 | NM_002422.3 | matrix metallopeptidase-3 |
| MMP7 | NM_002423.3 | matrix metallopeptidase-7 (matrilysin) |
| MMP8 | NM_002424.2 | matrix metallopeptidase-8 |
| MMP9 | NM_004994.2 | matrix metallopeptidase-9 (gelatinase B) |
| MMP10 | NM_002425.1 | matrix metallopeptidase-10 |
| MMP12 | NM_002426.3 | matrix metallopeptidase-12 |
| MMP13 | NM_002427.2 | matrix metallopeptidase-13 |
| MMP14 | NM_004995.2 | matrix metallopeptidase-14 |
| MMP28 | NM_001032278.1 | matrix metallopeptidase-28 |
| MT1A | NM_005946.2 | metallothionein 1A |
| MT2A | NM_005953.3 | metallothionein 2A |
| MYC | NM_002467.3 | MYC proto-oncogene, BHLH transcription factor |
| NF1 | NM_000267.2 | neurofibromin 1 |
| NGF | NM_002506.2 | nerve growth factor |
| PDGFA | NM_002607.5 | platelet-derived growth factor alpha polypeptide |
| PF4 | NM_002619.2 | platelet factor 4 |
| PI3 | NM_002638.3 | peptidase inhibitor 3 (elafin) |
| PI3K | NM_006218.2 | phosphatidylinositide 3-kinase |
| PLAT | NM_000931.2 | plasminogen activator, tissue |
| PLAU | NM_002658.2 | plasminogen activator, urokinase |
| PLAUR | NM_001005376.2 | plasminogen activator, urokinase receptor |
| POLR1B | NM_019014.3 | polymerase (RNA) I polypeptide B, 128 kDa |
| PTGS2 | NM_000963.1 | prostaglandin-endoperoxide synthase 2 |
| RAC1 | NM_006908.4 | ras-related C3 botulinum toxin substrate 1 (rho family) |
| RHOA | NM_001664.2 | ras homolog family member A |
| S100A7 | NM_002963.3 | psoriasin |
| S100A8 | NM_002964.3 | S100 calcium binding protein A8 |
| S100A9 | NM_002965.2 | S100 calcium binding protein A9 |
| SERPINB3 | NM_006919.2 | serpin family B member 3 (serine proteinase inhibitor) |
| SERPINE1 | NM_000602.2 | serpin peptidase inhibitor, clade E (plasminogen activator inhibitor type 1) |
| SLC30A1 | NM_021194.2 | solute carrier family 30 member 1 (ZnT1) |
| SLC39A2 | NM_014579.1 | solute carrier family 39 member 2 (ZIP2) |
| SLC39A4 | NM_017767.2 | solute carrier family 39 member 4 (ZIP4) |
| SOD1 | NM_000454.4 | superoxide dismutase [Cu-Zn] |
| STAT3 | NM_003150.3 | signal transducer and activator of transcription 3 |
| TFPI | NM_001032281.2 | tissue factor pathway inhibitor |
| TGFA | NM_003236.2 | transforming growth factor, alpha |
| TGFB1 | NM_000660.3 | transforming growth factor, beta 1 |
| TGFB2 | NM_003238.2 | transforming growth factor, beta 2 |
| TGFBR3 | NM_003243.3 | transforming growth factor, beta receptor III |
| THBD | NM_000361.2 | thrombomodulin |
| TIMP1 | NM_003254.2 | TIMP metallopeptidase inhibitor 1 |
| TIMP2 | NM_003255.4 | TIMP metallopeptidase inhibitor 2 |
| TIMP3 | NM_000362.4 | TIMP metallopeptidase inhibitor 3 |
| TLR4 | NM_138554.2 | toll-like receptor 4 |
| TMPRSS6 | NM_153609.2 | transmembrane protease, serine 6 |
| TNF | NM_000594.2 | tumor necrosis factor |
| VEGFA | NM_001025366.1 | vascular endothelial growth factor A |
| VTN | NM_000638.3 | vitronectin |
| VWF | NM_000552.3 | von Willebrand factor |
| WAS | NM_000377.2 | WASP actin nucleation promoting factor |
| WISP1 | NM_080838.1 | WNT1 inducible signaling pathway protein 1 |
| WNT5A | NM_003392.3 | wingless-type MMTV integration site family, member 5A |
| ABCF11 | NM_001090.2 | ATP-binding cassette, sub-family F (GCN20), member 1 |
| GUSB1 | NM_000181.3 | glucuronidase, beta |
| HPRT11 | NM_000194.1 | hypoxanthine phosphoribosyltransferase 1 |
| LDHA1 | NM_001165414.1 | lactate dehydrogenase A |
| PTEN1 | NM_000314.3 | phosphatase and tensin homolog |
| RPLP0 1 | NM_001002.3 | ribosomal protein, large, P0 |
1 Housekeeping Gene.
References
- Ågren, M.S.; Chafranska, L.; Eriksen, J.O.; Forman, J.L.; Bjerrum, M.J.; Schjerling, P.; Larsen, H.F.; Cottarelli, E.; Jorgensen, L.N.; Gjerdrum, L.M.R. Spatial expression of metallothionein, matrix metalloproteinase-1 and Ki-67 in human epidermal wounds treated with zinc and determined by quantitative immunohistochemistry: A randomised double-blind trial. Eur. J. Cell Biol. 2021, 100, 151147. [Google Scholar] [CrossRef] [PubMed]
- Michopoulou, A.; Rousselle, P. How do epidermal matrix metalloproteinases support re-epithelialization during skin healing? Eur. J. Dermatol. 2015, 25 (Suppl. 1), 33–42. [Google Scholar] [CrossRef] [PubMed]
- Nuutila, K.; Siltanen, A.; Peura, M.; Bizik, J.; Kaartinen, I.; Kuokkanen, H.; Nieminen, T.; Harjula, A.; Aarnio, P.; Vuola, J.; et al. Human skin transcriptome during superficial cutaneous wound healing. Wound Repair Regen. 2012, 20, 830–839. [Google Scholar] [CrossRef] [PubMed]
- Greco, J.A., 3rd; Pollins, A.C.; Boone, B.E.; Levy, S.E.; Nanney, L.B. A microarray analysis of temporal gene expression profiles in thermally injured human skin. Burns 2010, 36, 192–204. [Google Scholar] [CrossRef]
- Deonarine, K.; Panelli, M.C.; Stashower, M.E.; Jin, P.; Smith, K.; Slade, H.B.; Norwood, C.; Wang, E.; Marincola, F.M.; Stroncek, D.F. Gene expression profiling of cutaneous wound healing. J. Transl. Med. 2007, 5, 11. [Google Scholar] [CrossRef]
- Kutlu, N.; Svedman, P. The superficial dermal microcirculation in suction blister wounds on healthy volunteers. Vasc. Endovasc. Surg. 1992, 26, 200–212. [Google Scholar] [CrossRef]
- Oikarinen, A.; Kylmaniemi, M.; Autio-Harmainen, H.; Autio, P.; Salo, T. Demonstration of 72-kDa and 92-kDa forms of type IV collagenase in human skin: Variable expression in various blistering diseases, induction during re-epithelialization, and decrease by topical glucocorticoids. J. Investig. Dermatol. 1993, 101, 205–210. [Google Scholar] [CrossRef][Green Version]
- Lévy, J.J.; von Rosen, J.; Gassmuller, J.; Kleine Kuhlmann, R.; Lange, L. Validation of an in vivo wound healing model for the quantification of pharmacological effects on epidermal regeneration. Dermatology 1995, 190, 136–141. [Google Scholar] [CrossRef]
- Glaser, R.; Kiecolt-Glaser, J.K.; Marucha, P.T.; MacCallum, R.C.; Laskowski, B.F.; Malarkey, W.B. Stress-related changes in proinflammatory cytokine production in wounds. Arch. Gen. Psychiatry 1999, 56, 450–456. [Google Scholar] [CrossRef]
- Leivo, T.; Kiistala, U.; Vesterinen, M.; Owaribe, K.; Burgeson, R.E.; Virtanen, I.; Oikarinen, A. Re-epithelialization rate and protein expression in the suction-induced wound model: Comparison between intact blisters, open wounds and calcipotriol-pretreated open wounds. Br. J. Dermatol. 2000, 142, 991–1002. [Google Scholar] [CrossRef]
- Ågren, M.S.; Mirastschijski, U.; Karlsmark, T.; Saarialho-Kere, U.K. Topical synthetic inhibitor of matrix metalloproteinases delays epidermal regeneration of human wounds. Exp. Dermatol. 2001, 10, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Koivukangas, V.; Oikarinen, A. Suction blister model of wound healing. Methods Mol. Med. 2003, 78, 255–261. [Google Scholar] [PubMed]
- Kottner, J.; Hillmann, K.; Fimmel, S.; Seite, S.; Blume-Peytavi, U. Characterisation of epidermal regeneration in vivo: A 60-day follow-up study. J. Wound Care 2013, 22, 395–400. [Google Scholar] [CrossRef]
- Larsen, H.F.; Ahlström, M.G.; Gjerdrum, L.M.R.; Mogensen, M.; Ghathian, K.; Calum, H.; Sørensen, A.L.; Forman, J.L.; Vandeven, M.; Holerca, M.N.; et al. Noninvasive measurement of reepithelialization and microvascularity of suction-blister wounds with benchmarking to histology. Wound Repair Regen. 2017, 25, 984–993. [Google Scholar] [CrossRef] [PubMed]
- Ahlström, M.G.; Gjerdrum, L.M.R.; Larsen, H.F.; Fuchs, C.; Sørensen, A.L.; Forman, J.L.; Ågren, M.S.; Mogensen, M. Suction blister lesions and epithelialization monitored by optical coherence tomography. Skin Res. Technol. 2018, 24, 65–72. [Google Scholar] [CrossRef]
- Kjaer, M.; Frederiksen, A.K.S.; Nissen, N.I.; Willumsen, N.; van Hall, G.; Jorgensen, L.N.; Andersen, J.R.; Ågren, M.S. Multi-nutrient supplementation increases collagen synthesis during early wound repair in a randomized controlled trial in patients with inguinal hernia. J. Nutr. 2020, 150, 792–799. [Google Scholar] [CrossRef]
- Burian, E.A.; Sabah, L.; Kirketerp-Møller, K.; Gundersen, G.; Ågren, M.S. Effect of stabilized hypochlorous acid on re-epithelialization and bacterial bioburden in acute wounds: A randomized controlled trial in healthy volunteers. Acta Derm. Venereol. 2022, 102, adv00727. [Google Scholar] [CrossRef]
- Reis, P.P.; Waldron, L.; Goswami, R.S.; Xu, W.; Xuan, Y.; Perez-Ordonez, B.; Gullane, P.; Irish, J.; Jurisica, I.; Kamel-Reid, S. mRNA transcript quantification in archival samples using multiplexed, color-coded probes. BMC Biotechnol. 2011, 11, 46. [Google Scholar] [CrossRef]
- Fitzpatrick, T.B. The validity and practicality of sun-reactive skin types I through VI. Arch. Dermatol. 1988, 124, 869–871. [Google Scholar] [CrossRef]
- Kuhns, D.B.; DeCarlo, E.; Hawk, D.M.; Gallin, J.I. Dynamics of the cellular and humoral components of the inflammatory response elicited in skin blisters in humans. J. Clin. Investig. 1992, 89, 1734–1740. [Google Scholar] [CrossRef]
- McDaniel, J.C.; Belury, M.; Ahijevych, K.; Blakely, W. Omega-3 fatty acids effect on wound healing. Wound Repair Regen. 2008, 16, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Mihara, M.; Moriya, Y.; Ohsugi, Y. IL-6-soluble IL-6 receptor complex inhibits the proliferation of dermal fibroblasts. Int. J. Immunopharmacol. 1996, 18, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Ågren, M.S.; Steenfos, H.H.; Dabelsteen, S.; Hansen, J.B.; Dabelsteen, E. Proliferation and mitogenic response to PDGF-BB of fibroblasts isolated from chronic venous leg ulcers is ulcer-age dependent. J. Investig. Dermatol. 1999, 112, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Türlü, C.; Willumsen, N.; Marando, D.; Schjerling, P.; Biskup, E.; Hannibal, J.; Jorgensen, L.N.; Ågren, M.S. A human cellular model for colorectal anastomotic repair: The effect of localization and transforming growth factor-β1 treatment on collagen deposition and biomarkers. Int. J. Mol. Sci. 2021, 22, 1616. [Google Scholar] [CrossRef] [PubMed]
- Utani, A.; Momota, Y.; Endo, H.; Kasuya, Y.; Beck, K.; Suzuki, N.; Nomizu, M.; Shinkai, H. Laminin alpha 3 LG4 module induces matrix metalloproteinase-1 through mitogen-activated protein kinase signaling. J. Biol. Chem. 2003, 278, 34483–34490. [Google Scholar] [CrossRef] [PubMed]
- Inoue, M.; Kratz, G.; Haegerstrand, A.; Ståhle-Bäckdahl, M. Collagenase expression is rapidly induced in wound-edge keratinocytes after acute injury in human skin, persists during healing, and stops at re-epithelialization. J. Investig. Dermatol. 1995, 104, 479–483. [Google Scholar] [CrossRef]
- Vincenti, M.P.; Brinckerhoff, C.E. Transcriptional regulation of collagenase (MMP-1, MMP-13) genes in arthritis: Integration of complex signaling pathways for the recruitment of gene-specific transcription factors. Arthritis Res. Ther. 2002, 4, 157–164. [Google Scholar] [CrossRef]
- Saarialho-Kere, U.K.; Vaalamo, M.; Airola, K.; Niemi, K.M.; Oikarinen, A.I.; Parks, W.C. Interstitial collagenase is expressed by keratinocytes that are actively involved in reepithelialization in blistering skin disease. J. Investig. Dermatol. 1995, 104, 982–988. [Google Scholar] [CrossRef]
- Kitanaka, N.; Nakano, R.; Sugiura, K.; Kitanaka, T.; Namba, S.; Konno, T.; Nakayama, T.; Sugiya, H. Interleukin-1beta promotes interleulin-6 expression via ERK1/2 signaling pathway in canine dermal fibroblasts. PLoS ONE 2019, 14, e0220262. [Google Scholar] [CrossRef]
- Dumin, J.A.; Dickeson, S.K.; Stricker, T.P.; Bhattacharyya-Pakrasi, M.; Roby, J.D.; Santoro, S.A.; Parks, W.C. Pro-collagenase-1 (matrix metalloproteinase-1) binds the alpha(2)beta(1) integrin upon release from keratinocytes migrating on type I collagen. J. Biol. Chem. 2001, 276, 29368–29374. [Google Scholar] [CrossRef]
- Li, W.; Henry, G.; Fan, J.; Bandyopadhyay, B.; Pang, K.; Garner, W.; Chen, M.; Woodley, D.T. Signals that initiate, augment, and provide directionality for human keratinocyte motility. J. Investig. Dermatol. 2004, 123, 622–633. [Google Scholar] [CrossRef] [PubMed]
- Ågren, M.S.; auf dem Keller, U. Matrix metalloproteinases: How much can they do? Int. J. Mol. Sci. 2020, 21, 2678. [Google Scholar] [CrossRef] [PubMed]
- Saus, J.; Quinones, S.; Otani, Y.; Nagase, H.; Harris, E.D., Jr.; Kurkinen, M. The complete primary structure of human matrix metalloproteinase-3. Identity with stromelysin. J. Biol. Chem. 1988, 263, 6742–6745. [Google Scholar] [CrossRef] [PubMed]
- Ågren, M.S.; Schnabel, R.; Christensen, L.H.; Mirastschijski, U. Tumor necrosis factor-alpha-accelerated degradation of type I collagen in human skin is associated with elevated matrix metalloproteinase (MMP)-1 and MMP-3 ex vivo. Eur. J. Cell Biol. 2015, 94, 12–21. [Google Scholar] [CrossRef]
- Chakraborty, S.; Sampath, D.; Yu Lin, M.O.; Bilton, M.; Huang, C.K.; Nai, M.H.; Njah, K.; Goy, P.A.; Wang, C.C.; Guccione, E.; et al. Agrin-Matrix Metalloproteinase-12 axis confers a mechanically competent microenvironment in skin wound healing. Nat. Commun. 2021, 12, 6349. [Google Scholar] [CrossRef]
- Salonurmi, T.; Parikka, M.; Kontusaari, S.; Pirilä, E.; Munaut, C.; Salo, T.; Tryggvason, K. Overexpression of TIMP-1 under the MMP-9 promoter interferes with wound healing in transgenic mice. Cell Tissue Res. 2004, 315, 27–37. [Google Scholar] [CrossRef]
- Aplin, A.C.; Zhu, W.H.; Fogel, E.; Nicosia, R.F. Vascular regression and survival are differentially regulated by MT1-MMP and TIMPs in the aortic ring model of angiogenesis. Am. J. Physiol. Cell Physiol. 2009, 297, C471–C480. [Google Scholar] [CrossRef]
- Emonard, H.; Munaut, C.; Melin, M.; Lortat-Jacob, H.; Grimaud, J.A. Interleukin-6 does not regulate interstitial collagenase, stromelysin and tissue inhibitor of metalloproteinases synthesis by cultured human fibroblasts. Matrix 1992, 12, 471–474. [Google Scholar] [CrossRef]
- Han, R.; Smith, T.J. Induction by IL-1 beta of tissue inhibitor of metalloproteinase-1 in human orbital fibroblasts: Modulation of gene promoter activity by IL-4 and IFN-gamma. J. Immunol. 2005, 174, 3072–3079. [Google Scholar] [CrossRef]
- Rømer, J.; Bugge, T.H.; Pyke, C.; Lund, L.R.; Flick, M.J.; Degen, J.L.; Dano, K. Impaired wound healing in mice with a disrupted plasminogen gene. Nat. Med. 1996, 2, 287–292. [Google Scholar] [CrossRef]
- Lund, L.R.; Rømer, J.; Bugge, T.H.; Nielsen, B.S.; Frandsen, T.L.; Degen, J.L.; Stephens, R.W.; Danø, K. Functional overlap between two classes of matrix-degrading proteases in wound healing. EMBO J. 1999, 18, 4645–4656. [Google Scholar] [CrossRef] [PubMed]
- Rømer, J.; Lund, L.R.; Eriksen, J.; Ralfkiaer, E.; Zeheb, R.; Gelehrter, T.D.; Danø, K.; Kristensen, P. Differential expression of urokinase-type plasminogen activator and its type-1 inhibitor during healing of mouse skin wounds. J. Investig. Dermatol. 1991, 97, 803–811. [Google Scholar] [CrossRef] [PubMed]
- D’Alessio, S.; Gerasi, L.; Blasi, F. uPAR-deficient mouse keratinocytes fail to produce EGFR-dependent laminin-5, affecting migration in vivo and in vitro. J. Cell. Sci. 2008, 121 Pt 23, 3922–3932. [Google Scholar] [CrossRef] [PubMed]
- Simone, T.M.; Longmate, W.M.; Law, B.K.; Higgins, P.J. Targeted inhibition of PAI-1 activity impairs epithelial migration and wound closure following cutaneous injury. Adv. Wound Care 2015, 4, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, E.; Toksoy, A.; Goebeler, M.; Debus, S.; Brocker, E.B.; Gillitzer, R. Chemokines IL-8, GROalpha, MCP-1, IP-10, and Mig are sequentially and differentially expressed during phase-specific infiltration of leukocyte subsets in human wound healing. Am. J. Pathol. 1998, 153, 1849–1860. [Google Scholar] [CrossRef]
- Devalaraja, R.M.; Nanney, L.B.; Du, J.; Qian, Q.; Yu, Y.; Devalaraja, M.N.; Richmond, A. Delayed wound healing in CXCR2 knockout mice. J. Investig. Dermatol. 2000, 115, 234–244. [Google Scholar] [CrossRef]
- Müller-Decker, K.; Scholz, K.; Neufang, G.; Marks, F.; Furstenberger, G. Localization of prostaglandin-H synthase-1 and -2 in mouse skin: Implications for cutaneous function. Exp. Cell Res. 1998, 242, 84–91. [Google Scholar] [CrossRef]
- Stoll, S.W.; Rittie, L.; Johnson, J.L.; Elder, J.T. Heparin-binding EGF-like growth factor promotes epithelial-mesenchymal transition in human keratinocytes. J. Investig. Dermatol. 2012, 132, 2148–2157. [Google Scholar] [CrossRef]
- Du, H.; Zhou, Y.; Suo, Y.; Liang, X.; Chai, B.; Duan, R.; Huang, X.; Li, Q. CCN1 accelerates re-epithelialization by promoting keratinocyte migration and proliferation during cutaneous wound healing. Biochem. Biophys. Res. Commun. 2018, 505, 966–972. [Google Scholar] [CrossRef]
- Chmielowiec, J.; Borowiak, M.; Morkel, M.; Stradal, T.; Munz, B.; Werner, S.; Wehland, J.; Birchmeier, C.; Birchmeier, W. c-Met is essential for wound healing in the skin. J. Cell Biol. 2007, 177, 151–162. [Google Scholar] [CrossRef]
- Kiwanuka, E.; Hackl, F.; Caterson, E.J.; Nowinski, D.; Junker, J.P.; Gerdin, B.; Eriksson, E. CCN2 is transiently expressed by keratinocytes during re-epithelialization and regulates keratinocyte migration in vitro by the ras-MEK-ERK signaling pathway. J. Surg. Res. 2013, 185, e109–e119. [Google Scholar] [CrossRef]
- Marchese, C.; Chedid, M.; Dirsch, O.R.; Csaky, K.G.; Santanelli, F.; Latini, C.; LaRochelle, W.J.; Torrisi, M.R.; Aaronson, S.A. Modulation of keratinocyte growth factor and its receptor in reepithelializing human skin. J. Exp. Med. 1995, 182, 1369–1376. [Google Scholar] [CrossRef] [PubMed]
- Tsuboi, R.; Sato, C.; Kurita, Y.; Ron, D.; Rubin, J.S.; Ogawa, H. Keratinocyte growth factor (FGF-7) stimulates migration and plasminogen activator activity of normal human keratinocytes. J. Investig. Dermatol. 1993, 101, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Nauroy, P.; Nyström, A. Kallikreins: Essential epidermal messengers for regulation of the skin microenvironment during homeostasis, repair and disease. Matrix Biol. Plus 2020, 6, 100019. [Google Scholar] [CrossRef] [PubMed]
- Kishibe, M.; Bando, Y.; Tanaka, T.; Ishida-Yamamoto, A.; Iizuka, H.; Yoshida, S. Kallikrein-related peptidase 8-dependent skin wound healing is associated with upregulation of kallikrein-related peptidase 6 and PAR2. J. Investig. Dermatol. 2012, 132, 1717–1724. [Google Scholar] [CrossRef] [PubMed]
- Ågren, M.S.; Phothong, N.; Burian, E.A.; Mogensen, M.; Haedersdal, M.; Jorgensen, L.N. Topical zinc oxide assessed in two human wound-healing models. Acta Derm. Venereol. 2021, 101, adv00465. [Google Scholar] [CrossRef]
- Nyström, A.; Velati, D.; Mittapalli, V.R.; Fritsch, A.; Kern, J.S.; Bruckner-Tuderman, L. Collagen VII plays a dual role in wound healing. J. Clin. Investig. 2013, 123, 3498–3509. [Google Scholar] [CrossRef]
- Saarialho-Kere, U.; Kerkela, E.; Jahkola, T.; Suomela, S.; Keski-Oja, J.; Lohi, J. Epilysin (MMP-28) expression is associated with cell proliferation during epithelial repair. J. Investig. Dermatol. 2002, 119, 14–21. [Google Scholar] [CrossRef]
- Inoue, Y.; Hasegawa, S.; Ban, S.; Yamada, T.; Date, Y.; Mizutani, H.; Nakata, S.; Tanaka, M.; Hirashima, N. ZIP2 protein, a zinc transporter, is associated with keratinocyte differentiation. J. Biol. Chem. 2014, 289, 21451–21462. [Google Scholar] [CrossRef]
- Sandilands, A.; Sutherland, C.; Irvine, A.D.; McLean, W.H. Filaggrin in the frontline: Role in skin barrier function and disease. J. Cell Sci. 2009, 122 Pt 9, 1285–1294. [Google Scholar] [CrossRef]
- Makino, T.; Mizawa, M.; Yamakoshi, T.; Takaishi, M.; Shimizu, T. Expression of filaggrin-2 protein in the epidermis of human skin diseases: A comparative analysis with filaggrin. Biochem. Biophys. Res. Commun. 2014, 449, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.H.; Choi, D.; Chun, Y.J.; Noh, M. Keratinocyte-derived IL-24 plays a role in the positive feedback regulation of epidermal inflammation in response to environmental and endogenous toxic stressors. Toxicol. Appl. Pharmacol. 2014, 280, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Kirketerp-Møller, K.; Doerfler, P.; Schoeffmann, N.; Wolff-Winiski, B.; Niazi, O.; Pless, V.; Karlsmark, T.; Ågren, M.S. Biomarkers of skin graft healing in venous leg ulcers. Acta Derm. Venereol. 2022, 102, adv00749. [Google Scholar] [CrossRef] [PubMed]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).