Role of EZH2 in Uterine Gland Development
Abstract
1. Introduction
2. Results
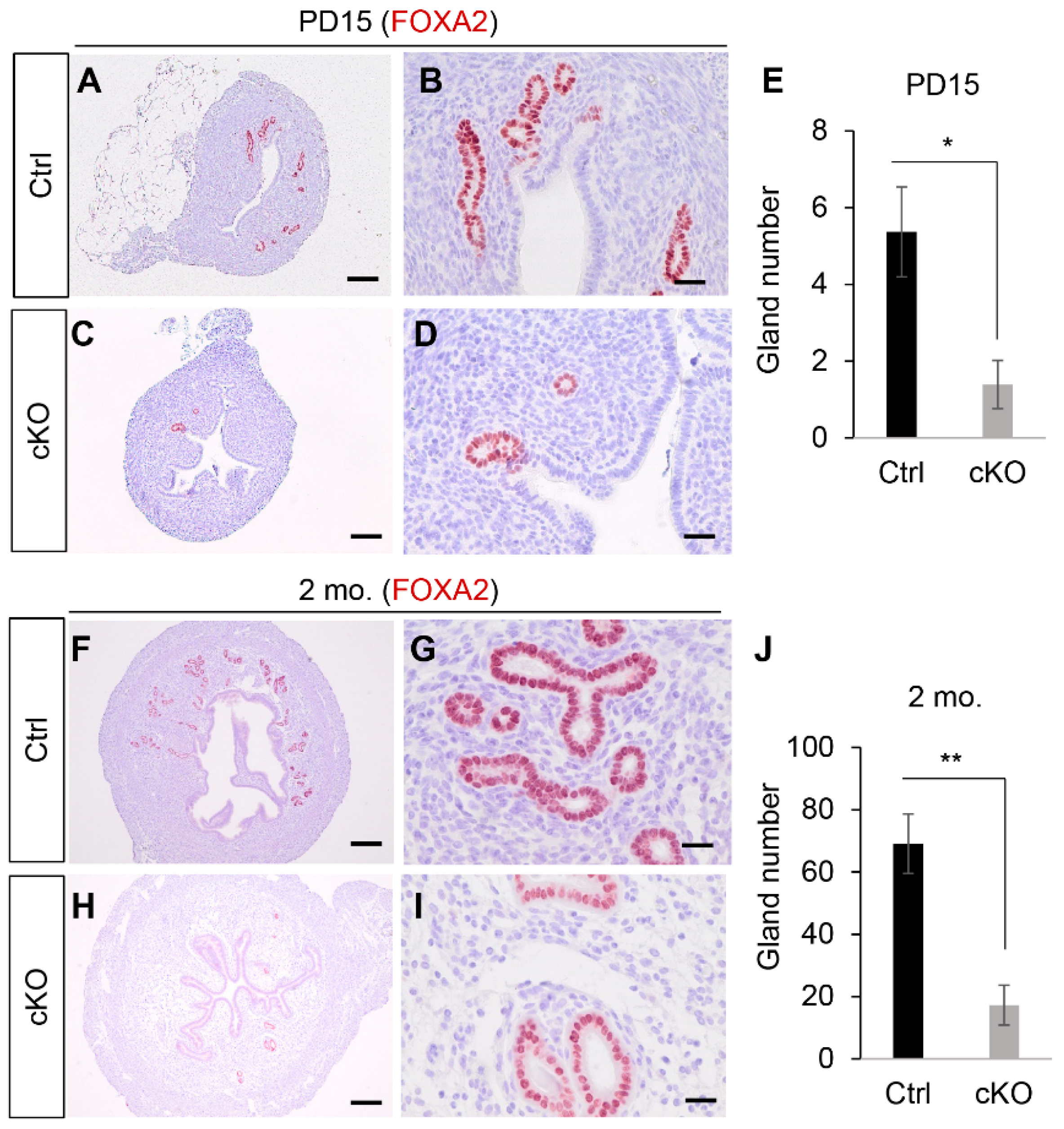
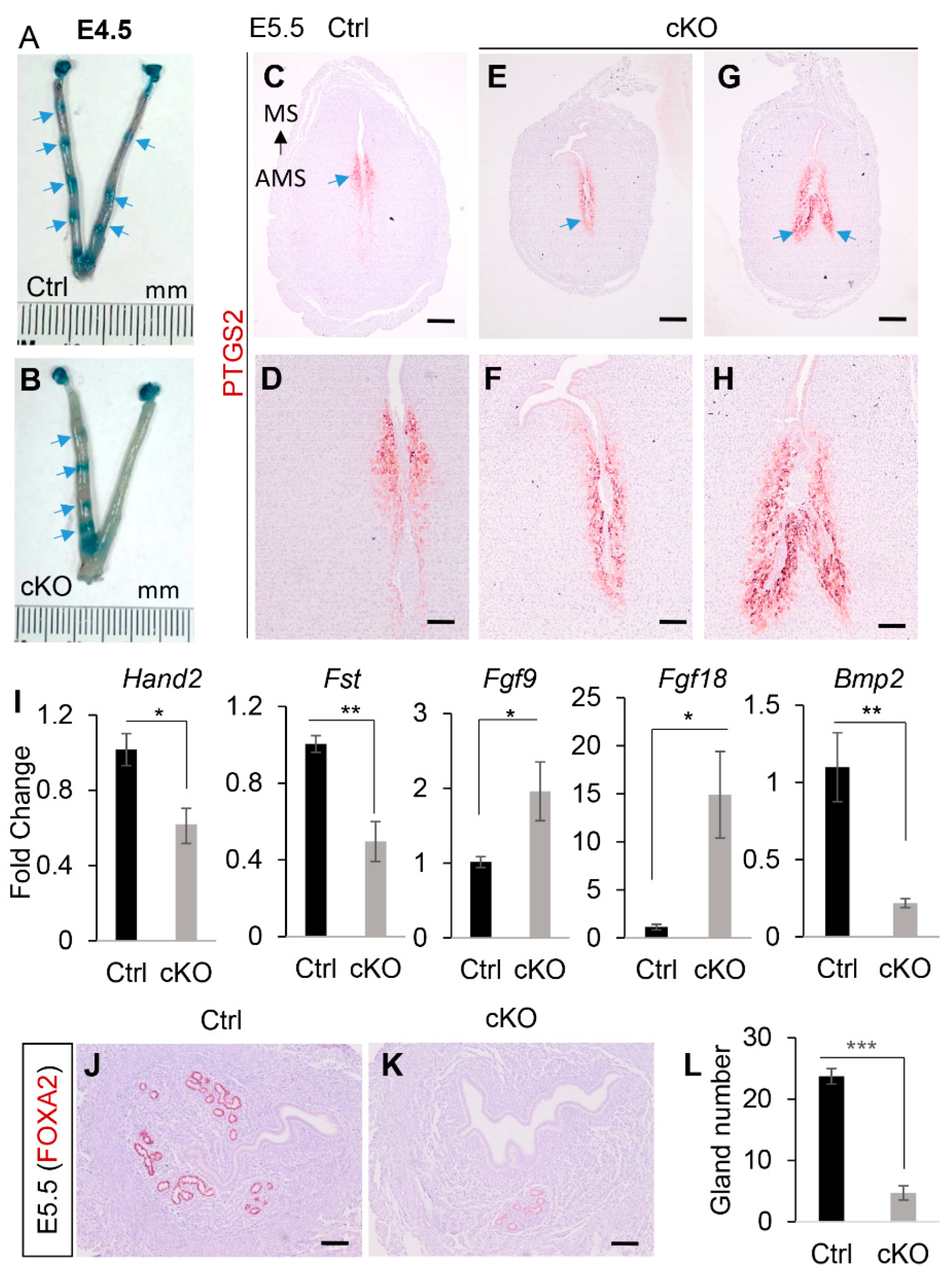
2.1. Conditional Deletion of Ezh2 Using Amhr2-Cre Recombinase Causes Defective Uterine Gland Formation
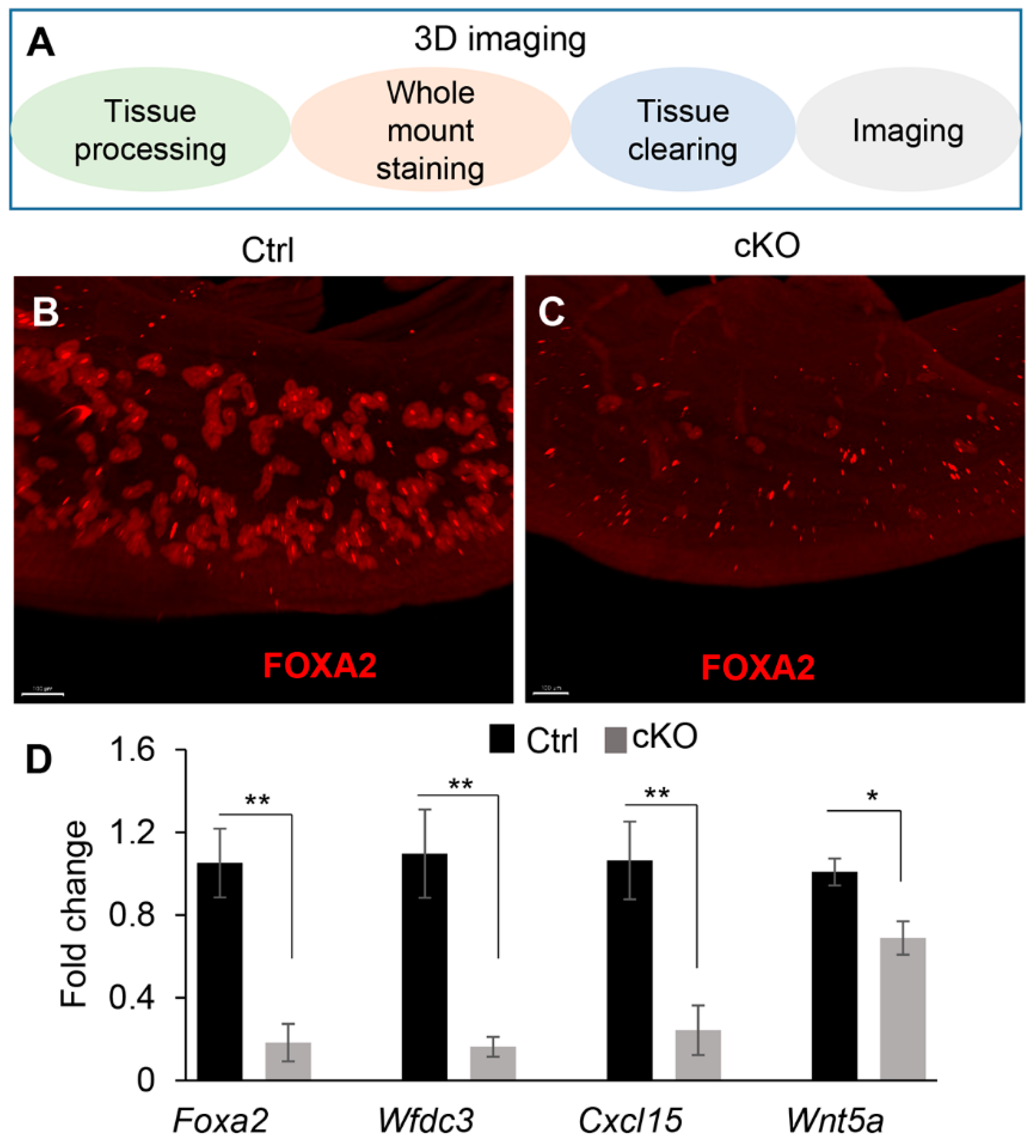
2.2. Conditional Deletion of Ezh2 Using Amhr2-Cre Does Not Cause Basal-like Cell Phenotype in the Uterus
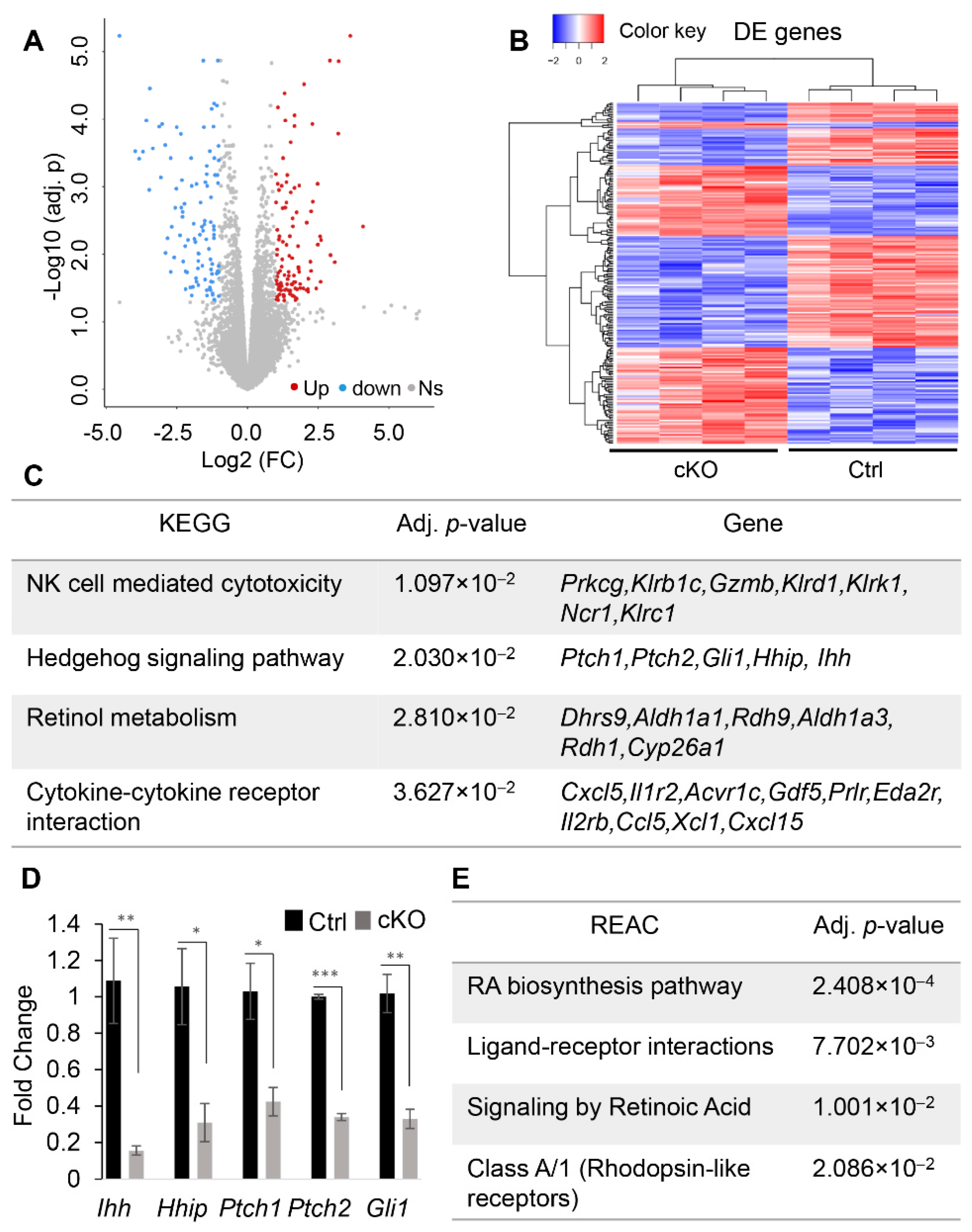
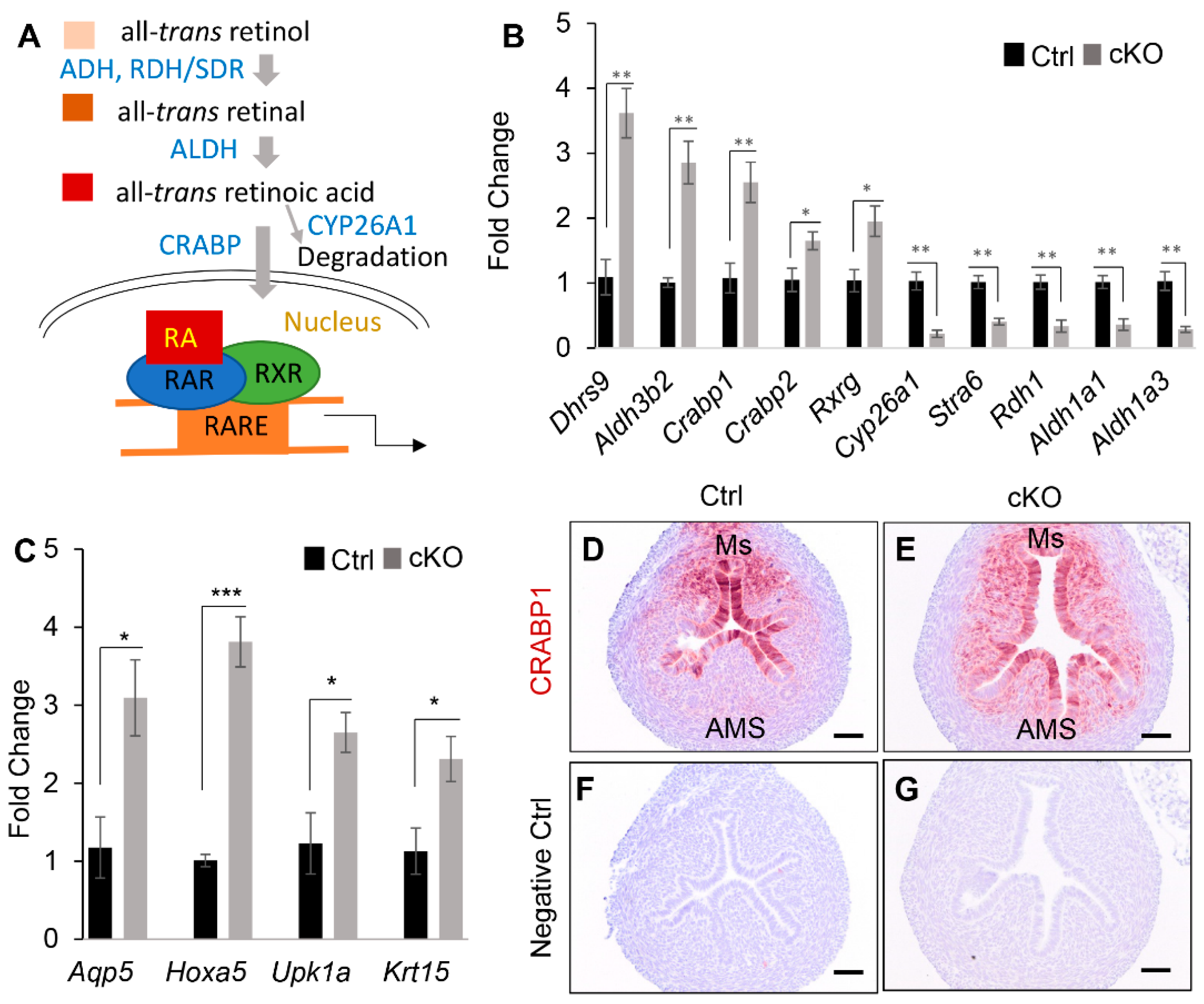
2.3. Transcriptomic Profiling to Identify Molecular Changes Associated with Uterine Gland Abnormalities in Ezh2 Conditionally Deleted Mice
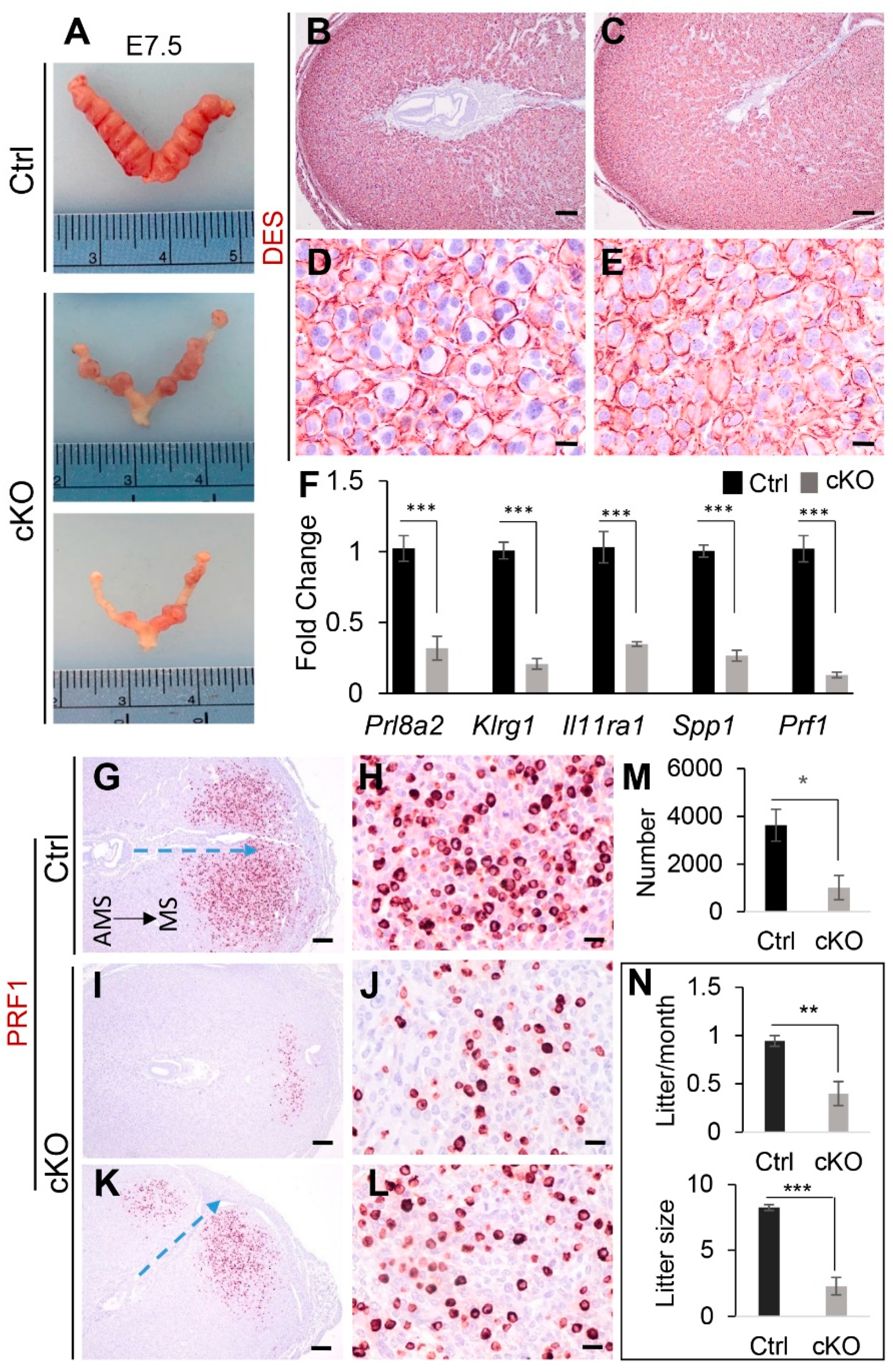
2.4. Conditional Deletion of Ezh2 Leads to Impaired Uterine Function
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Animal Models, Treatments, and Tissue Collections
4.3. Histological Analysis and Immunostaining
4.4. Quantitative Reverse Transcription (qRT)-PCR
4.5. Whole-Mount Immunofluorescent Staining and 3D Imaging of the Uterus
4.6. Hormone Assay
4.7. RNA-Sequencing and Bioinformatics Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schwartz, Y.B.; Pirrotta, V. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 2007, 8, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Morey, L.; Helin, K. Polycomb group protein-mediated repression of transcription. Trends Biochem. Sci. 2010, 35, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Sauvageau, M.; Sauvageau, G. Polycomb group proteins: Multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 2010, 7, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Chou, R.H.; Yu, Y.L.; Hung, M.C. The roles of EZH2 in cell lineage commitment. Am. J. Transl. Res. 2011, 3, 243–250. [Google Scholar] [PubMed]
- Kim, K.H.; Roberts, C.W. Targeting EZH2 in cancer. Nat. Med. 2016, 22, 128–134. [Google Scholar] [CrossRef]
- Eskander, R.N.; Ji, T.; Huynh, B.; Wardeh, R.; Randall, L.M.; Hoang, B. Inhibition of enhancer of zeste homolog 2 (EZH2) expression is associated with decreased tumor cell proliferation, migration, and invasion in endometrial cancer cell lines. Int. J. Gynecol. Cancer 2013, 23, 997–1005. [Google Scholar] [CrossRef]
- Zhang, Q.; Dong, P.; Liu, X.; Sakuragi, N.; Guo, S.W. Enhancer of Zeste homolog 2 (EZH2) induces epithelial-mesenchymal transition in endometriosis. Sci. Rep. 2017, 7, 6804. [Google Scholar] [CrossRef]
- Oki, S.; Sone, K.; Oda, K.; Hamamoto, R.; Ikemura, M.; Maeda, D.; Takeuchi, M.; Tanikawa, M.; Mori-Uchino, M.; Nagasaka, K.; et al. Oncogenic histone methyltransferase EZH2: A novel prognostic marker with therapeutic potential in endometrial cancer. Oncotarget 2017, 8, 40402–40411. [Google Scholar] [CrossRef]
- Gu, Y.; Zhang, J.; Guan, H. Expression of EZH2 in endometrial carcinoma and its effects on proliferation and invasion of endometrial carcinoma cells. Oncol. Lett. 2017, 14, 7191–7196. [Google Scholar] [CrossRef][Green Version]
- Roh, J.W.; Choi, J.E.; Han, H.D.; Hu, W.; Matsuo, K.; Nishimura, M.; Lee, J.S.; Kwon, S.Y.; Cho, C.H.; Kim, J.; et al. Clinical and biological significance of EZH2 expression in endometrial cancer. Cancer Biol. Ther. 2020, 21, 147–156. [Google Scholar] [CrossRef]
- Kikuchi, J.; Koyama, D.; Wada, T.; Izumi, T.; Hofgaard, P.O.; Bogen, B.; Furukawa, Y. Phosphorylation-mediated EZH2 inactivation promotes drug resistance in multiple myeloma. J. Clin. Investig. 2015, 125, 4375–4390. [Google Scholar] [CrossRef]
- Mallen-St Clair, J.; Soydaner-Azeloglu, R.; Lee, K.E.; Taylor, L.; Livanos, A.; Pylayeva-Gupta, Y.; Miller, G.; Margueron, R.; Reinberg, D.; Bar-Sagi, D. EZH2 couples pancreatic regeneration to neoplastic progression. Genes Dev. 2012, 26, 439–444. [Google Scholar] [CrossRef]
- Fang, X.; Ni, N.; Wang, X.; Tian, Y.; Ivanov, I.; Rijnkels, M.; Bayless, K.J.; Lydon, J.P.; Li, Q. EZH2 and endometrial cancer development: Insights from a mouse model. Cells 2022, 11, 909. [Google Scholar] [CrossRef]
- Wang, Y.; Hou, N.; Cheng, X.; Zhang, J.; Tan, X.; Zhang, C.; Tang, Y.; Teng, Y.; Yang, X. Ezh2 acts as a tumor suppressor in kras-driven lung adenocarcinoma. Int. J. Biol. Sci. 2017, 13, 652–659. [Google Scholar] [CrossRef]
- O’Carroll, D.; Erhardt, S.; Pagani, M.; Barton, S.C.; Surani, M.A.; Jenuwein, T. The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 2001, 21, 4330–4336. [Google Scholar] [CrossRef]
- Grimaldi, G.; Christian, M.; Steel, J.H.; Henriet, P.; Poutanen, M.; Brosens, J.J. Down-regulation of the histone methyltransferase EZH2 contributes to the epigenetic programming of decidualizing human endometrial stromal cells. Mol. Endocrinol. 2011, 25, 1892–1903. [Google Scholar] [CrossRef]
- Osokine, I.; Siewiera, J.; Rideaux, D.; Ma, S.; Tsukui, T.; Erlebacher, A. Gene silencing by EZH2 suppresses TGF-beta activity within the decidua to avert pregnancy-adverse wound healing at the maternal-fetal interface. Cell Rep. 2022, 38, 110329. [Google Scholar] [CrossRef]
- Soyal, S.M.; Mukherjee, A.; Lee, K.Y.; Li, J.; Li, H.G.; DeMayo, F.J.; Lydon, J.P. Cre-mediated recombination in cell lineages that express the progesterone receptor. Genesis 2005, 41, 58–66. [Google Scholar] [CrossRef]
- Fang, X.; Ni, N.; Lydon, J.P.; Ivanov, I.; Bayless, K.J.; Rijnkels, M.; Li, Q. Enhancer of Zeste 2 polycomb repressive complex 2 subunit is required for uterine epithelial integrity. Am. J. Pathol. 2019, 189, 1212–1225. [Google Scholar] [CrossRef]
- Nanjappa, M.K.; Mesa, A.M.; Medrano, T.I.; Jefferson, W.N.; DeMayo, F.J.; Williams, C.J.; Lydon, J.P.; Levin, E.R.; Cooke, P.S. The histone methyltransferase EZH2 is required for normal uterine development and function in mice. Biol. Reprod. 2019, 101, 306–317. [Google Scholar] [CrossRef]
- Mesa, A.M.; Mao, J.D.; Nanjappa, M.K.; Medrano, T.I.; Tevosian, S.; Yu, F.H.; Kinkade, J.; Lyu, Z.; Liu, Y.; Joshi, T.; et al. Mice lacking uterine enhancer of zeste homolog 2 have transcriptomic changes associated with uterine epithelial proliferation. Physiol. Genom. 2020, 52, 81–95. [Google Scholar] [CrossRef]
- Mesa, A.M.; Mao, J.; Medrano, T.I.; Bivens, N.J.; Jurkevich, A.; Tuteja, G.; Cooke, P.S.; Rosenfeld, C.S. Spatial transcriptomics analysis of uterine gene expression in enhancer of zeste homolog 2 conditional knockout micedagger. Biol. Reprod. 2021, 105, 1126–1139. [Google Scholar] [CrossRef]
- Spencer, T.E. Biological roles of uterine glands in pregnancy. Semin. Reprod. Med. 2014, 32, 346–357. [Google Scholar] [CrossRef]
- Burton, G.J.; Scioscia, M.; Rademacher, T.W. Endometrial secretions: Creating a stimulatory microenvironment within the human early placenta and implications for the aetiopathogenesis of preeclampsia. J. Reprod. Immunol. 2011, 89, 118–125. [Google Scholar] [CrossRef]
- Stewart, C.A.; Fisher, S.J.; Wang, Y.; Stewart, M.D.; Hewitt, S.C.; Rodriguez, K.F.; Korach, K.S.; Behringer, R.R. Uterine gland formation in mice is a continuous process, requiring the ovary after puberty, but not after parturition. Biol. Reprod. 2011, 85, 954–964. [Google Scholar] [CrossRef][Green Version]
- Gray, C.A.; Bartol, F.F.; Tarleton, B.J.; Wiley, A.A.; Johnson, G.A.; Bazer, F.W.; Spencer, T.E. Developmental biology of uterine glands. Biol. Reprod. 2001, 65, 1311–1323. [Google Scholar] [CrossRef]
- Dunlap, K.A.; Filant, J.; Hayashi, K.; Rucker, E.B., 3rd; Song, G.; Deng, J.M.; Behringer, R.R.; DeMayo, F.J.; Lydon, J.; Jeong, J.W.; et al. Postnatal deletion of Wnt7a inhibits uterine gland morphogenesis and compromises adult fertility in mice. Biol. Reprod. 2011, 85, 386–396. [Google Scholar] [CrossRef]
- Mericskay, M.; Kitajewski, J.; Sassoon, D. Wnt5a is required for proper epithelial-mesenchymal interactions in the uterus. Development 2004, 131, 2061–2072. [Google Scholar] [CrossRef]
- Stewart, C.A.; Wang, Y.; Bonilla-Claudio, M.; Martin, J.F.; Gonzalez, G.; Taketo, M.M.; Behringer, R.R. CTNNB1 in mesenchyme regulates epithelial cell differentiation during Mullerian duct and postnatal uterine development. Mol. Endocrinol. 2013, 27, 1442–1454. [Google Scholar] [CrossRef]
- Deutscher, E.; Yao, H.H.-C. Essential roles of mesenchyme-derived beta-catenin in mouse Mullerian duct morphogenesis. Dev. Biol. 2007, 307, 227–236. [Google Scholar] [CrossRef][Green Version]
- Jamin, S.P.; Arango, N.A.; Mishina, Y.; Hanks, M.C.; Behringer, R.R. Requirement of Bmpr1a for Mullerian duct regression during male sexual development. Nat. Genet. 2002, 32, 408–410. [Google Scholar] [CrossRef]
- Arango, N.A.; Kobayashi, A.; Wang, Y.; Jamin, S.P.; Lee, H.H.; Orvis, G.D.; Behringer, R.R. A mesenchymal perspective of Mullerian duct differentiation and regression in Amhr2-lacZ mice. Mol. Reprod. Dev. 2008, 75, 1154–1162. [Google Scholar] [CrossRef]
- Pangas, S.A.; Li, X.; Umans, L.; Zwijsen, A.; Huylebroeck, D.; Gutierrez, C.; Wang, D.; Martin, J.F.; Jamin, S.P.; Behringer, R.R.; et al. Conditional deletion of Smad1 and Smad5 in somatic cells of male and female gonads leads to metastatic tumor development in mice. Mol. Cell. Biol. 2008, 28, 248–257. [Google Scholar] [CrossRef]
- Varjosalo, M.; Taipale, J. Hedgehog signaling. J. Cell Sci. 2007, 120, 3–6. [Google Scholar] [CrossRef]
- Metzler, M.A.; Raja, S.; Elliott, K.H.; Friedl, R.M.; Tran, N.Q.H.; Brugmann, S.A.; Larsen, M.; Sandell, L.L. RDH10-mediated retinol metabolism and RARalpha-mediated retinoic acid signaling are required for submandibular salivary gland initiation. Development 2018, 145, dev164822. [Google Scholar] [CrossRef]
- Wang, Y.A.; Shen, K.; Wang, Y.; Brooks, S.C. Retinoic acid signaling is required for proper morphogenesis of mammary gland. Dev. Dyn. 2005, 234, 892–899. [Google Scholar] [CrossRef]
- Duester, G. Retinoic acid synthesis and signaling during early organogenesis. Cell 2008, 134, 921–931. [Google Scholar] [CrossRef]
- Napoli, J.L. Physiological insights into all-trans-retinoic acid biosynthesis. Biochim. Biophys. Acta 2012, 1821, 152–167. [Google Scholar] [CrossRef]
- Nomura, J.; Horie, I.; Seto, M.; Nagai, K.; Hisatsune, A.; Miyata, T.; Isohama, Y. All-trans retinoic acid increases expression of aquaporin-5 and plasma membrane water permeability via transactivation of Sp1 in mouse lung epithelial cells. Biochem. Biophys. Res. Commun. 2006, 351, 1048–1053. [Google Scholar] [CrossRef]
- Packer, A.I.; Mailutha, K.G.; Ambrozewicz, L.A.; Wolgemuth, D.J. Regulation of the Hoxa4 and Hoxa5 genes in the embryonic mouse lung by retinoic acid and TGFbeta1: Implications for lung development and patterning. Dev. Dyn. 2000, 217, 62–74. [Google Scholar] [CrossRef]
- Zupancic, D.; Korac-Prlic, J.; Kreft, M.E.; Frankovic, L.; Vilovic, K.; Jeruc, J.; Romih, R.; Terzic, J. Vitamin A rich diet diminishes early urothelial carcinogenesis by altering retinoic acid signaling. Cancers 2020, 12, 1712. [Google Scholar] [CrossRef]
- Kim, D.; Chen, R.; Sheu, M.; Kim, N.; Kim, S.; Islam, N.; Wier, E.M.; Wang, G.; Li, A.; Park, A.; et al. Noncoding dsRNA induces retinoic acid synthesis to stimulate hair follicle regeneration via TLR3. Nat. Commun. 2019, 10, 2811. [Google Scholar] [CrossRef]
- Goad, J.; Ko, Y.A.; Kumar, M.; Syed, S.M.; Tanwar, P.S. Differential Wnt signaling activity limits epithelial gland development to the anti-mesometrial side of the mouse uterus. Dev. Biol. 2017, 423, 138–151. [Google Scholar] [CrossRef]
- Amengual, J.; Zhang, N.; Kemerer, M.; Maeda, T.; Palczewski, K.; Von Lintig, J. STRA6 is critical for cellular vitamin A uptake and homeostasis. Hum. Mol. Genet. 2014, 23, 5402–5417. [Google Scholar] [CrossRef]
- Kelleher, A.M.; Milano-Foster, J.; Behura, S.K.; Spencer, T.E. Uterine glands coordinate on-time embryo implantation and impact endometrial decidualization for pregnancy success. Nat. Commun. 2018, 9, 2435. [Google Scholar] [CrossRef]
- Chakraborty, I.; Das, S.K.; Wang, J.; Dey, S.K. Developmental expression of the cyclo-oxygenase-1 and cyclo-oxygenase-2 genes in the peri-implantation mouse uterus and their differential regulation by the blastocyst and ovarian steroids. J. Mol. Endocrinol. 1996, 16, 107–122. [Google Scholar] [CrossRef]
- Li, Q.; Kannan, A.; DeMayo, F.J.; Lydon, J.P.; Cooke, P.S.; Yamagishi, H.; Srivastava, D.; Bagchi, M.K.; Bagchi, I.C. The antiproliferative action of progesterone in uterine epithelium is mediated by Hand2. Science 2011, 331, 912–916. [Google Scholar] [CrossRef]
- Fullerton, P.T., Jr.; Monsivais, D.; Kommagani, R.; Matzuk, M.M. Follistatin is critical for mouse uterine receptivity and decidualization. Proc. Natl. Acad. Sci. USA 2017, 114, E4772–E4781. [Google Scholar] [CrossRef]
- Monsivais, D.; Clementi, C.; Peng, J.; Titus, M.M.; Barrish, J.P.; Creighton, C.J.; Lydon, J.P.; DeMayo, F.J.; Matzuk, M.M. Uterine ALK3 is essential during the window of implantation. Proc. Natl. Acad. Sci. USA 2016, 113, E387–E395. [Google Scholar] [CrossRef]
- Oliveira, S.F.; Greca, C.P.; Abrahamsohn, P.A.; Reis, M.G.; Zorn, T.M. Organization of desmin-containing intermediate filaments during differentiation of mouse decidual cells. Histochem. Cell Biol. 2000, 113, 319–327. [Google Scholar] [CrossRef]
- Croy, B.A.; Chantakru, S.; Esadeg, S.; Ashkar, A.A.; Wei, Q. Decidual natural killer cells: Key regulators of placental development (a review). J. Reprod. Immunol. 2002, 57, 151–168. [Google Scholar] [CrossRef]
- Kelleher, A.M.; Behura, S.K.; Burns, G.W.; Young, S.L.; DeMayo, F.J.; Spencer, T.E. Integrative analysis of the forkhead box A2 (FOXA2) cistrome for the human endometrium. FASEB J. 2019, 33, 8543–8554. [Google Scholar] [CrossRef] [PubMed]
- Spencer, T.E.; Dunlap, K.A.; Filant, J. Comparative developmental biology of the uterus: Insights into mechanisms and developmental disruption. Mol. Cell. Endocrinol. 2012, 354, 34–53. [Google Scholar] [CrossRef] [PubMed]
- Gaynor, L.M.; Colucci, F. Uterine Natural Killer Cells: Functional Distinctions and Influence on Pregnancy in Humans and Mice. Front. Immunol. 2017, 8, 467. [Google Scholar] [CrossRef]
- Ashkar, A.A.; Croy, B.A. Functions of uterine natural killer cells are mediated by interferon gamma production during murine pregnancy. Semin. Immunol. 2001, 13, 235–241. [Google Scholar] [CrossRef]
- Nakajima, T.; Iguchi, T.; Sato, T. Hedgehog signaling plays roles in epithelial cell proliferation in neonatal mouse uterus and vagina. Cell Tissue Res. 2012, 348, 239–247. [Google Scholar] [CrossRef]
- Cooke, P.S.; Spencer, T.E.; Bartol, F.F.; Hayashi, K. Uterine glands: Development, function and experimental model systems. Mol. Hum. Reprod. 2013, 19, 547–558. [Google Scholar] [CrossRef]
- Filant, J.; Zhou, H.J.; Spencer, T.E. Progesterone inhibits uterine gland development in the neonatal mouse uterus. Biol. Reprod. 2012, 86, 146. [Google Scholar] [CrossRef]
- Migone, F.F.; Ren, Y.; Cowan, R.G.; Harman, R.M.; Nikitin, A.Y.; Quirk, S.M. Dominant activation of the hedgehog signaling pathway alters development of the female reproductive tract. Genesis 2012, 50, 28–40. [Google Scholar] [CrossRef]
- Das, B.C.; Thapa, P.; Karki, R.; Das, S.; Mahapatra, S.; Liu, T.C.; Torregroza, I.; Wallace, D.P.; Kambhampati, S.; Van Veldhuizen, P.; et al. Retinoic acid signaling pathways in development and diseases. Bioorg. Med. Chem. 2014, 22, 673–683. [Google Scholar] [CrossRef]
- Nakajima, T.; Iguchi, T.; Sato, T. Retinoic acid signaling determines the fate of uterine stroma in the mouse Mullerian duct. Proc. Natl. Acad. Sci. USA 2016, 113, 14354–14359. [Google Scholar] [CrossRef]
- Yin, Y.; Haller, M.E.; Chadchan, S.B.; Kommagani, R.; Ma, L. Signaling through retinoic acid receptors is essential for mammalian uterine receptivity and decidualization. JCI Insight 2021, 6, e150254. [Google Scholar] [CrossRef]
- Nakajima, T.; Sato, T.; Iguchi, T.; Takasugi, N. Retinoic acid signaling determines the fate of the uterus from the mouse Müllerian duct. Reprod. Toxicol. 2019, 86, 56–61. [Google Scholar] [CrossRef]
- Jetten, A.M.; De Luca, L.M.; Nelson, K.; Schroeder, W.; Burlingame, S.; Fujimoto, W. Regulation of cornifin alpha expression in the vaginal and uterine epithelium by estrogen and retinoic acid. Mol. Cell. Endocrinol. 1996, 123, 7–15. [Google Scholar] [CrossRef]
- Wu, B.; An, C.; Li, Y.; Yin, Z.; Gong, L.; Li, Z.; Liu, Y.; Heng, B.C.; Zhang, D.; Ouyang, H.; et al. Reconstructing lineage hierarchies of mouse uterus epithelial development using single-cell analysis. Stem Cell Rep. 2017, 9, 381–396. [Google Scholar] [CrossRef]
- Molotkov, A.; Duester, G. Genetic evidence that retinaldehyde dehydrogenase Raldh1 (Aldh1a1) functions downstream of alcohol dehydrogenase Adh1 in metabolism of retinol to retinoic acid. J. Biol. Chem. 2003, 278, 36085–36090. [Google Scholar] [CrossRef]
- Cunha, G.R. Stromal induction and specification of morphogenesis and cytodifferentiation of the epithelia of the Mullerian ducts and urogenital sinus during development of the uterus and vagina in mice. J. Exp. Zool. 1976, 196, 361–370. [Google Scholar] [CrossRef]
- Rossant, J.; Zirngibl, R.; Cado, D.; Shago, M.; Giguère, V. Expression of a retinoic acid response element-hsplacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes Dev. 1991, 5, 1333–1344. [Google Scholar] [CrossRef]
- Kelleher, A.M.; DeMayo, F.J.; Spencer, T.E. Uterine Glands: Developmental Biology and Functional Roles in Pregnancy. Endocr. Rev. 2019, 40, 1424–1445. [Google Scholar] [CrossRef]
- Li, Q.; Pangas, S.A.; Jorgez, C.J.; Graff, J.M.; Weinstein, M.; Matzuk, M.M. Redundant roles of SMAD2 and SMAD3 in ovarian granulosa cells in vivo. Mol. Cell. Biol. 2008, 28, 7001–7011. [Google Scholar] [CrossRef]
- Gao, Y.; Vincent, D.F.; Davis, A.J.; Sansom, O.J.; Bartholin, L.; Li, Q. Constitutively active transforming growth factor β receptor 1 in the mouse ovary promotes tumorigenesis. Oncotarget 2016, 7, 40904–40918. [Google Scholar] [CrossRef] [PubMed]
- Bankhead, P.; Loughrey, M.B.; Fernandez, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Duran, S.; Lydon, J.P.; DeMayo, F.J.; Burghardt, R.C.; Bayless, K.J.; Bartholin, L.; Li, Q. Constitutive activation of transforming growth factor Beta receptor 1 in the mouse uterus impairs uterine morphology and function. Biol. Reprod. 2015, 92, 34. [Google Scholar] [CrossRef] [PubMed]
- Vo, N.K.; Dalton, R.P.; Liu, N.; Olson, E.N.; Goodman, R.H. Affinity purification of microRNA-133a with the cardiac transcription factor, Hand2. Proc. Natl. Acad. Sci. USA 2010, 107, 19231–19236. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Li, S.; Li, Q. Uterine epithelial cell proliferation and endometrial hyperplasia: Evidence from a mouse model. Mol. Hum. Reprod. 2014, 20, 776–786. [Google Scholar] [CrossRef]
- Naganuma, T.; Takagi, S.; Kanetake, T.; Kitamura, T.; Hattori, S.; Miyakawa, T.; Sassa, T.; Kihara, A. Disruption of the sjogren-larsson syndrome gene aldh3a2 in mice increases keratinocyte growth and retards skin barrier recovery. J. Biol. Chem. 2016, 291, 11676–11688. [Google Scholar] [CrossRef]
- Monsivais, D.; Clementi, C.; Peng, J.; Fullerton, P.T., Jr.; Prunskaite-Hyyrylainen, R.; Vainio, S.J.; Matzuk, M.M. BMP7 induces uterine receptivity and blastocyst attachment. Endocrinology 2017, 158, 979–992. [Google Scholar] [CrossRef]
- Wang, X.; Spandidos, A.; Wang, H.; Seed, B. PrimerBank: A PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 2012, 40, D1144–D1149. [Google Scholar] [CrossRef]
- Spandidos, A.; Wang, X.W.; Wang, H.J.; Seed, B. PrimerBank: A resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010, 38, D792–D799. [Google Scholar] [CrossRef]
- Renier, N.; Adams, E.L.; Kirst, C.; Wu, Z.; Azevedo, R.; Kohl, J.; Autry, A.E.; Kadiri, L.; Umadevi Venkataraju, K.; Zhou, Y.; et al. Mapping of Brain Activity by Automated Volume Analysis of Immediate Early Genes. Cell 2016, 165, 1789–1802. [Google Scholar] [CrossRef]
- Jalufka, F.L.; Min, S.W.; Platt, M.E.; Pritchard, A.L.; Margo, T.E.; Vernino, A.O.; Kirchhoff, M.A.; Massopust, R.T.; McCreedy, D.A. Hydrophobic and Hydrogel-Based Methods for Passive Tissue Clearing. Methods Mol. Biol. 2022, 2440, 197–209. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]

| Name | Catalog No. | Manufacturer | Host | Immunostaining |
|---|---|---|---|---|
| EZH2 | 5246 | Cell signaling, Danvers, MA | Rabbit | 1:200 |
| FOXA2 | Ab108422 | Abcam, Waltham, MA USA | Rabbit | 1:250 (IHC) 1:400 (WMS) |
| FOXA2 | 8186 | Cell signaling | Rabbit | 1:200 |
| ACTA2 | 19245 | Cell signaling | Rabbit | 1:500 |
| KRT14 | PA5-16722 | Thermo Fisher | Rabbit | 1:400 |
| TRP63 | 13109 | Cell signaling | Rabbit | 1:200 |
| CRABP1 | 13163 | Cell signaling | Rabbit | 1:400 |
| PRF1 | 31647 | Cell signaling | Rabbit | 1:200 |
| VIM | 5741 | Cell signaling | Rabbit | 1:200 |
| KRT8 | TROMA-I | DSHB, Iowa City, USA | Rat | 1:200 |
| PTGS2 | 12282 | Cell signaling | Rabbit | 1:600 |
| Desmin | Ab32362 | Abcam | Rabbit | 1:500 |
| Gene | F/R | Sequence (5′-3′) | Reference |
|---|---|---|---|
| Hoxa5 | F | CTCATTTTGCGGTCGCTATCC | PrimerBank ID: 6754232a1 |
| R | ATCCATGCCATTGTAGCCGTA | ||
| Upk1a | F | GGGCAACATCATTATTTTGCTGT | PrimerBank ID: 12835021a1 |
| R | CGTGAGGATCATGTACCGACG | ||
| Krt15 | F | AGCTATTGCAGAGAAAAACCGT | PrimerBank ID: 6680602a1 |
| R | GGTCCGTCTCAGGTCTGTG | ||
| Aqp5 | F | AGAAGGAGGTGTGTTCAGTTGC | PrimerBank ID: 6857757a1 |
| R | GCCAGAGTAATGGCCGGAT | ||
| Ihh | F | CTCTTGCCTACAAGCAGTTCA | PrimerBank ID: 14149643a1 |
| R | CCGTGTTCTCCTCGTCCTT | ||
| Hhip | F | TGAAGATGCTCTCGTTTAAGCTG | PrimerBank ID: 34328503a1 |
| R | CCACCACACAGGATCTCTCC | ||
| Ptch1 | F | AAAGAACTGCGGCAAGTTTTTG | PrimerBank ID: 6679519a1 |
| R | CTTCTCCTATCTTCTGACGGGT | ||
| Ptch2 | F | CTCCGCACCTCATATCCTAGC | PrimerBank ID: 6679517a1 |
| R | TCCCAGGAAGAGCACTTTGC | ||
| Spp1 | F | AGCAAGAAACTCTTCCAAGCAA | PrimerBank ID: 6678113a1 |
| R | GTGAGATTCGTCAGATTCATCCG | ||
| Rdh1 | F | GTCATGGGCCGAATGTCTTTC | PrimerBank ID: 20147789a1 |
| R | CACAAGTCTTGAAGCCTCCAG | ||
| Dhrs9 | F | ATGCTGTTTTGGTTGTTGGCT | PrimerBank ID: 30425272a1 |
| R | GTTCTGGCTGCTAAGTTTCCA | ||
| Crabp1 | F | CAGCAGCGAGAATTTCGACGA | PrimerBank ID: 7304975a1 |
| R | CGCACAGTAGTGGATGTCTTGA | ||
| Crabp2 | F | ATGCCTAACTTTTCTGGCAACT | PrimerBank ID: 33469075a1 |
| R | GCACAGTGGTGGAGGTTTTGA | ||
| Rxrg | F | CATGAGCCCTTCAGTAGCCTT | PrimerBank ID: 6677829a1 |
| R | CGGAGAGCCAAGAGCATTGAG | ||
| Stra6 | F | CTGGTACATCGAGGAACCTCT | PrimerBank ID: 6678171a1 |
| R | CCAGGAACGACAGTGAAGCC | ||
| Cyp26a1 | F | AAGCTCTGGGACCTGTACTGT | PrimerBank ID: 6681101a1 |
| R | CTCCGCTGAAGCACCATCT | ||
| Aldh1a1 | F | ATACTTGTCGGATTTAGGAGGCT | PrimerBank ID: 7304881a1 |
| R | GGGCCTATCTTCCAAATGAACA | ||
| Aldh1a3 | F | GGGTCACACTGGAGCTAGGA | PrimerBank ID: 31542123a1 |
| R | CTGGCCTCTTCTTGGCGAA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ni, N.; Jalufka, F.L.; Fang, X.; McCreedy, D.A.; Li, Q. Role of EZH2 in Uterine Gland Development. Int. J. Mol. Sci. 2022, 23, 15665. https://doi.org/10.3390/ijms232415665
Ni N, Jalufka FL, Fang X, McCreedy DA, Li Q. Role of EZH2 in Uterine Gland Development. International Journal of Molecular Sciences. 2022; 23(24):15665. https://doi.org/10.3390/ijms232415665
Chicago/Turabian StyleNi, Nan, Frank L. Jalufka, Xin Fang, Dylan A. McCreedy, and Qinglei Li. 2022. "Role of EZH2 in Uterine Gland Development" International Journal of Molecular Sciences 23, no. 24: 15665. https://doi.org/10.3390/ijms232415665
APA StyleNi, N., Jalufka, F. L., Fang, X., McCreedy, D. A., & Li, Q. (2022). Role of EZH2 in Uterine Gland Development. International Journal of Molecular Sciences, 23(24), 15665. https://doi.org/10.3390/ijms232415665




