CcMYB12 Positively Regulates Flavonoid Accumulation during Fruit Development in Carya cathayensis and Has a Role in Abiotic Stress Responses
Abstract
1. Introduction
2. Results
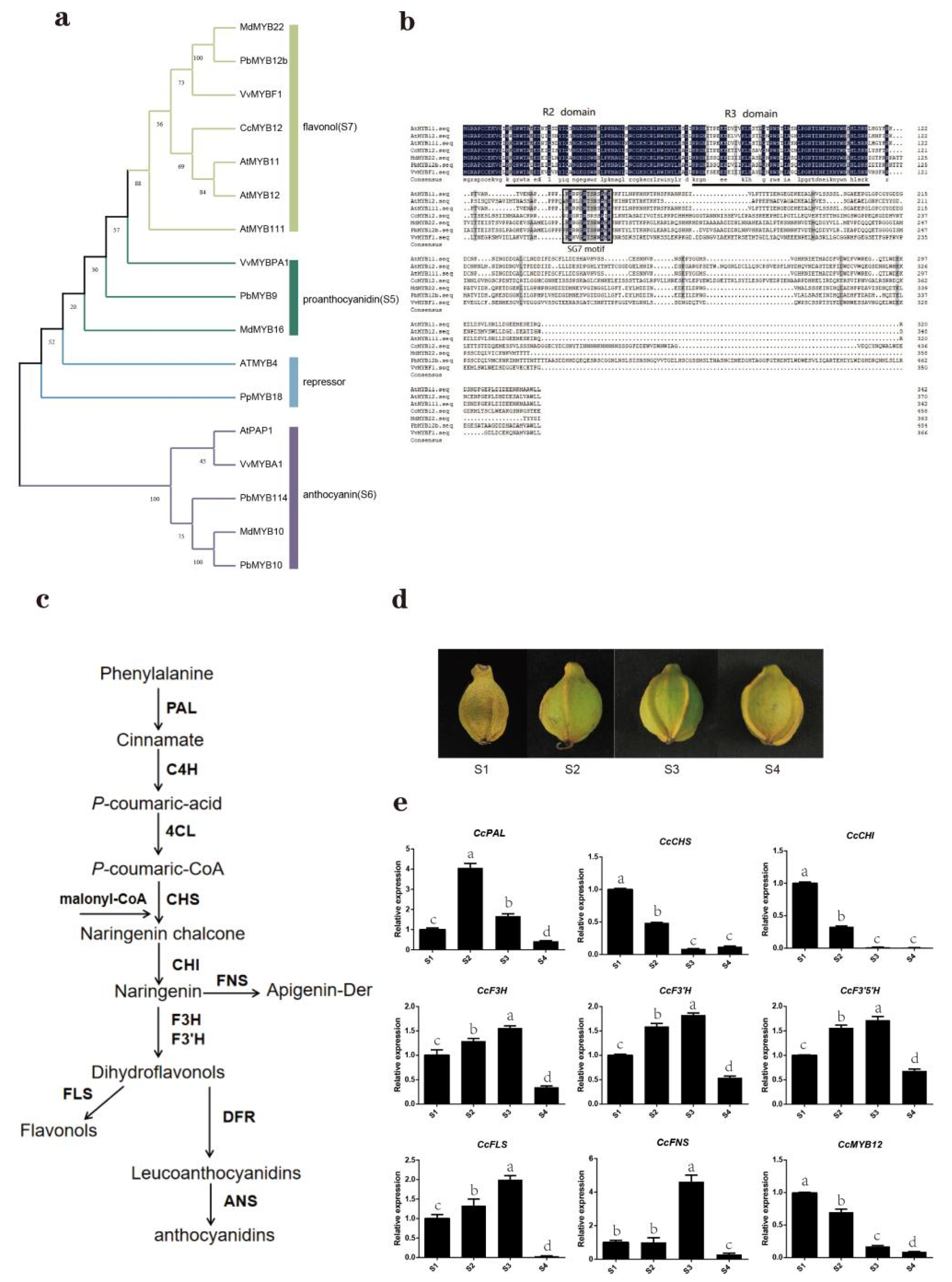
2.1. Identification, Phylogenetic Analysis, and Classification of R2R3-MYB Gene Family in Carya Cathayensis
2.2. Gene Structure and Protein Motif Assay of R2R3-MYB Family Members in Carya Cathayensis
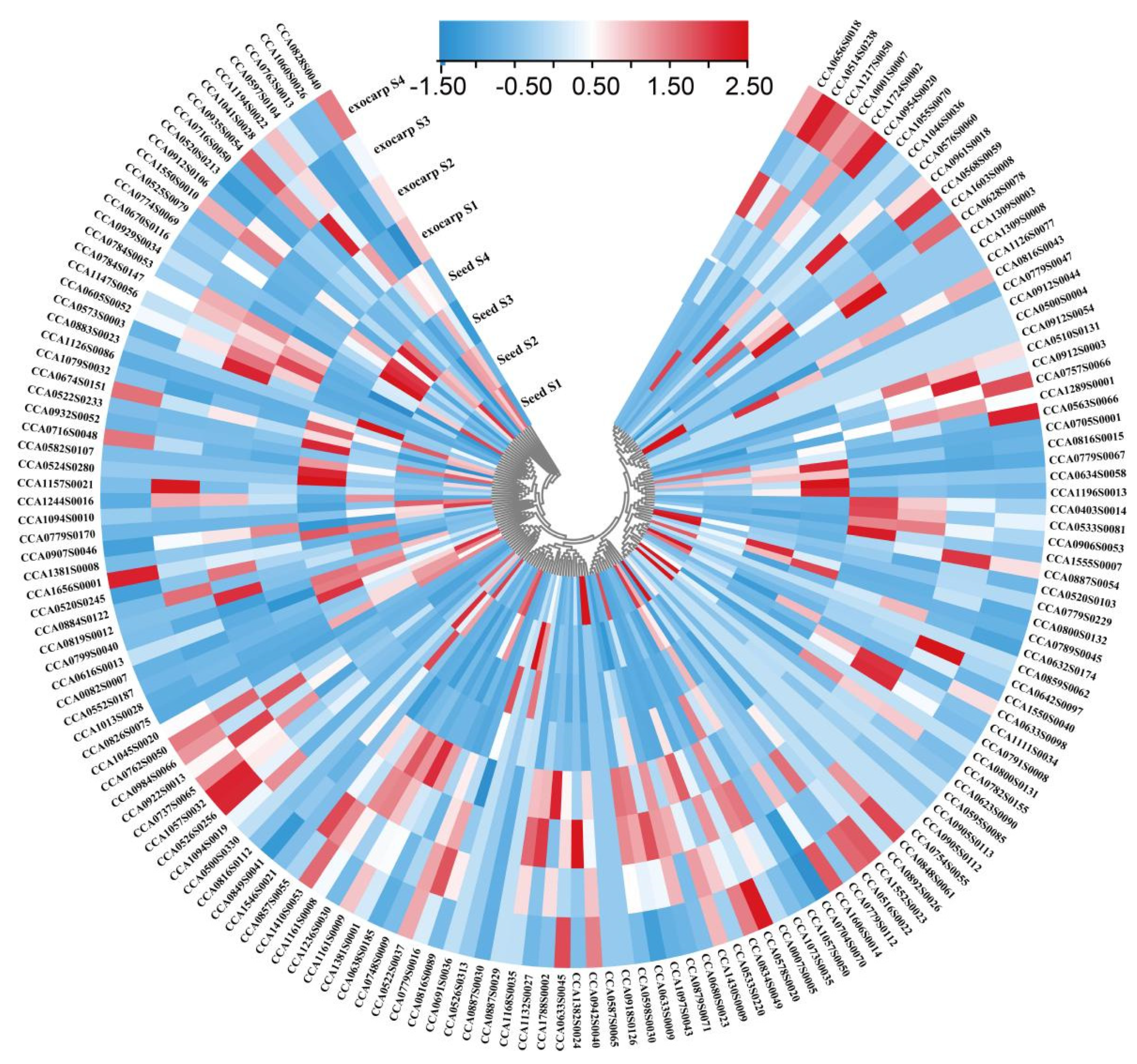
2.3. Expression Profiling of R2R3-MYB Family Members during Fruit Development in Carya Cathayensis
2.4. Identification and Characterization of CcMYB12 and Its Potential Relationship with Flavonol Synthesis
2.5. CcMYB12 Is Localized in the Nucleus and Has Self-Activation Ability
2.6. CcMYB12 Activates the Promoter of Flavonoid Biosynthesis Genes
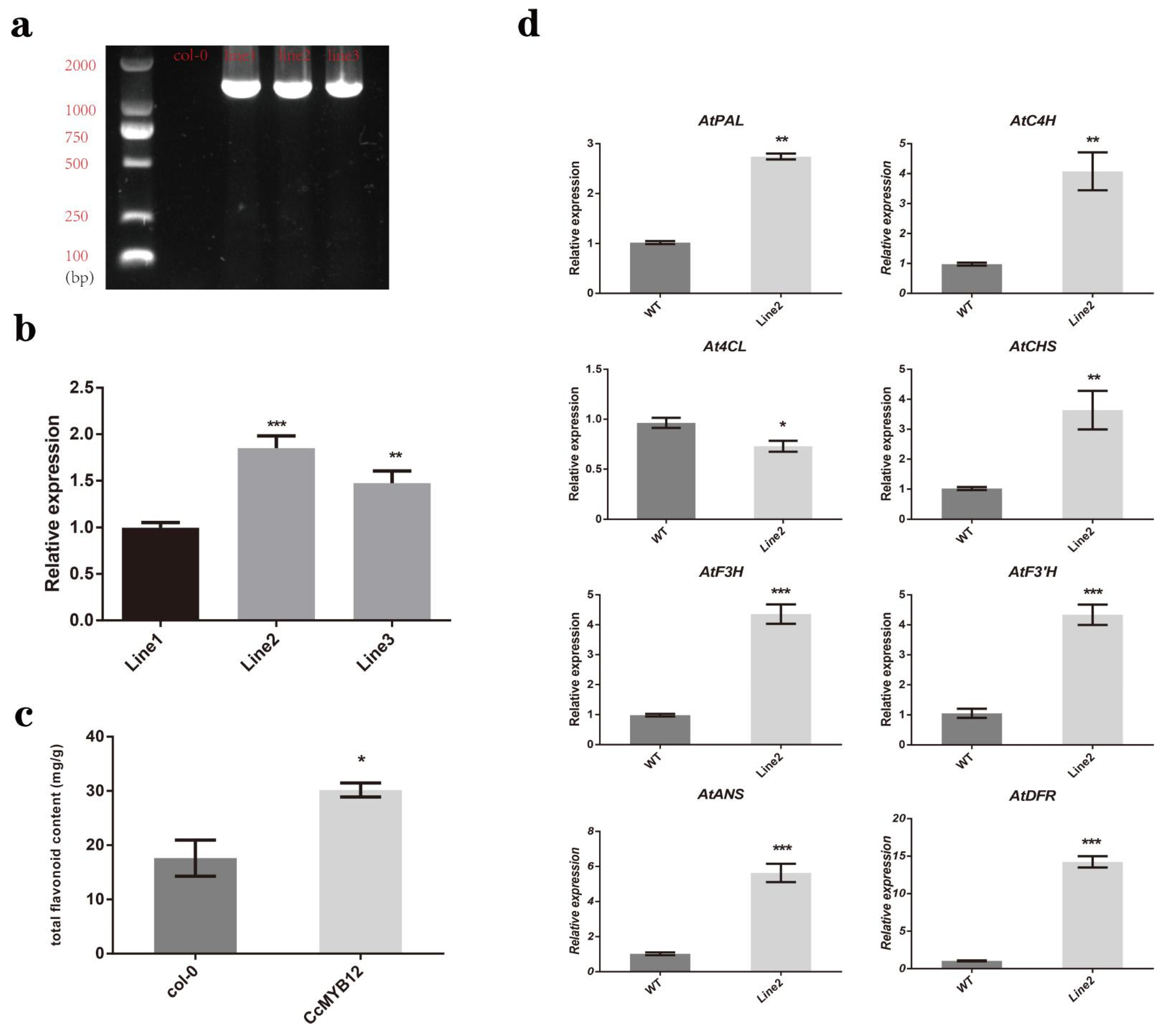
2.7. CcMYB12 Promoted the Expression of a Series of Flavonoid Biosynthesis Genes and Flavonoid Accumulation in Transgenic Arabidopsis Thaliana
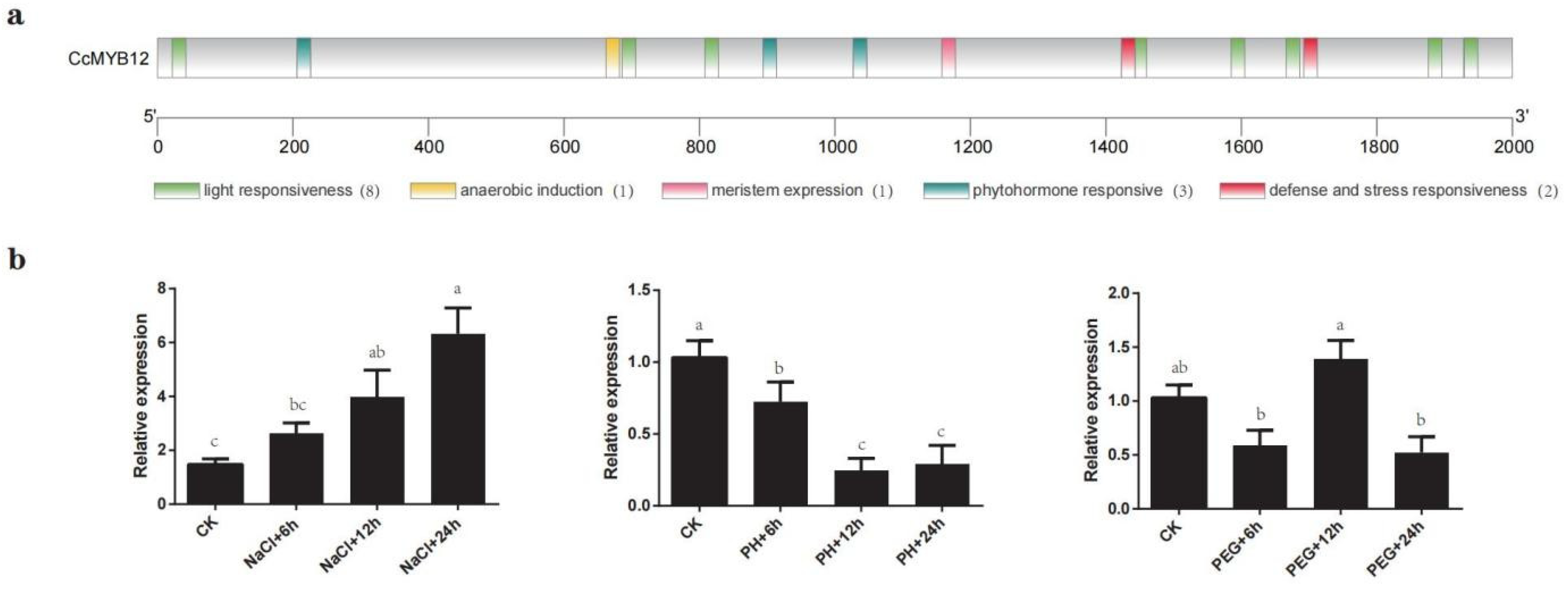
2.8. CcMYB12 May Participate in Response to Abiotic Stress
3. Discussion
4. Materials and Methods
4.1. Identification, Phylogenetic Analysis, and Sequence Alignment of R2R3-MYBs
4.2. Analysis of Gene Structures, Protein Motifs, Promoter Cis-Regulatory Elements, and Physiochemical Properties
4.3. Plasmid Construction
4.4. Plant Material and Treatment
4.5. RNA Extraction and Real-Time Quantitative PCR (RT-qPCR) Analysis
4.6. Determination of Total Flavonoid Content
4.7. Subcellular Localization Analysis of CcMYB12
4.8. Transactivation Activity of CcMYB12
4.9. Dual-Luciferase Reporter Assay
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Winkel-Shirley, B. Flavonoid Biosynthesis. A Colorful Model for Genetics, Biochemistry, Cell Biology, and Biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 2006, 57, 405–430. [Google Scholar] [CrossRef] [PubMed]
- Pourcel, L.; Routaboul, J.; Cheynier, V.; Lepinice, L.; Debeaujon, I. Flavonoid oxidation in plants: From biochemical properties to physiological functions. Trends Plant Sci. 2007, 12, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Grotewold, E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006, 57, 761–780. [Google Scholar] [CrossRef] [PubMed]
- Cavaiuolo, M.; Cocetta, G.; Ferrante, A. The Antioxidants Changes in Ornamental Flowers during Development and Senescence. Antioxidants 2013, 2, 132–155. [Google Scholar] [CrossRef]
- Zhang, P.; Du, H.; Wang, J.; Pu, Y.; Yang, C.; Yan, R.; Yang, H.; Cheng, H.; Yu, D. Multiplex CRISPR/Cas9-mediated metabolic engineering increases soya bean isoflavone content and resistance to soya bean mosaic virus. Plant Biotechnol. J. 2020, 18, 1384–1395. [Google Scholar] [CrossRef]
- Hosseinzadeh, E.; Hassanzadeh, A.; Marofi, F.; Solali, S. Flavonoid-Based Cancer Therapy: An Updated Review. Anti-Cancer Agents Med. Chem. 2020, 20, 1398–1414. [Google Scholar] [CrossRef]
- Broun, P. Transcriptional control of flavonoid biosynthesis: A complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis Pierre Broun. Curr. Opin. Plant Biol. 2005, 3, 272–279. [Google Scholar] [CrossRef]
- Zhao, L.; Gao, L.; Wang, H.; Chen, X.; Wang, Y.; Yang, H.; Wei, C.; Wan, X.; Xia, T. The R2R3-MYB, bHLH, WD40, and related transcription factors in flavonoid biosynthesis. Funct. Integr. Genom. 2013, 13, 75–98. [Google Scholar] [CrossRef]
- Yang, C.; Fang, X.; Wu, X.; Mao, Y.; Wang, L.; Chen, X. Transcriptional Regulation of Plant Secondary MetabolismF. J. Integr. Plant Biol. 2012, 54, 703–712. [Google Scholar] [CrossRef]
- Liu, J.; Osbourn, A.; Ma, P. MYB Transcription Factors as Regulators of Phenylpropanoid Metabolism in Plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef]
- Millard, P.S.; Kragelund, B.B.; Burow, M. R2R3 MYB Transcription Factors—Functions outside the DNA-Binding Domain. Trends Plant Sci. 2019, 24, 934–946. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, I.M.; Heim, M.A.; Weisshaar, B.; Uhrig, J.F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004, 40, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Cong, L.; Qu, Y.; Sha, G.; Zhang, S.; Ma, Y.; Chen, M.; Zhai, R.; Yang, C.; Xu, L.; Wang, Z. PbWRKY75 promotes anthocyanin synthesis by activating PbDFR, PbUFGT, and PbMYB10b in pear. Physiol. Plant. 2021, 173, 1841–1849. [Google Scholar] [CrossRef]
- Wang, W.; Wang, Y.; Li, H.; Liu, Z.; Cui, X.; Zhuang, J. Two MYB transcription factors (CsMYB2 and CsMYB26) are involved in flavonoid biosynthesis in tea plant [Camellia sinensis (L.) O. Kuntze]. BMC Plant Biol. 2018, 18, 288. [Google Scholar] [CrossRef]
- Greco, M.; Chiappetta, A.; Bruno, L.; Bitonti, M.B. In Posidonia oceanica cadmium induces changes in DNA methylation and chromatin patterning. J. Exp. Bot. 2012, 63, 695–709. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.C.; Wu, J.; Guan, M.L.; Zhao, C.H.; Geng, P.; Zhao, Q. Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J. 2019, 101, 637–652. [Google Scholar] [CrossRef] [PubMed]
- Anwar, M.; Yu, W.; Yao, H.; Zhou, P.; Allan, A.C.; Zeng, L. NtMYB3, an R2R3-MYB from Narcissus, Regulates Flavonoid Biosynthesis. Int. J. Mol. Sci. 2019, 20, 5456. [Google Scholar] [CrossRef] [PubMed]
- Youjun, H.; Lihong, X.; Zhongren, Z.; Rui, Z.; Zhengjia, W.; Chunying, H.; Ren, H.; Yumeng, L.; Tongqiang, F.; Jianhua, W.; et al. The genomes of pecan and Chinese hickory provide insights into Carya evolution and nut nutrition. Gigascience 2019, 8, giz036. [Google Scholar]
- Huang, C.Y.; Li, Y.; Wang, K.T.; Xi, J.W.; Xu, Y.F.; Si, X.L.; Pei, D.; Lu, S.H.; Xia, G.H.; Wang, J.H.; et al. Analysis of lipidomics profile of Carya cathayensis nuts and lipid dynamic changes during embryonic development. Food Chem. 2022, 370, 130975. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, J.H.; Wang, K.T.; Lu, S.H.; Ren, L.Y.; Huang, C.Y.; Pei, D.; Xing, Y.L.; Wang, Y.G.; Xu, Y.F.; et al. Comparison analysis of widely-targeted metabolomics revealed the variation of potential astringent ingredients and their dynamic accumulation in the seed coats of both Carya cathayensis and Carya illinoinensis. Food Chem. 2022, 374, 131688. [Google Scholar] [CrossRef] [PubMed]
- Editorial, S.A.O.T. Chinese Materia Medica: One Version; Shanghai Scientific and Technical Publishers: Shanghai, China, 1999; pp. 369–370. [Google Scholar]
- George, V.C.; Dellaire, G.; Rupasinghe, H.P.V. Plant flavonoids in cancer chemoprevention: Role in genome stability. J. Nutr. Biochem. 2017, 45, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Hou, N.; Xv, X.; Zhang, D.; Fan, T.; Zhang, Q.; Huang, Y. Flavonoid Synthesis and Metabolism During the Fruit Development in Hickory (Carya cathayensis). Front. Plant Sci. 2022, 13, 896421. [Google Scholar] [CrossRef]
- Mehrtens, F.; Kranz, H.; Bednarek, P.; Weisshaar, B. The Arabidopsis Transcription Factor MYB12 Is a Flavonol-Specific Regulator of Phenylpropanoid Biosynthesis. Plant Physiol. 2005, 138, 1083–1096. [Google Scholar] [CrossRef]
- Cao, Y.; Jia, H.; Xing, M.; Jin, R.; Grierson, D.; Gao, Z.; Sun, C.; Chen, K.; Xu, C.; Li, X. Genome-Wide Analysis of MYB Gene Family in Chinese Bayberry (Morella rubra) and Identification of Members Regulating Flavonoid Biosynthesis. Front. Plant Sci. 2021, 12, 1244. [Google Scholar] [CrossRef]
- Lin, N.; Liu, X.Y.; Zhu, W.F.; Cheng, X.; Wang, X.H.; Wan, X.C. Ambient Ultraviolet B Signal Modulates Tea Flavor Characteristics via Shifting a Metabolic Flux in Flavonoid Biosynthesis. J. Agric. Food Chem. 2021, 11, 3401–3414. [Google Scholar] [CrossRef]
- Yamagishi, M.; Sakai, M. The MicroRNA828/MYB12 Module Mediates Bicolor Pattern Development in Asiatic Hybrid Lily (Lilium spp.) Flowers. Front. Plant Sci. 2020, 11, 590791. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Zhong, R.; Lee, C.; Zhou, J.; McCarthy, R.L.; Ye, Z. A Battery of Transcription Factors Involved in the Regulation of Secondary Cell Wall Biosynthesis inArabidopsis. Plant Cell 2008, 20, 2763–2782. [Google Scholar] [CrossRef]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.R.; D’Amico, E.; Preuss, A.; Carbone, F.; Deiml, B.; Mourgues, F.; Perrotta, G.; Fischer, T.C.; Bovy, A.G.; Martens, S.; et al. Characterization of major enzymes and genes involved in flavonoid and proanthocyanidin biosynthesis during fruit development in strawberry (Fragaria xananassa). Arch. Biochem. Biophys. 2007, 1, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Nagaoka, S.; Takano, T. Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J. Exp. Bot. 2003, 54, 2231–2237. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, Y.; Wang, Y.; Liu, Z.; Gao, C. Comprehensive Analysis of MYB Gene Family and Their Expressions Under Abiotic Stresses and Hormone Treatments in Tamarix hispida. Front. Plant Sci. 2018, 9, 1303. [Google Scholar] [CrossRef]
- Tan, H.; Man, C.; Xie, Y.; Yan, J.; Chu, J.; Huang, J. A Crucial Role of GA-Regulated Flavonol Biosynthesis in Root Growth of Arabidopsis. Mol. Plant. 2019, 12, 521–537. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.; Shi, C.; Peng, Y.; Tan, H.; Xin, P.; Yang, Y.; Wang, F.; Li, X.; Chu, J.; Huang, J.; et al. Brassinosteroid-Activated BRI1-EMS-SUPPRESSOR 1 Inhibits Flavonoid Biosynthesis and Coordinates Growth and UV-B Stress Responses in Plants. Plant Cell 2020, 32, 3224–3239. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D. Basic local alignment search tool. J. Mol. Biol. 1990, 3, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-Wide Analysis of the ERF Gene Family in Arabidopsis and Rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Zhang, X.; Henriques, R.; Lin, S.; Niu, Q.; Chua, N. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef] [PubMed]
- De la Rosa, L.A.; Vazquez-Flores, A.A.; Alvarez-Parrilla, E.; Rodrigo-García, J.; Medina-Campos, O.N.; Ávila-Nava, A.; González-Reyes, S.; Pedraza-Chaverri, J. Content of major classes of polyphenolic compounds, antioxidant, antiproliferative, and cell protective activity of pecan crude extracts and their fractions. J. Funct. Foods 2014, 7, 219–228. [Google Scholar] [CrossRef]
- Yuan, M.; Xu, C.Y. BiFC Assay for Detecting Protein-protein Interaction in Tobacco Leaves. Bio-Protocol. 2018, 101, e1010133. [Google Scholar]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Ye, H.; Wang, K.; Huang, C.; Si, X.; Wang, J.; Xu, Y.; Huang, Y.; Huang, J.; Li, Y. CcMYB12 Positively Regulates Flavonoid Accumulation during Fruit Development in Carya cathayensis and Has a Role in Abiotic Stress Responses. Int. J. Mol. Sci. 2022, 23, 15618. https://doi.org/10.3390/ijms232415618
Wang Y, Ye H, Wang K, Huang C, Si X, Wang J, Xu Y, Huang Y, Huang J, Li Y. CcMYB12 Positively Regulates Flavonoid Accumulation during Fruit Development in Carya cathayensis and Has a Role in Abiotic Stress Responses. International Journal of Molecular Sciences. 2022; 23(24):15618. https://doi.org/10.3390/ijms232415618
Chicago/Turabian StyleWang, Yige, Hongyu Ye, Ketao Wang, Chunying Huang, Xiaolin Si, Jianhua Wang, Yifan Xu, Youjun Huang, Jianqin Huang, and Yan Li. 2022. "CcMYB12 Positively Regulates Flavonoid Accumulation during Fruit Development in Carya cathayensis and Has a Role in Abiotic Stress Responses" International Journal of Molecular Sciences 23, no. 24: 15618. https://doi.org/10.3390/ijms232415618
APA StyleWang, Y., Ye, H., Wang, K., Huang, C., Si, X., Wang, J., Xu, Y., Huang, Y., Huang, J., & Li, Y. (2022). CcMYB12 Positively Regulates Flavonoid Accumulation during Fruit Development in Carya cathayensis and Has a Role in Abiotic Stress Responses. International Journal of Molecular Sciences, 23(24), 15618. https://doi.org/10.3390/ijms232415618





