BioMThermDB 1.0: Thermophysical Database of Proteins in Solutions
Abstract
1. Introduction
2. Results and Discussion
3. Materials and Methods
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zidar, M.; Rozman, P.; Belko-Perkel, K.; Ravnik, M. Control of viscosity in biopharmaceutical protein formulations. J. Colloid Interface Sci. 2020, 580, 308–317. [Google Scholar] [CrossRef] [PubMed]
- Wang, W. Protein aggregation and its inhibition in biopharmaceutics. Int. J. Pharm. 2005, 289, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Nikam, R.; Kulandaisamy, A.; Harini, K.; Sharma, D.; Gromiha, M.M. ProThermDB: Thermodynamic database for proteins and mutants revisited after 15 years. Nucleic Acids Res. 2021, 49, D420–D424. [Google Scholar] [CrossRef] [PubMed]
- Kulandaisamy, A.; Sakthivel, R.; Gromiha, M.M. MPTherm: Database for membrane protein thermodynamics for understanding folding and stability. Brief. Bioinform. 2021, 22, 2119–2125. [Google Scholar] [CrossRef] [PubMed]
- Jemimah, S.; Yugandhar, K.; Gromiha, M.M. PROXiMATE: A database of mutant protein–protein complex thermodynamics and kinetics. Bioinformatics 2017, 33, 2787–2788. [Google Scholar] [CrossRef]
- Kumar, M.D.S.; Gromiha, M.M. PINT: Protein–protein Interactions Thermodynamic Database. Nucleic Acids Res. 2006, 34, D195–D198. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- McManus, J.J.; Charbonneau, P.; Zaccarelli, E.; Asherie, N. The physics of protein self-assembly. Curr. Opin. Colloid Interface Sci. 2016, 22, 73–79. [Google Scholar] [CrossRef]
- Mason, B.D.; van Enk, J.Z.; Zhang, L.; Remmele, R.L., Jr.; Zhang, J. Liquid-liquid phase separation of a monoclonal antibody and nonmonotonic influence of hofmeister anions. Biophys. J. 2010, 99, 3792–3800. [Google Scholar] [CrossRef]
- Kastelic, M.; Kalyuzhnyi, Y.V.; Hribar-Lee, B.; Dill, K.A.; Vlachy, V. Protein aggregation in salt solutions. Proc. Natl. Aacd. Sci. USA 2015, 112, 6766–6770. [Google Scholar] [CrossRef] [PubMed]
- Kastelic, M.; Dill, K.A.; Kalyuzhnyi, Y.V.; Vlachy, V. Controlling the viscosities of antibody solutions through control of their binding sites. J. Mol. Liq. 2018, 270, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Janc, T.; Kastelic, M.; Bončina, M.; Vlachy, V. Salt-specific effects in lysozyme solutions. Condens. Matter Phys. 2016, 19, 1–12. [Google Scholar] [CrossRef]
- Brudar, S.; Hribar-Lee, B. Effect of Buffer on Protein Stability in Aqueous Solutions: A Simple Protein Aggregation Model. J. Phys. Chem. B 2021, 125, 2504–2512. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; Scherer, T.M.; Shire, S.J.; Kalonia, D.S. Use of dynamic light scattering to determine second virial coefficient in a semidilute concentration regime. Anal. Biochem. 2011, 411, 292–296. [Google Scholar] [CrossRef] [PubMed]
- Salis, A.; Monduzzi, M. Not only pH. Specific buffer effects in biological systems. Curr. Opin. Colloid Interface Sci. 2016, 23, 1–9. [Google Scholar] [CrossRef]
- Jaklin, M.; Hritz, J.; Hribar-Lee, B. A new fibrillization mechanism of β-lactoglobulin in glycine solutions. Int. J. Biol. Macromol. 2022, 216, 414–425. [Google Scholar] [CrossRef] [PubMed]
- Brudar, S.; Hribar-Lee, B. The Role of Buffers in Wild-Type HEWL Amyloid Fibril Formation Mechanism. Biomolecules 2019, 9, 65. [Google Scholar] [CrossRef] [PubMed]
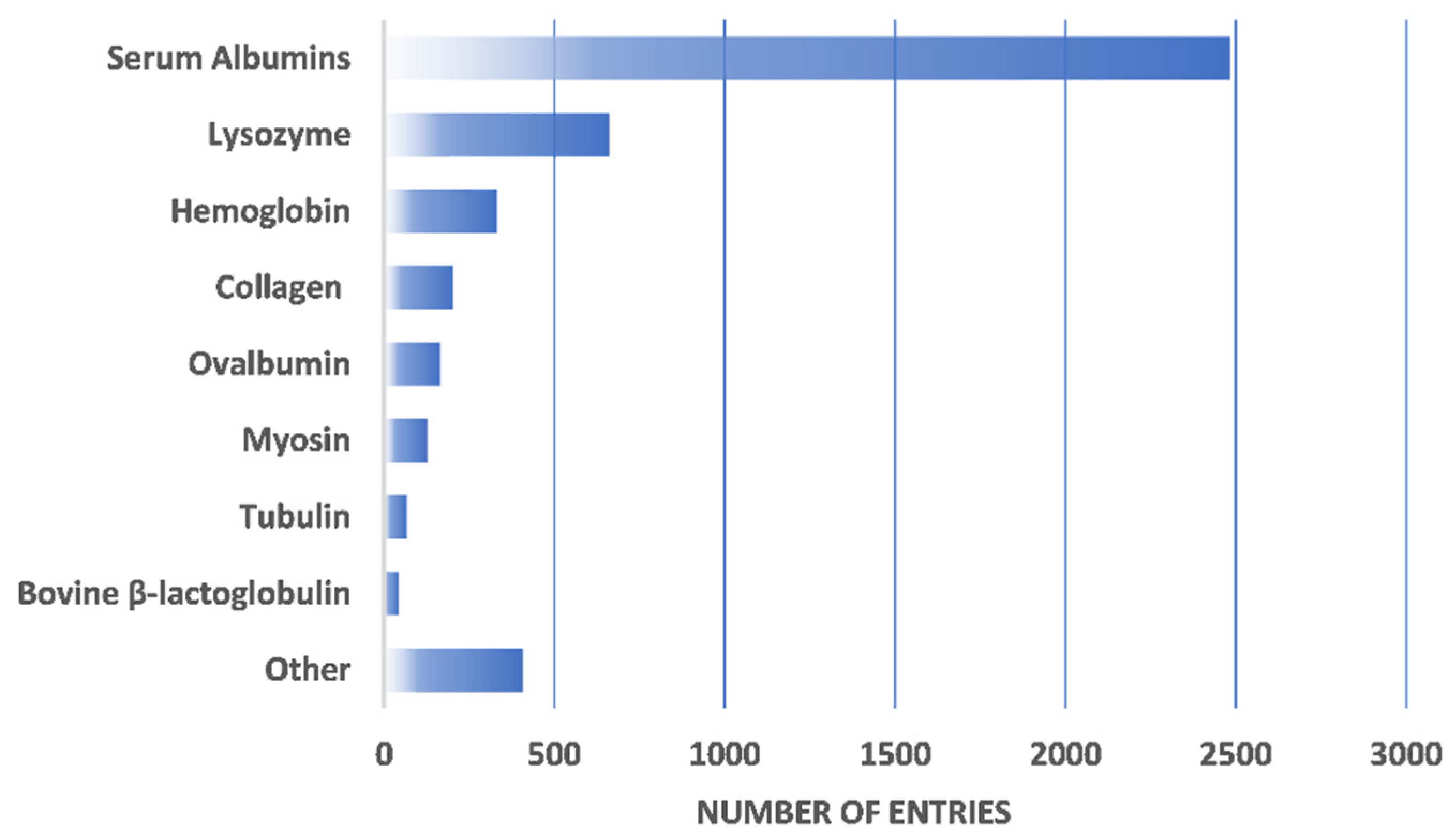
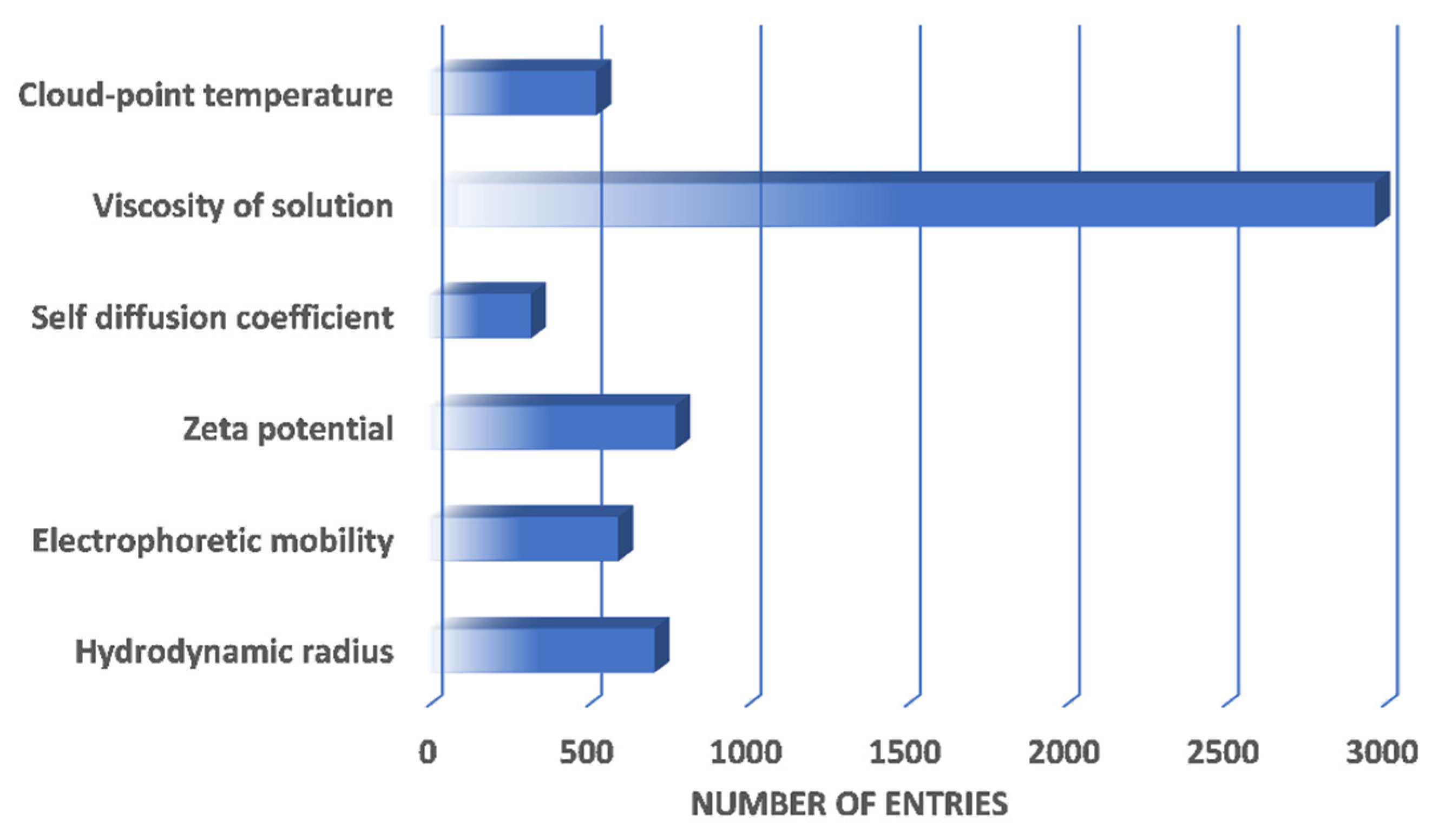
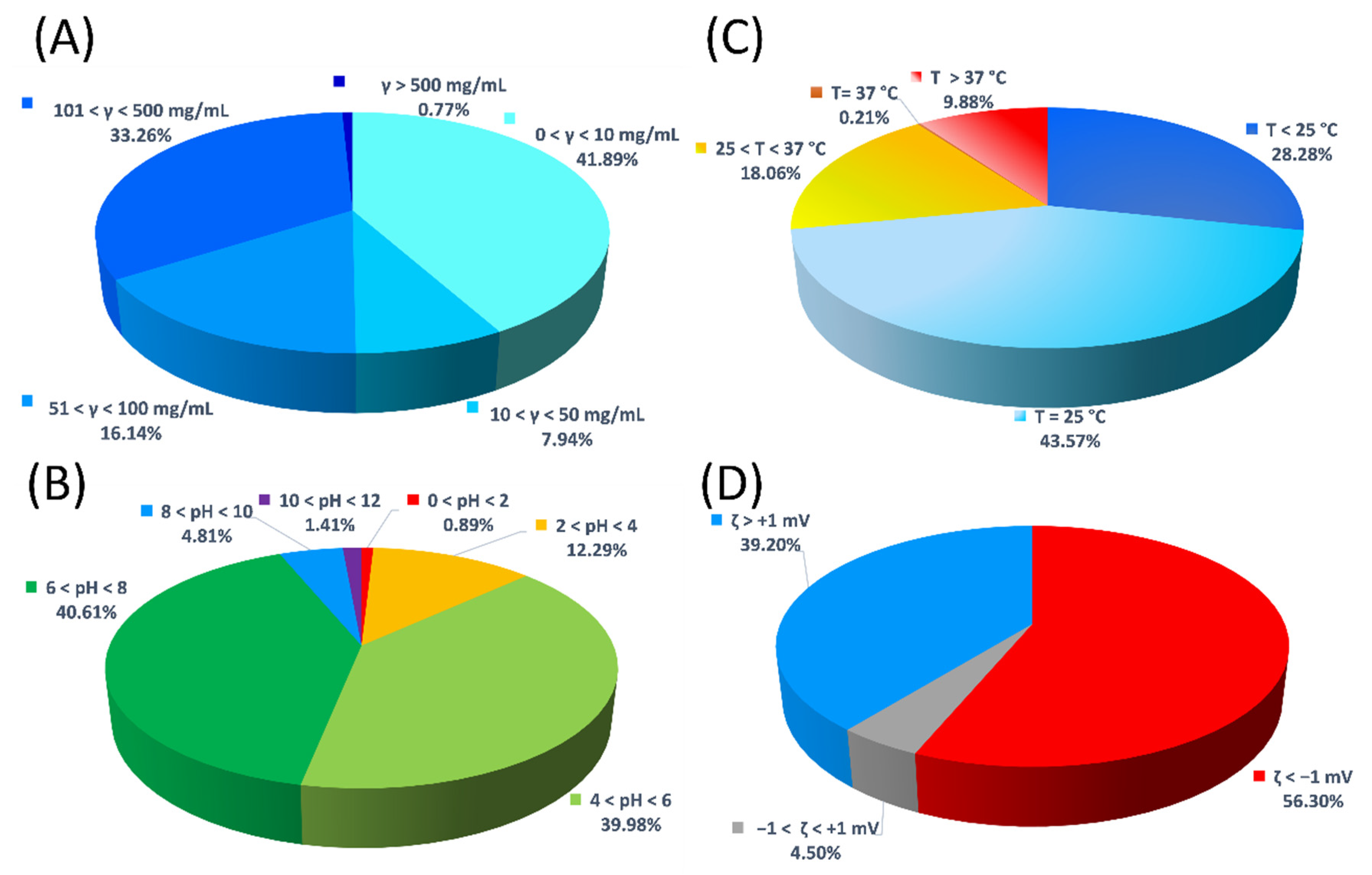
| Protein | Number of Entries |
|---|---|
| Soy-protein isolate | 107 |
| Rice-flour proteins | 42 |
| Erythrocytes | 30 |
| Horse globulins | 26 |
| CP12C75S (C-terminal disulfide bridge mutant) | 26 |
| Globulin | 24 |
| Conalbumin | 19 |
| CP12C31S (N-terminal disulfide bridge mutant) | 18 |
| Microtubule-associated Proteins (MAPs) | 14 |
| 4S α2–β1–glycoprotein | 13 |
| Recombinant p53 (1–93) | 13 |
| Neurofilaments | 11 |
| Serum orosomucoid | 10 |
| Fibrinogen | 10 |
| Lipoprotein ([4-14C] cholesterol-labeled) in dog’s blood serum | 10 |
| Wild-type CP12 protein | 7 |
| Ovomucoid O | 7 |
| Nuclease | 6 |
| βL-crystallin | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nikolić, M.; Brudar, S.; Coutsias, E.; Dill, K.A.; Lukšič, M.; Simmerling, C.; Hribar-Lee, B. BioMThermDB 1.0: Thermophysical Database of Proteins in Solutions. Int. J. Mol. Sci. 2022, 23, 15371. https://doi.org/10.3390/ijms232315371
Nikolić M, Brudar S, Coutsias E, Dill KA, Lukšič M, Simmerling C, Hribar-Lee B. BioMThermDB 1.0: Thermophysical Database of Proteins in Solutions. International Journal of Molecular Sciences. 2022; 23(23):15371. https://doi.org/10.3390/ijms232315371
Chicago/Turabian StyleNikolić, Mina, Sandi Brudar, Evangelos Coutsias, Ken A. Dill, Miha Lukšič, Carlos Simmerling, and Barbara Hribar-Lee. 2022. "BioMThermDB 1.0: Thermophysical Database of Proteins in Solutions" International Journal of Molecular Sciences 23, no. 23: 15371. https://doi.org/10.3390/ijms232315371
APA StyleNikolić, M., Brudar, S., Coutsias, E., Dill, K. A., Lukšič, M., Simmerling, C., & Hribar-Lee, B. (2022). BioMThermDB 1.0: Thermophysical Database of Proteins in Solutions. International Journal of Molecular Sciences, 23(23), 15371. https://doi.org/10.3390/ijms232315371







