Longevity-Associated Variant of BPIFB4 Confers Neuroprotection in the STHdh Cell Model of Huntington Disease
Abstract
1. Introduction
2. Results
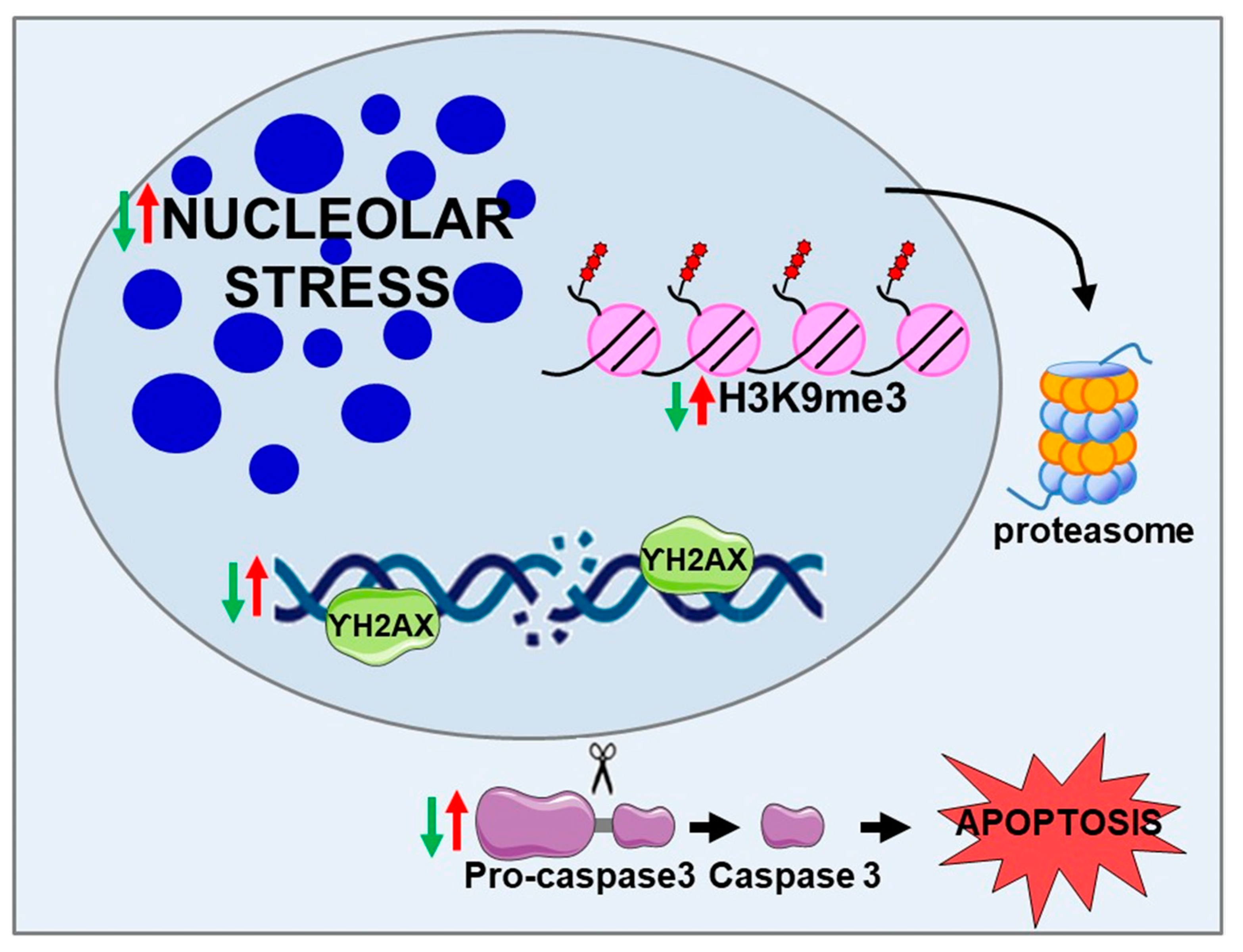
2.1. BPIFB4 Isoforms Confer Resistance to Nucleolar Stress
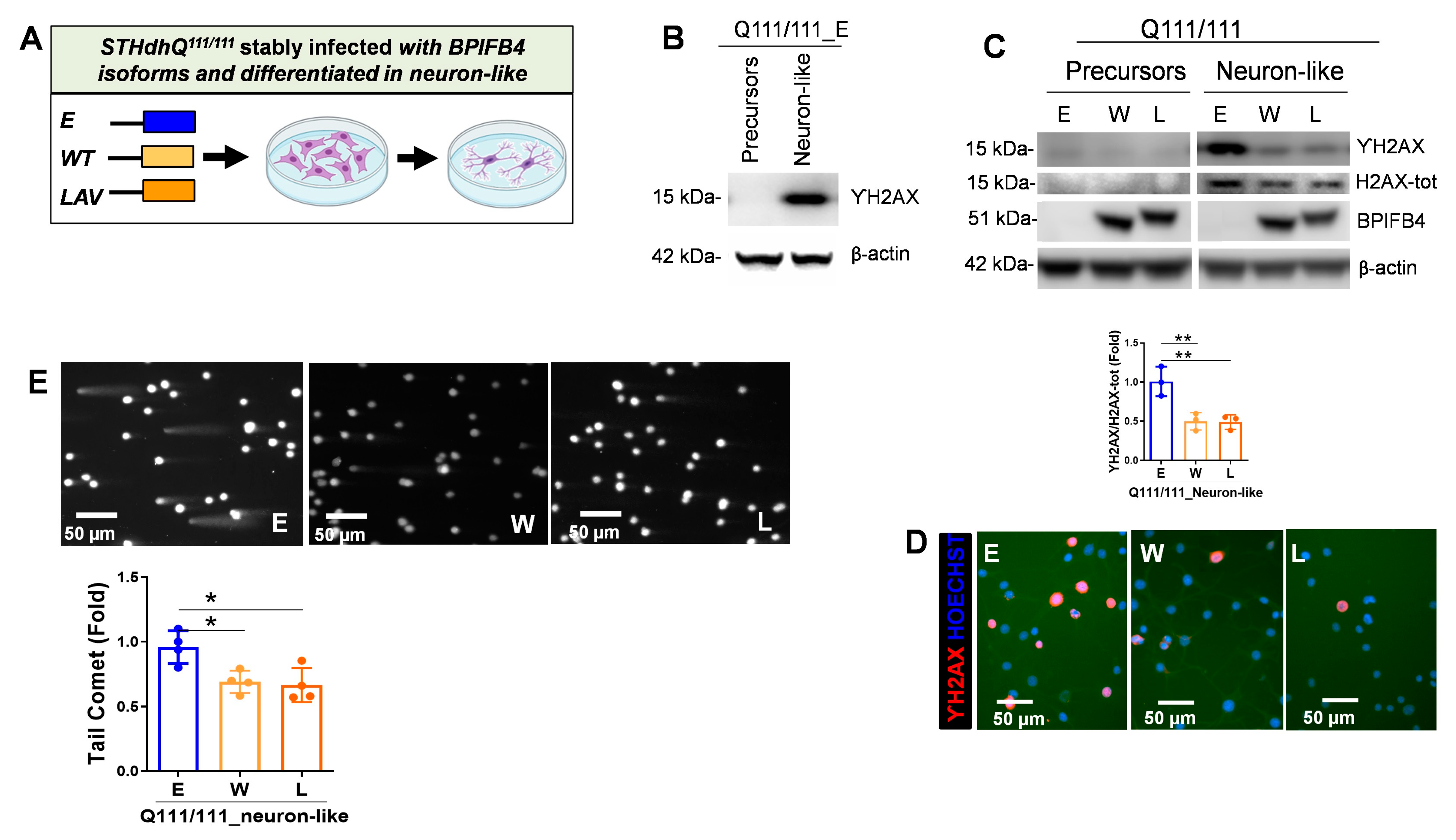
2.2. BPIFB4 Isoforms Decrease DNA Damage
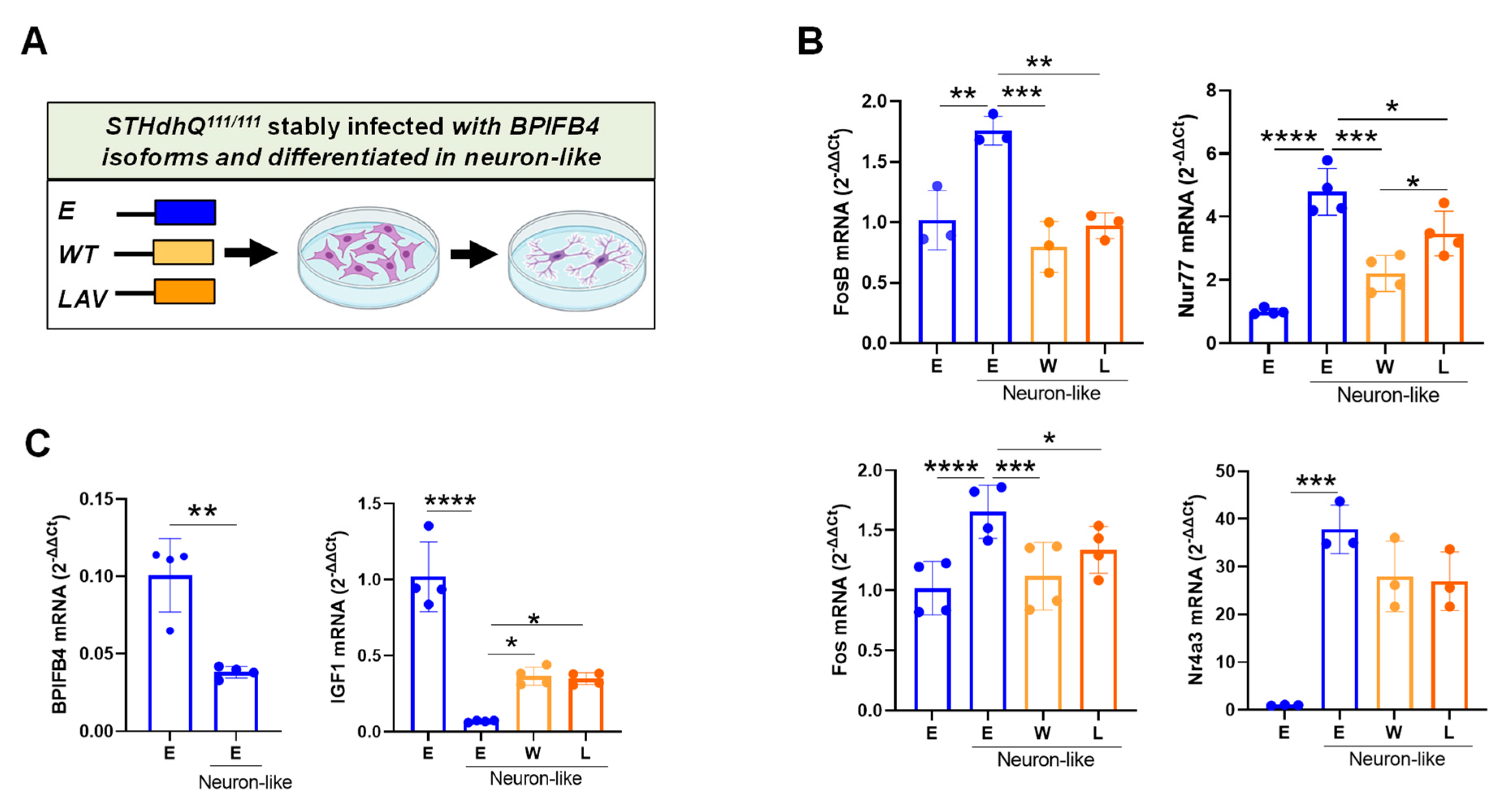
2.3. BPIFB4 Isoforms Partially Preserve Cellular Identity
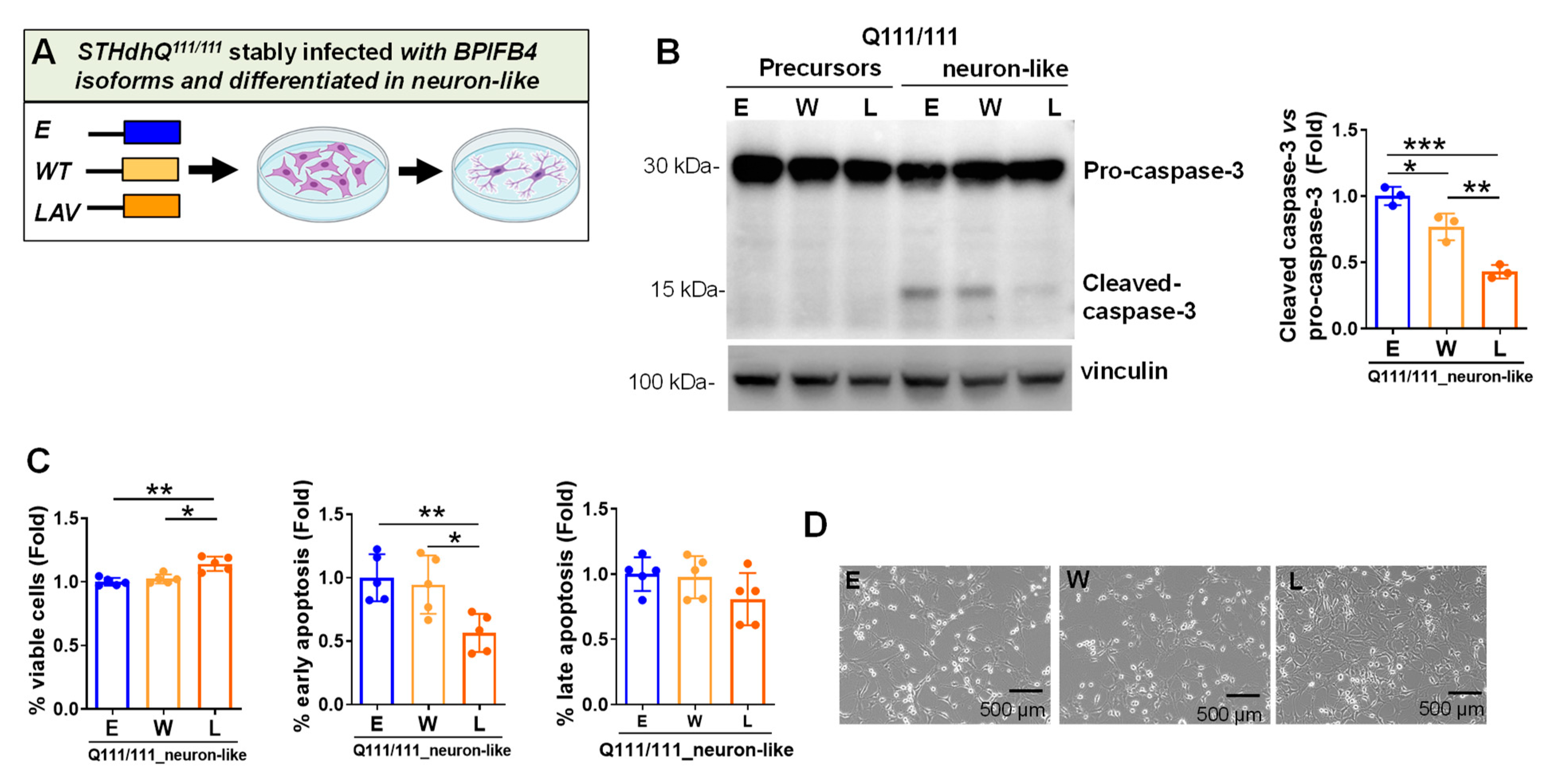
2.4. LAV-BPIFB4 Isoform Protects against Apoptosis through the Deregulation of Caspase Cascade
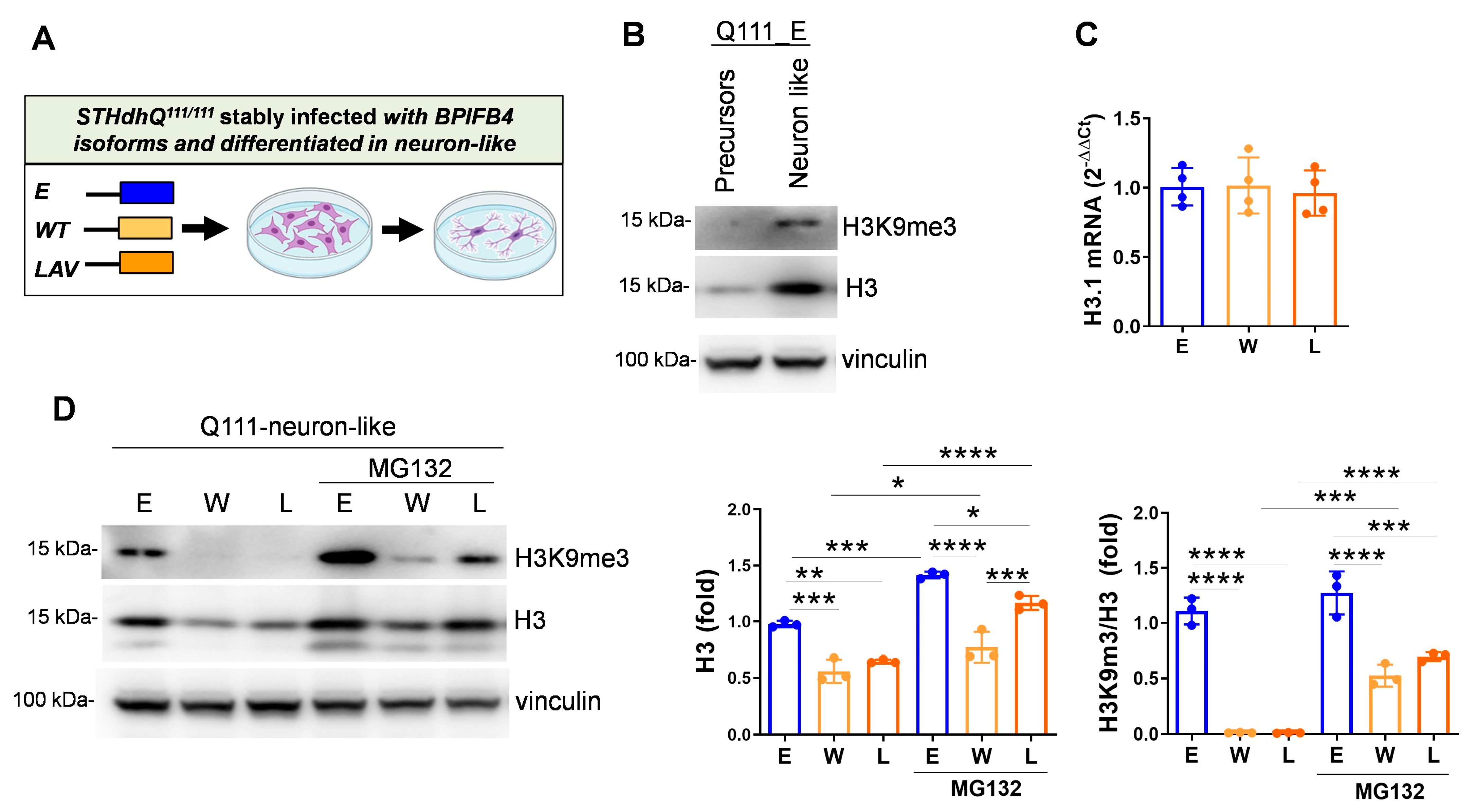
2.5. BPIFB4 Isoforms Decrease the Level of Trimethylated Histone 3 on Lysine 9 and Remodel Chromatin
- Nucleolar stress trigged by NCL sequestration in CAG RNA foci
- DNA damage and DNA repair dysfunction
- Caspase 3-dependent cell-death pathway
- H3K9me3-dependent heterochromatin condensation
- Decreases nucleolar stress
- Counteracts DNA damage and enhances DNA repair efficiency
- Inhibits the cleavage of pro-caspase 3, counteracting cell death in favor of cell viability
- Reduces the level of H3K9me3 through the ubiquitin–proteasome system
3. Discussion
3.1. LAV-BPIFB4 Counteracts the Nucleolar Stress
3.2. LAV-BPIFB4 Protects HD Neuron-like Cells from DNA Damage and Apoptosis via Inhibition of Caspase Signaling
3.3. BPIFB4 Isoforms Partially Counteract Neuronal Differentiation
3.4. BPIFB4 Reduces the Level of H3K9me3 in HD Neuron-like Cells
4. Material and Methods
4.1. Cell Maintenance, Treatment, and Differentiation
4.2. RNA Extraction and Quantitative Real-Time Analysis
4.3. Western Blotting
4.4. Immunofluorescence
4.5. Cell Viability
4.6. Comet Assay
4.7. Detection of Apoptosis by Flow-Cytometry
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- The Huntington’s Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 1993, 72, 971–983. [Google Scholar] [CrossRef]
- Vonsattel, J.P.; Myers, R.H.; Stevens, T.J.; Ferrante, R.J.; Bird, E.D.; Richardson, E.P., Jr. Neuropathological classification of Huntington’s disease. J. Neuropathol. Exp. Neurol. 1985, 44, 559–577. [Google Scholar] [CrossRef]
- Wang, C.E.; Tydlacka, S.; Orr, A.L.; Yang, S.H.; Graham, R.K.; Hayden, M.R.; Li, S.; Chan, A.W.; Li, X.J. Accumulation of N-terminal mutant huntingtin in mouse and monkey models implicated as a pathogenic mechanism in Huntington’s disease. Hum. Mol. Genet. 2008, 17, 2738–2751. [Google Scholar] [CrossRef]
- Leavitt, B.R.; van Raamsdonk, J.M.; Shehadeh, J.; Fernandes, H.; Murphy, Z.; Graham, R.K.; Wellington, C.L.; Raymond, L.A.; Hayden, M.R. Wild-type huntingtin protects neurons from excitotoxicity. J. Neurochem. 2006, 96, 1121–1129. [Google Scholar] [CrossRef]
- McKinstry, S.U.; Karadeniz, Y.B.; Worthington, A.K.; Hayrapetyan, V.Y.; Ozlu, M.I.; Serafin-Molina, K.; Risher, W.C.; Ustunkaya, T.; Dragatsis, I.; Zeitlin, S.; et al. Huntingtin is required for normal excitatory synapse development in cortical and striatal circuits. J. Neurosci. 2014, 34, 9455–9472. [Google Scholar] [CrossRef] [PubMed]
- Dragatsis, I.; Dietrich, P.; Ren, H.; Deng, Y.P.; Del Mar, N.; Wang, H.B.; Johnson, I.M.; Jones, K.R.; Reiner, A. Effect of early embryonic deletion of huntingtin from pyramidal neurons on the development and long-term survival of neurons in cerebral cortex and striatum. Neurobiol. Dis. 2018, 111, 102–117. [Google Scholar] [CrossRef] [PubMed]
- Mehler, M.F.; Petronglo, J.R.; Arteaga-Bracho, E.E.; Gulinello, M.E.; Winchester, M.L.; Pichamoorthy, N.; Young, S.K.; DeJesus, C.D.; Ishtiaq, H.; Gokhan, S.; et al. Loss-of-Huntingtin in Medial and Lateral Ganglionic Lineages Differentially Disrupts Regional Interneuron and Projection Neuron Subtypes and Promotes Huntington’s Disease-Associated Behavioral, Cellular, and Pathological Hallmarks. J. Neurosci. 2019, 39, 1892–1909. [Google Scholar] [CrossRef]
- Abdelmohsen, K.; Gorospe, M. RNA-binding protein nucleolin in disease. RNA Biol. 2012, 9, 799–808. [Google Scholar] [CrossRef]
- Tsoi, H.; Lau, T.C.; Tsang, S.Y.; Lau, K.F.; Chan, H.Y. CAG expansion induces nucleolar stress in polyglutamine diseases. Proc. Natl. Acad. Sci. USA 2012, 109, 13428–13433. [Google Scholar] [CrossRef] [PubMed]
- Stack, E.C.; Del Signore, S.J.; Luthi-Carter, R.; Soh, B.Y.; Goldstein, D.R.; Matson, S.; Goodrich, S.; Markey, A.L.; Cormier, K.; Hagerty, S.W.; et al. Modulation of nucleosome dynamics in Huntington’s disease. Hum. Mol. Genet. 2007, 16, 1164–1175. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.; Lee, J.; Hagerty, S.W.; Soh, B.Y.; McAlpin, S.E.; Cormier, K.A.; Smith, K.M.; Ferrante, R.J. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington’s disease. Proc. Natl. Acad. Sci. USA 2006, 103, 19176–19181. [Google Scholar] [CrossRef]
- Hwang, Y.J.; Hyeon, S.J.; Kim, Y.; Lim, S.; Lee, M.Y.; Kim, J.; Londhe, A.M.; Gotina, L.; Kim, Y.; Pae, A.N.; et al. Modulation of SETDB1 activity by APQ ameliorates heterochromatin condensation, motor function, and neuropathology in a Huntington’s disease mouse model. J. Enzyme Inhib. Med. Chem. 2021, 36, 856–868. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, P.; De Cristofaro, T.; Affaitati, A.; Pizzulo, G.M.; Feliciello, A.; Criscuolo, C.; De Michele, G.; Filla, A.; Avvedimento, E.V.; Varrone, S. DNA damage induced by polyglutamine-expanded proteins. Hum. Mol Genet. 2003, 12, 2301–2309. [Google Scholar] [CrossRef] [PubMed]
- Illuzzi, J.; Yerkes, S.; Parekh-Olmedo, H.; Kmiec, E.B. DNA breakage and induction of DNA damage response proteins precede the appearance of visible mutant huntingtin aggregates. J. Neurosci. Res. 2009, 87, 733–747. [Google Scholar] [CrossRef] [PubMed]
- Enokido, Y.; Tamura, T.; Ito, H.; Arumughan, A.; Komuro, A.; Shiwaku, H.; Sone, M.; Foulle, R.; Sawada, H.; Ishiguro, H.; et al. Mutant huntingtin impairs Ku70-mediated DNA repair. J. Cell Biol. 2010, 189, 425–443. [Google Scholar] [CrossRef] [PubMed]
- Villa, F.; Carrizzo, A.; Ferrario, A.; Maciag, A.; Cattaneo, M.; Spinelli, C.C.; Montella, F.; Damato, A.; Ciaglia, E.; Puca, A.A. A Model of Evolutionary Selection: The Cardiovascular Protective Function of the Longevity Associated Variant of BPIFB4. Int. J. Mol. Sci. 2018, 19, 3229. [Google Scholar] [CrossRef] [PubMed]
- Villa, F.; Carrizzo, A.; Spinelli, C.C.; Ferrario, A.; Malovini, A.; Maciąg, A.; Damato, A.; Auricchio, A.; Spinetti, G.; Sangalli, E.; et al. Analysis Reveals a Longevity-Associated Protein Modulating Endothelial Function and Angiogenesis. Circ. Res. 2015, 117, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Villa, F.; Malovini, A.; Carrizzo, A.; Spinelli, C.C.; Ferrario, A.; Maciąg, A.; Madonna, M.; Bellazzi, R.; Milanesi, L.; Vecchione, C.; et al. Serum BPIFB4 levels classify health status in long-living individuals. Immun. Ageing 2015, 12, 27. [Google Scholar] [CrossRef]
- Puca, A.A.; Carrizzo, A.; Spinelli, C.; Damato, A.; Ambrosio, M.; Villa, F.; Ferrario, A.; Maciag, A.; Fornai, F.; Lenzi, P.; et al. Single systemic transfer of a human gene associated with exceptional longevity halts the progression of atherosclerosis and inflammation in ApoE knockout mice through a CXCR4-mediated mechanism. Eur. Heart J. 2020, 41, 2487–2497. [Google Scholar] [CrossRef] [PubMed]
- Dang, Z.; Avolio, E.; Thomas, A.C.; Faulkner, A.; Beltrami, A.P.; Cervellin, C.; Carrizzo, A.; Maciag, A.; Gu, Y.; Ciaglia, E.; et al. Transfer of a human gene variant associated with exceptional longevity improves cardiac function in obese type 2 diabetic mice through induction of the SDF-1/CXCR4 signalling pathway. Eur. J. Heart Fail. 2020, 22, 1568–1581. [Google Scholar] [CrossRef]
- Malavolta, M.; Dato, S.; Villa, F.; Rango, F.; Iannone, F.; Ferrario, A.; Maciag, A.; Ciaglia, E.; D’amato, A.; Carrizzo, A.; et al. LAV-BPIFB4 associates with reduced frailty in humans and its transfer prevents frailty progression in old mice. Aging 2019, 11, 6555–6568. [Google Scholar] [CrossRef]
- Di Pardo, A.; Ciaglia, E.; Cattaneo, M.; Maciag, A.; Montella, F.; Lopardo, V.; Ferrario, A.; Villa, F.; Madonna, M.; Amico, E.; et al. The longevity-associated variant of BPIFB4 improves a CXCR4-mediated striatum-microglia crosstalk preventing disease progression in a mouse model of Huntington’s disease. Cell Death Dis. 2020, 11, 546. [Google Scholar] [CrossRef]
- Ciaglia, E.; Lopardo, V.; Montella, F.; Carrizzo, A.; Di Pietro, P.; Malavolta, M.; Giacconi, R.; Orlando, F.; Cattaneo, M.; Madeddu, P.; et al. Transfer of the longevity-associated variant of BPIFB4 gene rejuvenates immune system and vasculature by a reduction of CD38+ macrophages and NAD+ decline. Cell Death Dis. 2022, 13, 86. [Google Scholar] [CrossRef]
- Spinelli, C.C.; Carrizzo, A.; Ferrario, A.; Villa, F.; Damato, A.; Ambrosio, M.; Madonna, M.; Frati, G.; Fucile, S.; Sciaccaluga, M.; et al. LAV-BPIFB4 isoform modulates eNOS signalling through Ca2+/PKC-alpha-dependent mechanism. Cardiovasc. Res. 2017, 113, 795–804. [Google Scholar] [CrossRef]
- Trettel, F.; Rigamonti, D.; Hilditch-Maguire, P.; Wheeler, V.C.; Sharp, A.H.; Persichetti, F.; Cattaneo, E.; MacDonald, M.E. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum. Mol. Genet. 2000, 9, 2799–2809. [Google Scholar] [CrossRef] [PubMed]
- Tsai, R.Y.; Pederson, T. Connecting the nucleolus to the cell cycle and human disease. FASEB J. 2014, 28, 3290–32963. [Google Scholar] [CrossRef]
- Nicolas, E.; Parisot, P.; Pinto-Monteiro, C.; de Walque, R.; De Vleeschouwer, C.; Lafontaine, D.L. Involvement of human ribosomal proteins in nucleolar structure and p53-dependent nucleolar stress. Nat. Commun. 2016, 7, 11390. [Google Scholar] [CrossRef] [PubMed]
- Hariharan, N.; Sussman, M.A. Stressing on the nucleolus in cardiovascular disease. Biochim. Biophys. Acta 2014, 1842, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Madabhushi, R.; Gao, F.; Pfenning, A.R.; Pan, L.; Yamakawa, S.; Seo, J.; Rueda, R.; Phan, T.X.; Yamakawa, H.; Pao, P.C.; et al. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell 2015, 161, 1592–1605. [Google Scholar] [CrossRef]
- Supeno, N.E.; Pati, S.; Hadi, R.A.; Ghani, A.R.; Mustafa, Z.; Abdullah, J.M.; Idris, F.M.; Han, X.; Jaafar, H. IGF-1 acts as controlling switch for long-term proliferation and maintenance of EGF/FGF-responsive striatal neural stem cells. Int. J. Med. Sci. 2013, 10, 522–531. [Google Scholar] [CrossRef]
- Kobayashi, J.; Fujimoto, H.; Sato, J.; Hayashi, I.; Burma, S.; Matsuura, S.; Chen, D.J.; Komatsu, K. Nucleolin participates in DNA double-strand break-induced damage response through MDC1-dependent pathway. PLoS ONE 2012, 7, e49245. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, M.; Derheimer, F.A.; Tait-Mulder, J.; Kastan, M.B. Nucleolin mediates nucleosome disruption critical for DNA double-strand break repair. Proc. Natl. Acad. Sci. USA 2013, 110, 16874–16879. [Google Scholar] [CrossRef] [PubMed]
- Ianov, L.; De Both, M.; Chawla, M.K.; Rani, A.; Kennedy, A.J.; Piras, I.; Day, J.J.; Siniard, A.; Kumar, A.; Sweatt, J.D.; et al. Hippocampal Transcriptomic Profiles: Subfield Vulnerability to Age and Cognitive Impairment. Front. Aging Neurosci. 2017, 9, 383. [Google Scholar] [CrossRef]
- Park, J.; Lee, K.; Kim, K.; Yi, S.J. The role of histone modifications: From neurodevelopment to neurodiseases. Signal Transduct. Target. Ther. 2022, 7, 217. [Google Scholar]
- Köhler, F.; Bormann, F.; Raddatz, G.; Gutekunst, J.; Corless, S.; Musch, T.; Lonsdorf, A.S.; Erhardt, S.; Lyko, F.; Rodríguez-Paredes, M. Epigenetic deregulation of lamina-associated domains in Hutchinson-Gilford progeria syndrome. Genome Med. 2020, 12, 46. [Google Scholar] [CrossRef]
- Kaneda, R.; Takada, S.; Yamashita, Y.; Choi, Y.L.; Nonaka-Sarukawa, M.; Soda, M.; Misawa, Y.; Isomura, T.; Shimada, K.; Mano, H. Genome-wide histone methylation profile for heart failure. Genes Cells 2009, 14, 69–77. [Google Scholar] [CrossRef]
- Papait, R.; Cattaneo, P.; Kunderfranco, P.; Greco, C.; Carullo, P.; Guffanti, A.; Viganò, V.; Stirparo, G.G.; Latronico, M.V.; Hasenfuss, G.; et al. Genome-wide analysis of histone marks identifying an epigenetic signature of promoters and enhancers underlying cardiac hypertrophy. Proc. Natl. Acad. Sci. USA 2013, 110, 20164–20169. [Google Scholar] [CrossRef]
- Lee, M.Y.; Lee, J.; Hyeon, S.J.; Cho, H.; Hwang, Y.J.; Shin, J.Y.; McKee, A.C.; Kowall, N.W.; Kim, J.I.; Stein, T.D.; et al. Epigenome signatures landscaped by histone H3K9me3 are associated with the synaptic dysfunction in Alzheimer’s disease. Aging Cell 2020, 19, e13153. [Google Scholar] [CrossRef]
- Song, W.; Zsindely, N.; Faragó, A.; Marsh, J.L.; Bodai, L. Systematic genetic interaction studies identify histone demethylase Utx as potential target for ameliorating Huntington’s disease. Hum. Mol. Genet. 2018, 27, 649–666. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cattaneo, M.; Maciag, A.; Milella, M.S.; Ciaglia, E.; Bruno, A.; Puca, A.A. Longevity-Associated Variant of BPIFB4 Confers Neuroprotection in the STHdh Cell Model of Huntington Disease. Int. J. Mol. Sci. 2022, 23, 15313. https://doi.org/10.3390/ijms232315313
Cattaneo M, Maciag A, Milella MS, Ciaglia E, Bruno A, Puca AA. Longevity-Associated Variant of BPIFB4 Confers Neuroprotection in the STHdh Cell Model of Huntington Disease. International Journal of Molecular Sciences. 2022; 23(23):15313. https://doi.org/10.3390/ijms232315313
Chicago/Turabian StyleCattaneo, Monica, Anna Maciag, Maria Serena Milella, Elena Ciaglia, Antonino Bruno, and Annibale Alessandro Puca. 2022. "Longevity-Associated Variant of BPIFB4 Confers Neuroprotection in the STHdh Cell Model of Huntington Disease" International Journal of Molecular Sciences 23, no. 23: 15313. https://doi.org/10.3390/ijms232315313
APA StyleCattaneo, M., Maciag, A., Milella, M. S., Ciaglia, E., Bruno, A., & Puca, A. A. (2022). Longevity-Associated Variant of BPIFB4 Confers Neuroprotection in the STHdh Cell Model of Huntington Disease. International Journal of Molecular Sciences, 23(23), 15313. https://doi.org/10.3390/ijms232315313






