Design, Synthesis and Antimicrobial Properties of New Tetracyclic Quinobenzothiazine Derivatives
Abstract
:1. Introduction
2. Results and Discussion
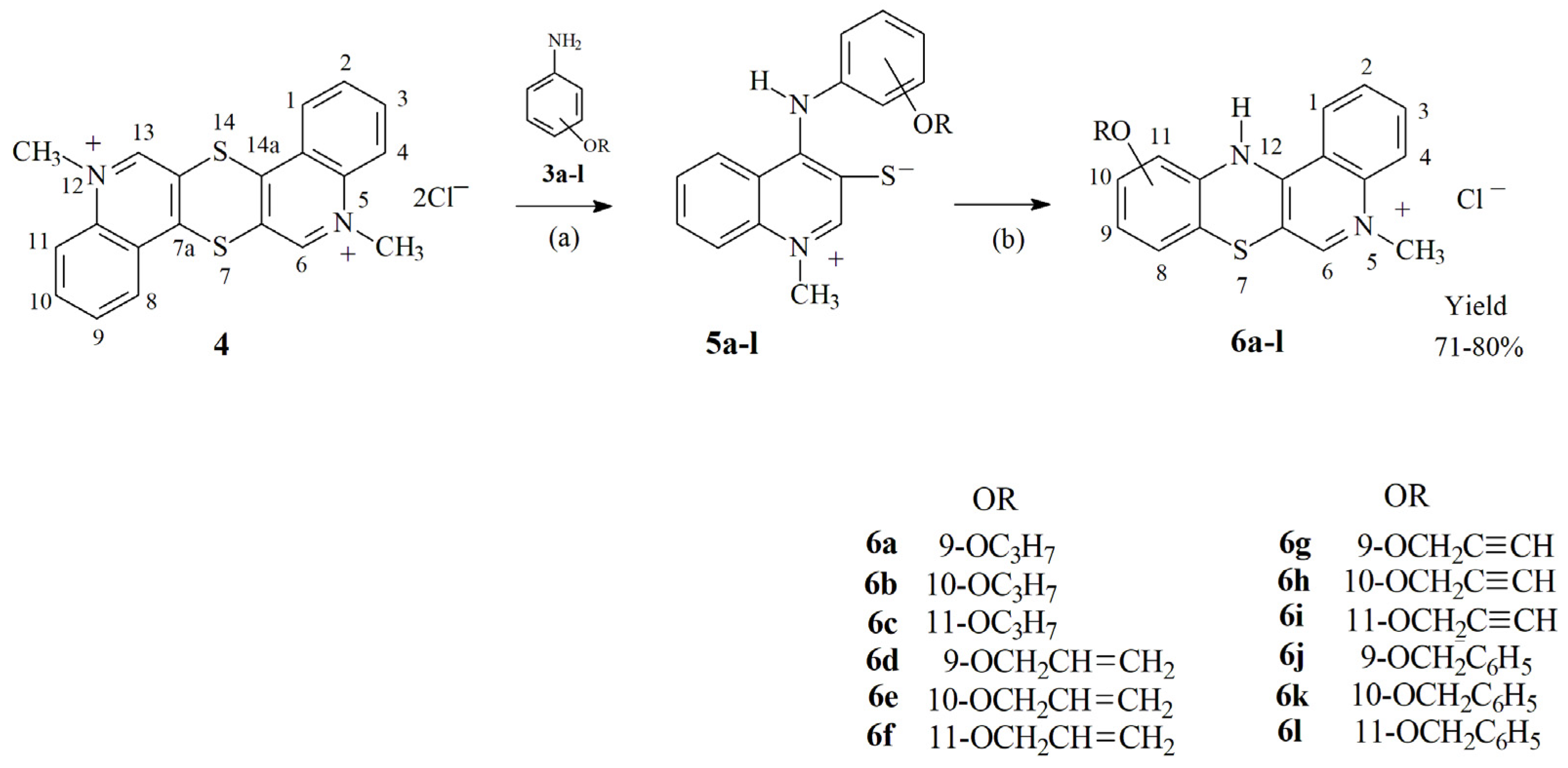
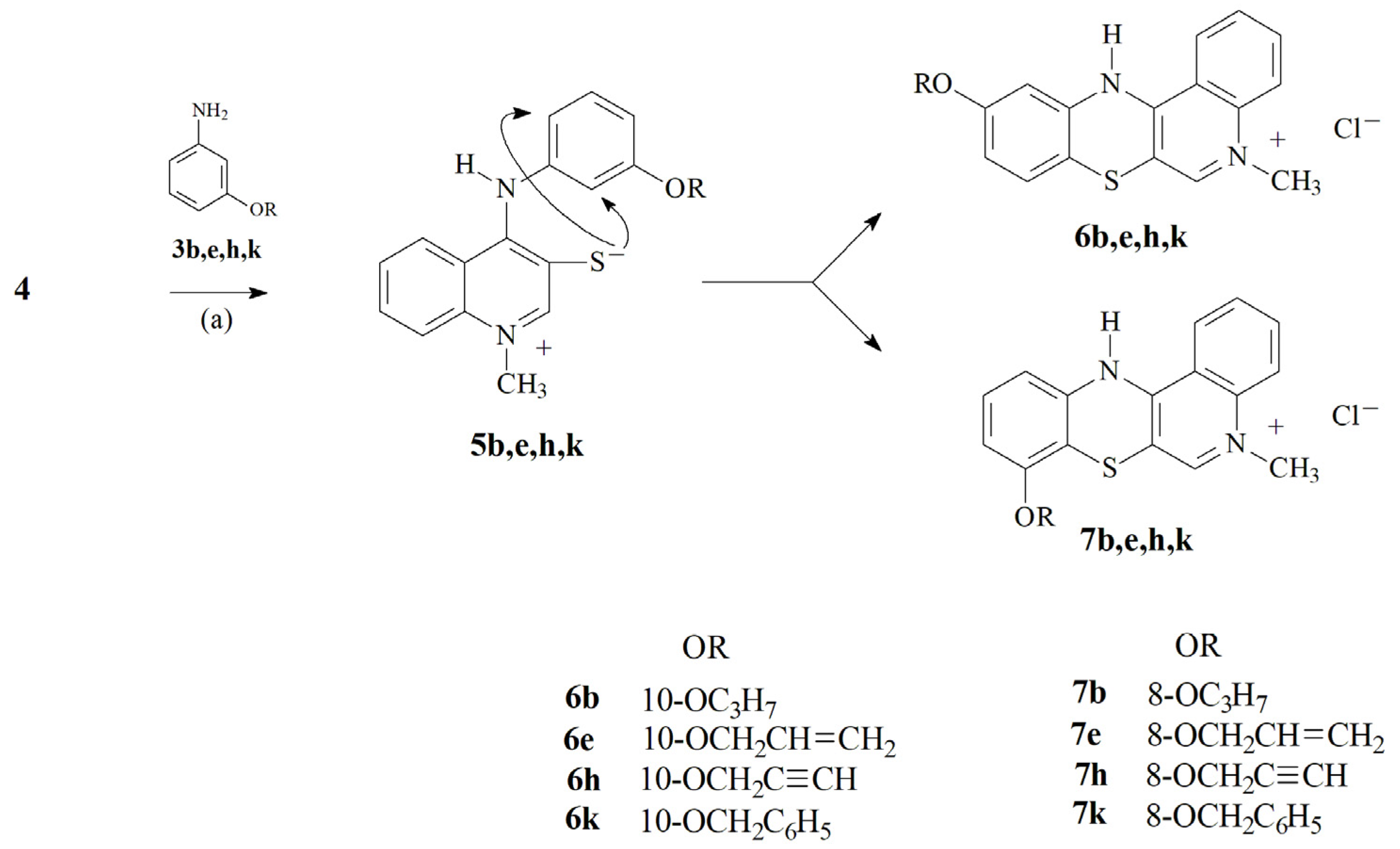
2.1. Chemistry—Design and Synthesis
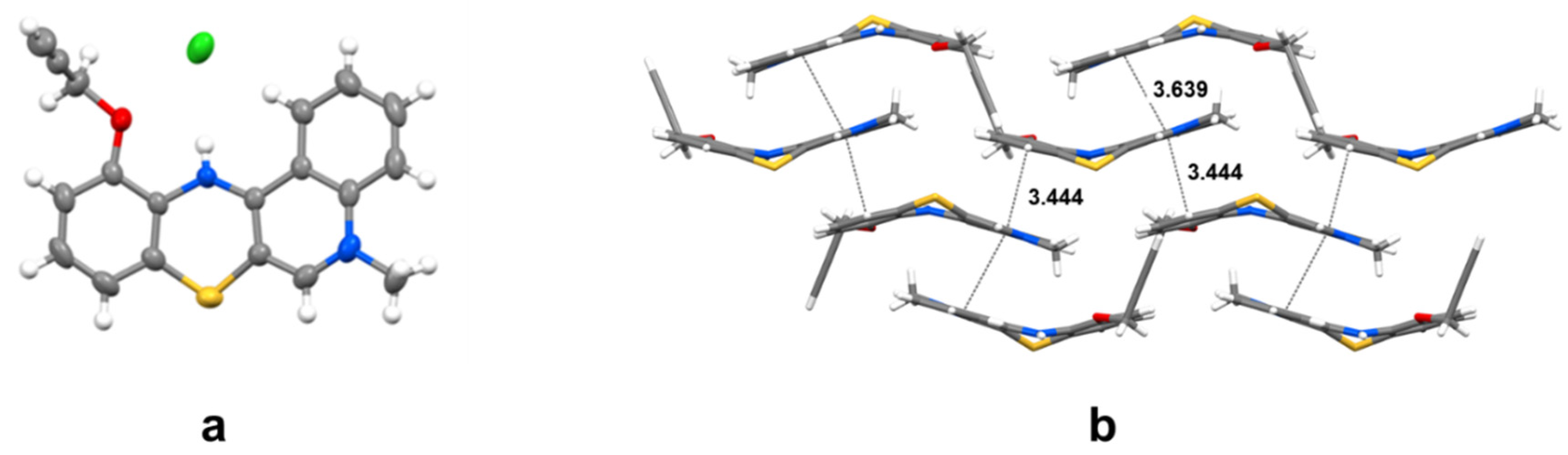
2.2. Structural Analysis
2.3. Biological Screening
2.3.1. In Vitro Antimicrobial Activity
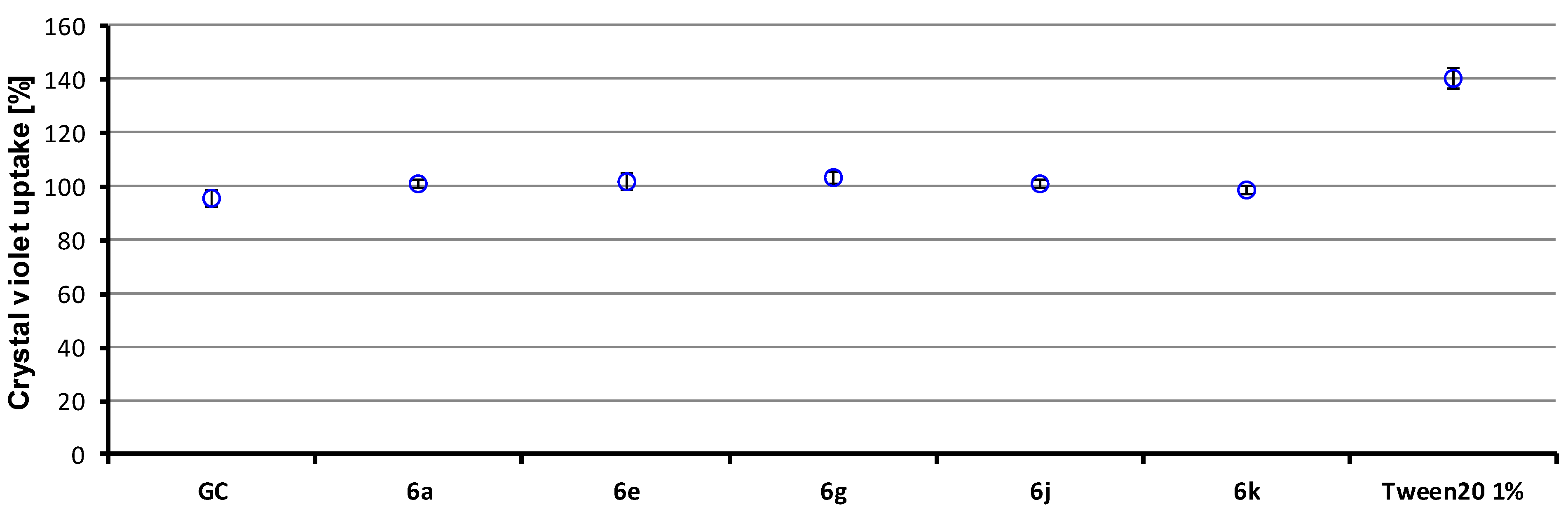
2.3.2. In Vitro Cell Viability
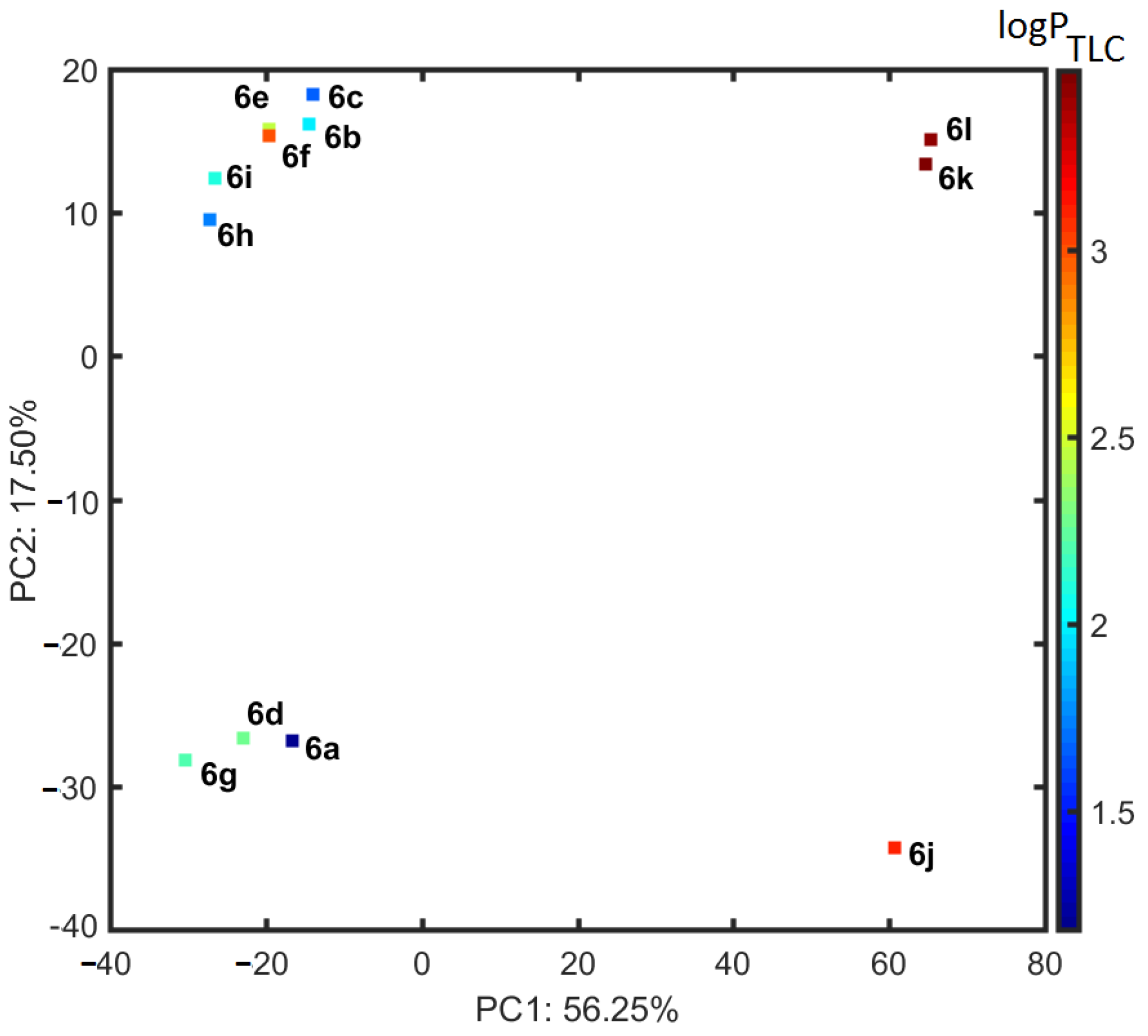
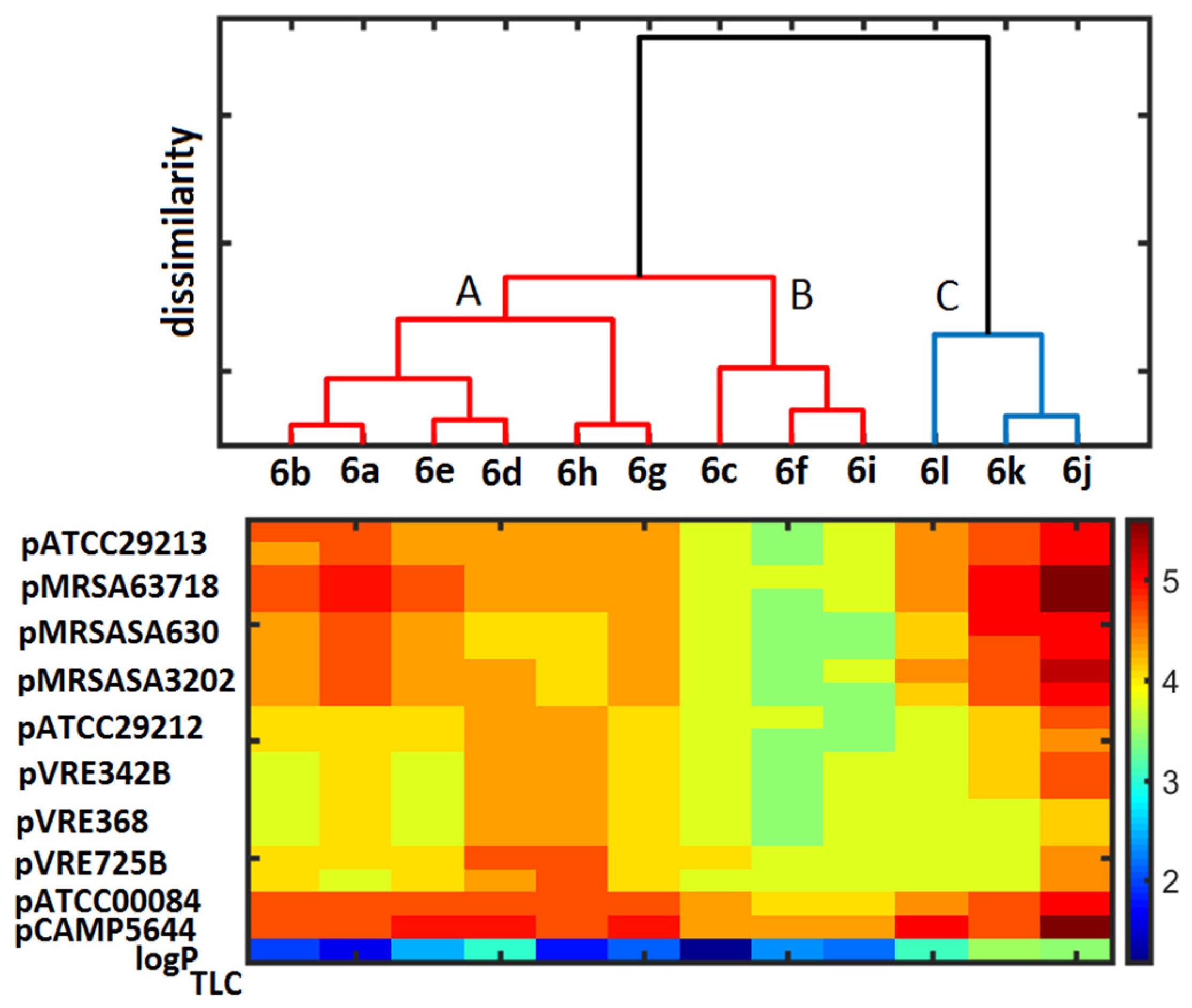
2.4. Similarity-Mediated Property Mapping
3. Materials and Methods
3.1. Chemisty
3.1.1. Synthesis of Acetanilide Derivatives 2a–l
3.1.2. Synthesis of Aniline Derivatives 3a–l
3.1.3. Synthesis of 5-Methyl-12H-Quino[3,4-b][1,4]Benzothiazine Chloride 6a–l
3.2. X-ray Structural Analysis
3.3. Thin-Layer Chromatography
3.4. Biological Evaluation
3.4.1. In Vitro Antibacterial Evaluation
3.4.2. Determination of Minimum Bactericidal Concentrations
3.4.3. MTT Assay
3.4.4. In Vitro Antimycobacterial Evaluation
3.4.5. In Vitro Cell Viability Analysis
3.5. Principal Component and Hierarchical Clustering Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Berntsen, A. Uber das Methylenblau, U. Ber. Dtsch. Chem. Ges. 1883, 16, 2896–2904. [Google Scholar]
- Gupta, R.R.; Kumar, M. Synthesis, properties and reactions of phenothiazines. In Phenothiazine and 1,4-Benzothiazines—Chemical and Biological Aspect; Gupta, R.R., Ed.; Elsevier: Amsterdam, The Netherlands, 1988; pp. 1–161. [Google Scholar]
- Silberg, I.A.; Cormos, G.; Oniciu, D.C. Retrosynthetic approach to the synthesis of phenothiazines. In Advances in Heterocyclic Chemistry; Katritzky, A.R., Ed.; Elsevier: New York, NY, USA, 2006; pp. 205–237. [Google Scholar]
- Mosnaim, A.D.; Ranade, V.V.; Wolf, M.E.; Puente, J.; Valenzuela, M.A. Phenothiazine molecule provides the basic chemical structure for various classes of pharmacotherapeutic agents. Am. J. Therapeut. 2006, 13, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Pluta, K.; Morak-Młodawska, B.; Jeleń, M. Recent progress in biological activities of synthesized phenothiazines. Eur. J. Med. Chem. 2011, 46, 3179–3189. [Google Scholar] [CrossRef]
- Aszczyszyn, A.J.; Gąsiorowski, K.; Świątek, P.; Malinka, W.; Cieślik-Boczula, K.; Petrus, J.; Matusewicz-Czarnik, B. Chemical structure of phenothiazines and their biological activity. Pharm. Rep. 2012, 64, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Aaron, J.J.; Gaye Seye, M.D.; Trajkovska, S.; Motohashi, N. Bioactive phenothiazines and benzo[a]phenothiazines: Spectroscopic studies and biological and biomedical properties and applications. In Topics in Heterocyclic Chemistry; Springer: Berlin, Germany, 2009; Volume 16, pp. 153–231. [Google Scholar]
- Pluta, K.; Jeleń, M.; Morak-Młodawska, B.; Zimecki, M.; Artym, J.; Kocięba, M.; Zaczyńska, E. Azaphenothiazines—Promising phenothiazine derivatives. An insight into nomenclature, synthesis, structure elucidation and biological properties. Eur. J. Med. Chem. 2017, 138, 774–806. [Google Scholar] [CrossRef]
- Zięba, A.; Latocha, M.; Sochanik, A. Synthesis and in vitro antiproliferative activity of novel 12(H)-quino [3,4-b][1,4]benzothiazine derivatives. Med. Chem. Res. 2013, 22, 4158–4163. [Google Scholar] [CrossRef] [Green Version]
- Zięba, A.; Czuba, Z.P.; Król, W. In vitro antimicrobial activity of novel azaphenothiazine derivatives. Acta Pol. Pharm. 2012, 69, 1149–1152. [Google Scholar]
- Barazarte, A.; Lobo, G.; Gamboa, N.; Rodrigues, J.R.; Capparelli, M.V.; Alvarez-Larena, A.; Lopez, S.E.; Charris, J.E. Synthesis and antimalarial activity of pyrazolo and pyrimido benzothiazine dioxide derivatives. Eur. J. Med. Chem. 2009, 44, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Matralis, A.N.; Kourounakis, A.P. Design of novel potent antihyperlipidemic agents with antioxidant/anti-inflammatory properties exploiting phenothiazine’s strong antioxidant activity. J. Med. Chem. 2014, 57, 2568–2581. [Google Scholar] [CrossRef]
- Wesołowska, O. Interaction of phenothiazines, stilbenes and flavonoids with multidrug resistance-associated transporters, P-glycoprotein and MRP1. Acta Biochim. Pol. 2011, 58, 433–448. [Google Scholar] [CrossRef]
- Gonzalez-Munoz, G.C.; Arce, M.P.; Lopez, B.; Perez, C.; Romero, A.; del Barrio, L.; Martín-de-Saavedra, M.D.; Egea, J.; Leon, R.; Villarroya, M.; et al. Old phenothiazine and dibenzothiadiazepine derivatives for tomorrow’s neuroprotective therapies against neurodegenerative diseases. Eur. J. Med. Chem. 2010, 45, 6152–6158. [Google Scholar] [CrossRef]
- Pluta, K.; Morak-Młodawska, B.; Jeleń, M. Synthesis and properties of diaza-, triaza- and tetraazaphenothiazines. J. Heterocycl. Chem. 2009, 45, 355–391. [Google Scholar] [CrossRef]
- Zięba, A.; Latocha, M.; Sochanik, A.; Nycz, A.; Kuśmierz, D. Synthesis and in vitro antiproliferative activity of novel phenyl ring-substituted 5-alkyl-12(H)quino [3,4-b][1,4]benzothiazine derivatives. Molecules 2016, 21, 1455. [Google Scholar] [CrossRef] [Green Version]
- Kaneko, T.; Clark, R.; Ohi, N.; Kawahara, T.; Akamatsu, H.; Ozaki, F.; Kamada, A.; Okano, K.; Yokohama, H.; Muramoto, K.; et al. Inhibitors of adhesion molecules expression; the synthesis and pharmacological properties of 10H-pyrazino [2,3-b][1,4]benzothiazine derivatives. Chem. Pharm. Bull. 2002, 50, 922–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaneko, T.; Clark, R.; Ohi, N.; Ozaki, F.; Kawahara, T.; Kamada, A.; Okano, K.; Yokohama, H.; Ohkuro, M.; Muramoto, K.; et al. Piperidine carboxylic acid derivatives of 10H-pyrazino [2,3-b][1,4]benzothiazine as orally-active adhesion molecule inhibitors. Chem. Pharm. Bull. 2004, 52, 675–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peltason, L.; Bajorath, J. Systematic computational analysis of structure-activity relationships: Concepts, challenges and recent advances. Future Med. Chem. 2009, 1, 451–466. [Google Scholar] [CrossRef] [PubMed]
- Holliday, J.D.; Salim, N.; Whittle, M.; Willett, P. Analysis and display of the size dependence of chemical similarity coefficients. J. Chem. Inf. Comput. Sci. 2003, 43, 819–828. [Google Scholar] [CrossRef]
- Lopez-Lopez, E.; Prieto-Martínez, F.D.; Medina-Franco, J.L. Activity landscape and molecular modeling to explore the SAR of dual epigenetic inhibitors: A focus on G9a and DNMT1. Molecules 2018, 23, 3282. [Google Scholar] [CrossRef] [Green Version]
- Zięba, A.; Maślankiewicz, A.; Suwińska, K. Azinyl Sulfides, Part LXIII, 1-Alkyl-4-(arylamino)quinolinium-3-thiolates and 7-Alkyl-12H-quino [3,4-b]-1,4-benzothiazinium salts. Eur. J. Org. Chem. 2000, 16, 2947–2953. [Google Scholar] [CrossRef]
- Zięba, A.; Suwińska, K. 1-Alkyl-4-(3-pyridinylamino)quinolinium-3-thiolates and their transformation into new diazaphenothiazine derivatives. Heterocycles 2006, 68, 495–503. [Google Scholar] [CrossRef]
- Zięba, A.; Sochanik, A.; Szurko, A.; Rams, M.; Mrozek, A.; Cmoch, P. Synthesis and in vitro antiproliferative activity of 5-alkyl-12(H)-quino [3,4-b][1,4]benzothiazinium salts. Eur. J. Med. Chem. 2010, 45, 4733–4739. [Google Scholar] [CrossRef] [PubMed]
- Empel, A.; Bak, A.; Kozik, V.; Latocha, M.; Cizek, A.; Jampilek, J.; Suwinska, K.; Sochanik, A.; Zieba, A. Towards property profiling: Synthesis and SAR probing of new tetracyclic diazaphenothiazine analogues. Int. J. Mol. Sci. 2021, 22, 12826. [Google Scholar] [CrossRef] [PubMed]
- Global Laboratory Standards for a Healthier World. Available online: https://clsi.org/ (accessed on 15 November 2022).
- Zadrazilova, I.; Pospisilova, S.; Pauk, K.; Imramovsky, A.; Vinsova, J.; Cizek, A.; Jampilek, J. In vitro bactericidal activity of 4- and 5-chloro-2-hydroxy-N-[1-oxo-1-(phenylamino)alkan-2-yl]benzamides against MRSA. BioMed Res. Int. 2015, 2015, 349534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oravcova, V.; Zurek, L.; Townsend, A.; Clark, A.B.; Ellis, J.C.; Cizek, A. American crows as carriers of vancomycin-resistant enterococci with van A gene. Environ. Microbiol. 2014, 16, 939–949. [Google Scholar] [CrossRef]
- Sundarsingh, J.A.T.; Ranjitha, J.; Rajan, A.; Shankar, V. Features of the biochemistry of Mycobacterium smegmatis, as a possible model for Mycobacterium tuberculosis. J. Inf. Public Health 2020, 13, 1255–1264. [Google Scholar]
- Luukinen, H.; Hammaren, M.M.; Vanha-Aho, L.M.; Parikka, M. Modeling tuberculosis in Mycobacterium marinum infected adult Zebrafish. J. Vis. Exp. 2018, 140, 58299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pankey, G.A.; Sabath, L.D. Clinical relevance of bacteriostatic versus bactericidal mechanisms of action in the treatment of Gram-positive bacterial infections. Clin. Infect. Dis. 2004, 38, 864–870. [Google Scholar] [CrossRef] [Green Version]
- Nubel, U.; Dordel, J.; Kurt, K.; Strommenger, B.; Westh, H.; Shukla, S.K.; Zemlickova, H.; Leblois, R.; Wirth, T.; Jombart, T.; et al. A timescale for evolution, population expansion, and spatial spread of an emerging clone of methicillin-resistant Staphylococcus aureus. PLoS Pathog. 2010, 6, e1000855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonec, T.; Zadrazilova, I.; Nevin, E.; Kauerova, T.; Pesko, M.; Kos, J.; Oravec, M.; Kollar, P.; Coffey, A.; O’Mahony, J.; et al. Synthesis and biological evaluation of N-alkoxyphenyl-3-hydroxynaphthalene-2-carboxanilides. Molecules 2015, 20, 9767–9787. [Google Scholar] [CrossRef] [Green Version]
- Gonec, T.; Pospisilova, T.S.; Kauerova, T.; Kos, J.; Dohanosova, J.; Oravec, M.; Kollar, P.; Coffey, A.; Liptaj, T.; Cizek, A.; et al. N-alkoxyphenylhydroxynaphthalenecarboxamides and their antimycobacterial activity. Molecules 2016, 21, 1068. [Google Scholar] [CrossRef] [Green Version]
- Imramovsky, A.; Pesko, M.; Kralova, K.; Vejsova, M.; Stolarikova, J.; Vinsova, J.; Jampilek, J. Investigating spectrum of biological activity of 4- and 5-chloro-2-hydroxy-N-[2-(arylamino)-1-alkyl-2-oxoethyl]benzamides. Molecules 2011, 16, 2414–2430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pauk, K.; Zadrazilova, I.; Imramovsky, A.; Vinsova, J.; Pokorna, M.; Masarikova, M.; Cizek, A.; Jampilek, J. New derivatives of salicylamides: Preparation and antimicrobial activity against various bacterial species. Bioorg. Med. Chem. 2013, 21, 6574–6581. [Google Scholar] [CrossRef] [PubMed]
- Pindjakova, D.; Pilarova, E.; Pauk, K.; Michnova, H.; Hosek, J.; Magar, P.; Cizek, A.; Imramovsky, A.; Jampilek, J. Study of biological activities and ADMET-related properties of salicylanilide-based peptidomimetics. Int. J. Mol. Sci. 2022, 23, 11648. [Google Scholar] [CrossRef] [PubMed]
- Portela, C.A.; Smart, K.F.; Tumanov, S.; Cook, G.M.; Villas-Boas, S.G. Global metabolic response of Enterococcus faecalis to oxygen. J. Bacteriol. 2014, 196, 2012–2022. [Google Scholar] [CrossRef] [Green Version]
- Gilmore, M.S.; Clewell, D.B.; Ike, Y.; Shankar, N. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. Available online: https://www.ncbi.nlm.nih.gov/books/NBK190432/ (accessed on 15 September 2022).
- Ramos, S.; Silva, V.; Dapkevicius, M.d.L.E.; Igrejas, G.; Poeta, P. Enterococci, from harmless bacteria to a pathogen. Microorganisms 2020, 8, 1118. [Google Scholar] [CrossRef]
- Gilmore, M.S.; Salamzade, R.; Selleck, E.; Bryan, N.; Mello, S.S.; Manson, A.L.; Earl, A.M. Genes contributing to the unique biology and intrinsic antibiotic resistance of Enterococcus faecalis. mBio 2020, 11, e02962-20. [Google Scholar] [CrossRef]
- Measuring Cell Viability/Cytotoxicity. Dojindo EU GmbH, Munich, Germany. Available online: https://www.dojindo.eu.com/Protocol/Dojindo-Cell-Proliferation-Protocol.pdf (accessed on 15 September 2022).
- Grela, E.; Kozłowska, J.; Grabowiecka, A. Current methodology of MTT assay in bacteria—A review. Acta Histochem. 2018, 120, 303–311. [Google Scholar] [CrossRef]
- Jampilek, J. Drug repurposing to overcome microbial resistance. Drug Discov. Today 2022, 27, 2028–2041. [Google Scholar] [CrossRef]
- Jampilek, J. Novel avenues for identification of new antifungal drugs and current challenges. Expert Opin. Drug Discov. 2022, 17, 949–968. [Google Scholar] [CrossRef]
- Bedaquilin. DrugBank. Available online: https://go.drugbank.com/drugs/DB08903 (accessed on 14 February 2022).
- Khan, S.R.; Singh, S.; Roy, K.K.; Akhtar, M.S.; Saxena, A.K.; Krishnan, M.Y. Biological evaluation of novel substituted chloroquinolines targeting mycobacterial ATP synthase. Int. J. Antimicrob. Agents 2013, 41, 41–46. [Google Scholar] [CrossRef]
- Kos, J.; Zadrazilova, I.; Nevin, E.; Soral, M.; Gonec, T.; Kollar, P.; Oravec, M.; Coffey, A.; O’Mahony, J.; Liptaj, T.; et al. Ring-substituted 8-hydroxyquinoline-2-carboxanilides as potential antimycobacterial agents. Bioorg. Med. Chem. 2015, 23, 4188–4196. [Google Scholar] [CrossRef] [PubMed]
- Mapari, M.; Bhole, R.P.; Khedekar, P.B.; Chikhale, R.V. Challenges in targeting mycobacterial ATP synthase: The known and beyond. J. Mol. Struct. 2022, 1247, 131331. [Google Scholar] [CrossRef]
- Paritala, H.; Carroll, K.S. New targets and inhibitors of Mycobacterial sulfur metabolism. Infect. Disord. Drug Targets 2013, 13, 85–115. [Google Scholar] [CrossRef] [PubMed]
- Mathew, B.; Ross, L.; Reynolds, R.C. A novel quinoline derivative that inhibits mycobacterial FtsZ. Tuberculosis 2013, 93, 398–400. [Google Scholar] [CrossRef] [Green Version]
- Warman, A.J.; Rito, T.S.; Fisher, N.E.; Moss, D.M.; Berry, N.G.; O’Neill, P.M.; Ward, S.A.; Biagini, G.A. Antitubercular pharmacodynamics of phenothiazines. J. Antimicrob. Chemother. 2013, 68, 869–880. [Google Scholar] [CrossRef] [Green Version]
- Kristiansen, J.E.; Dastidar, S.G.; Palchoudhuri, S.; Roy, D.S.; Das, S.; Hendricks, O.; Christensen, J.B. Phenothiazines as a solution for multidrug resistant tuberculosis: From the origin to present. Int. Microbiol. 2015, 18, 1–12. [Google Scholar]
- Suffness, M.; Douros, J. Current status of the NCI plant and animal product program. J. Nat. Prod. 1982, 45, 1–14. [Google Scholar] [CrossRef]
- Kruger, A.; Maltarollo, V.G.; Wrenger, C.; Kronenberger, T. ADME profiling in drug discovery and a new path paved on silica. In Drug Discovery and Development—New Advances; Gaitonde, V., Karmakar, P., Trivedi, A., Eds.; IntechOpen: Rijeka, Croatia, 2019; Available online: https://www.intechopen.com/chapters/66969 (accessed on 4 October 2022).
- Kerns, E.H.; Di, L. Drug-Like Properties: Concepts. Structure Design and Methods: From ADME to Toxicity Optimization; Academic Press: San Diego, CA, USA, 2008. [Google Scholar]
- Bak, A.; Polanski, J. Modeling robust QSAR 3: SOM-4D-QSAR with iterative variable elimination IVE-PLS: Application to steroid, azo dye, and benzoic acid series. J. Chem. Inf. Model. 2007, 47, 1469–1480. [Google Scholar] [CrossRef]
- Kos, J.; Kozik, V.; Pindjakova, D.; Jankech, T.; Smolinski, A.; Stepankova, S.; Hosek, J.; Oravec, M.; Jampilek, J.; Bak, A. Synthesis and hybrid SAR property modeling of novel cholinesterase inhibitors. Int. J. Mol. Sci. 2021, 22, 3444. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. SHELXT-integrated space-group and crystal-structure determination. Acta Cryst. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- National Committee for Clinical Laboratory Standards. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed.; M07; NCCLS: Wayne, PA, USA, 2018. [Google Scholar]
- Schwalbe, R.; Steele-Moore, L.; Goodwin, A.C. Antimicrobial Susceptibility Testing Protocols; CRC Press: Boca Raton, FL, USA, 2007. [Google Scholar]
- Scandorieiro, S.; de Camargo, L.C.; Lancheros, C.A.; Yamada-Ogatta, S.F.; Nakamura, C.V.; de Oliveira, A.G.; Andrade, C.G.; Duran, N.; Nakazato, G.; Kobayashi, R.K. Synergistic and additive effect of oregano essential oil and biological silver nanoparticles against multidrug-resistant bacterial strains. Front. Microbiol. 2016, 7, 760. [Google Scholar] [CrossRef] [PubMed]
- Guimaraes, A.C.; Meireles, L.M.; Lemos, M.F.; Guimaraes, M.C.C.; Endringer, D.C.; Fronza, M.; Scherer, R. Antibacterial activity of terpenes and terpenoids present in essential oils. Molecules 2019, 24, 2471. [Google Scholar] [CrossRef] [PubMed]

| No. | MIC [μM] MBC [μM] | IC50 [µM] NHDF | logPTLC | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SA | MRSA1 | MRSA2 | MRSA3 | EF | VRE1 | VRE2 | VRE3 | MS | MM | |||
| 6a | 22.3 22.3 | 22.3 22.3 | 22.3 22.3 | 11.1 11.1 | 89.1 89.1 | 89.1 89.1 | 89.1 89.1 | 89.1 178 | 22.3 – | 22.3 – | >100 | 1.65 |
| 6b | 22.3 44.6 | 44.6 44.6 | 44.6 44.6 | 22.3 22.3 | 89.1 89.1 | 178 178 | 178 178 | 89.1 89.1 | 22.3 – | 22.3 – | 65.5 | 1.99 |
| 6c | 178 178 | 178 178 | 178 178 | 178 178 | 178 178 | 178 178 | 178 178 | 89.1 178 | 44.6 – | 44.6 – | >100 | 1.18 |
| 6d | 44.8 44.8 | 44.8 44.8 | 89.6 89.6 | 44.8 44.8 | 44.8 44.8 | 44.8 44.8 | 44.8 44.8 | 22.4 44.8 | 22.4 – | 11.2 – | >100 | 2.98 |
| 6e | 44.8 44.8 | 44.8 44.8 | 44.8 44.8 | 22.4 22.4 | 89.6 89.6 | 179 179 | 179 179 | 89.6 89.6 | 22.4 – | 11.2 – | >100 | 2.47 |
| 6f | 359 359 | 359 359 | 359 359 | 179 359 | 179 359 | 359 359 | 359 359 | 179 179 | 89.6 – | 44.8 – | >100 | 2.29 |
| 6g | 45.1 45.1 | 45.1 45.1 | 45.1 45.1 | 45.1 45.1 | 90.1 90.1 | 90.1 90.1 | 90.1 90.1 | 90.1 90.1 | 22.5 – | 11.3 – | >100 | 2.11 |
| 6h | 45.1 45.1 | 90.1 90.1 | 90.1 90.1 | 45.1 45.1 | 45.1 45.1 | 45.1 45.1 | 45.1 45.1 | 22.5 22.5 | 22.5 – | 22.5 – | >100 | 1.74 |
| 6i | 180 180 | 180 361 | 361 361 | 180 180 | 361 361 | 180 180 | 180 180 | 180 180 | 90.1 – | 45.1 – | 47.5 | 2.20 |
| 6j | 9.83 9.83 | 4.91 9.83 | 9.83 9.83 | 2.46 2.46 | 19.7 39.3 | 19.7 19.7 | 78.6 78.6 | 39.3 39.3 | 9.83 – | 2.50 – | 37.7 | 3.41 |
| 6k | 19.7 19.7 | 19.7 19.7 | 9.83 19.7 | 9.83 9.83 | 78.6 78.6 | 78.6 78.6 | 157 157 | 157 157 | 19.7 – | 19.7 – | >100 | 3.48 |
| 6l | 39.3 39.3 | 39.3 78.6 | 78.6 78.6 | 39.3 39.3 | 157 157 | 157 157 | 157 157 | 157 157 | 39.3 – | 9.83 – | >100 | 3.10 |
| AMP | 5.72 5.72 | – | – | – | 2.81 2.81 | 11.5 11.5 | 11.5 11.5 | 11.5 11.5 | – | – | – | – |
| OXA | 1.25 1.25 | 79.8 79.8 | 29.7 29.7 | 9.9 9.9 | – | – | – | – | – | – | – | – |
| TTC | 4.52 NT | 36.1 NT | 36.1 NT | >72.1 NT | – | – | – | – | – | – | – | – |
| CPX | 1.51 NT | 48.3 NT | 24.2 NT | 24.2 NT | – | – | – | – | – | – | – | – |
| INH | – | – | – | – | – | – | – | – | 117 – | 467 – | – | – |
| RIF | – | – | – | – | – | – | – | – | 19.4 – | 2.43 – | – | – |
| No. | Conc. | S. aureus Respiration Inhibition [%] |
|---|---|---|
| 6a | 2× MIC (2× MBC) | 97.7 |
| 6e | 2× MIC (2× MBC) | 97.4 |
| 6g | 2× MIC (2× MBC) | 96.8 |
| 6j | 0.5× MIC (0.5× MBC) | 91.3 |
| 6k | 2× MIC (2× MBC) | 97.6 |
| APM | 16× MIC (>16× MBC) | 81.9 |
| CPX | 32× MIC (32× MBC) | 96.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kisiel-Nawrot, E.; Pindjakova, D.; Latocha, M.; Bak, A.; Kozik, V.; Suwinska, K.; Sochanik, A.; Cizek, A.; Jampilek, J.; Zięba, A. Design, Synthesis and Antimicrobial Properties of New Tetracyclic Quinobenzothiazine Derivatives. Int. J. Mol. Sci. 2022, 23, 15078. https://doi.org/10.3390/ijms232315078
Kisiel-Nawrot E, Pindjakova D, Latocha M, Bak A, Kozik V, Suwinska K, Sochanik A, Cizek A, Jampilek J, Zięba A. Design, Synthesis and Antimicrobial Properties of New Tetracyclic Quinobenzothiazine Derivatives. International Journal of Molecular Sciences. 2022; 23(23):15078. https://doi.org/10.3390/ijms232315078
Chicago/Turabian StyleKisiel-Nawrot, Ewa, Dominika Pindjakova, Malgorzata Latocha, Andrzej Bak, Violetta Kozik, Kinga Suwinska, Aleksander Sochanik, Alois Cizek, Josef Jampilek, and Andrzej Zięba. 2022. "Design, Synthesis and Antimicrobial Properties of New Tetracyclic Quinobenzothiazine Derivatives" International Journal of Molecular Sciences 23, no. 23: 15078. https://doi.org/10.3390/ijms232315078
APA StyleKisiel-Nawrot, E., Pindjakova, D., Latocha, M., Bak, A., Kozik, V., Suwinska, K., Sochanik, A., Cizek, A., Jampilek, J., & Zięba, A. (2022). Design, Synthesis and Antimicrobial Properties of New Tetracyclic Quinobenzothiazine Derivatives. International Journal of Molecular Sciences, 23(23), 15078. https://doi.org/10.3390/ijms232315078







