An Integration of Linkage Mapping and GWAS Reveals the Key Genes for Ear Shank Length in Maize
Abstract
:1. Introduction
2. Results
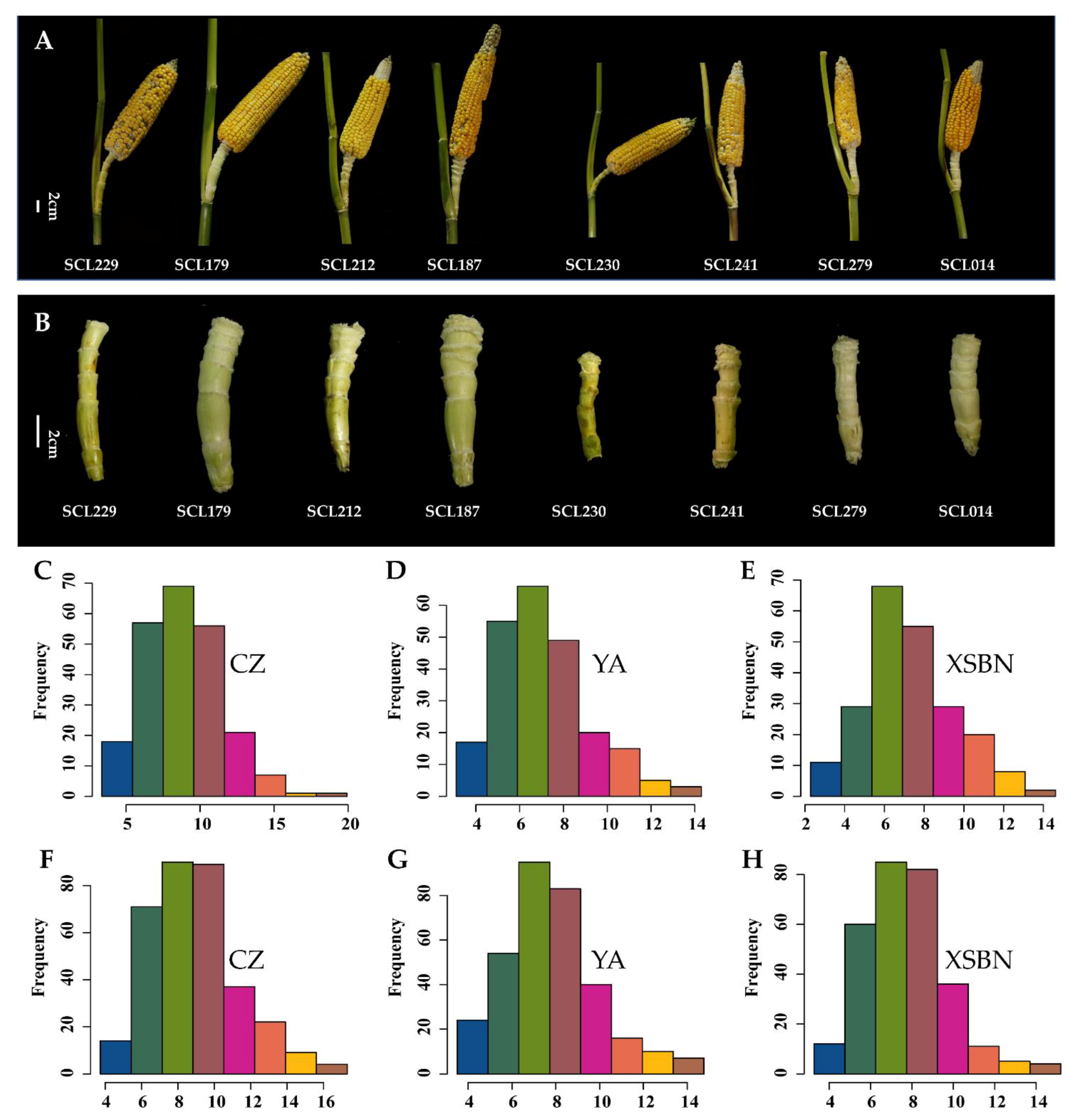
2.1. Evaluation of ESL Phenotype
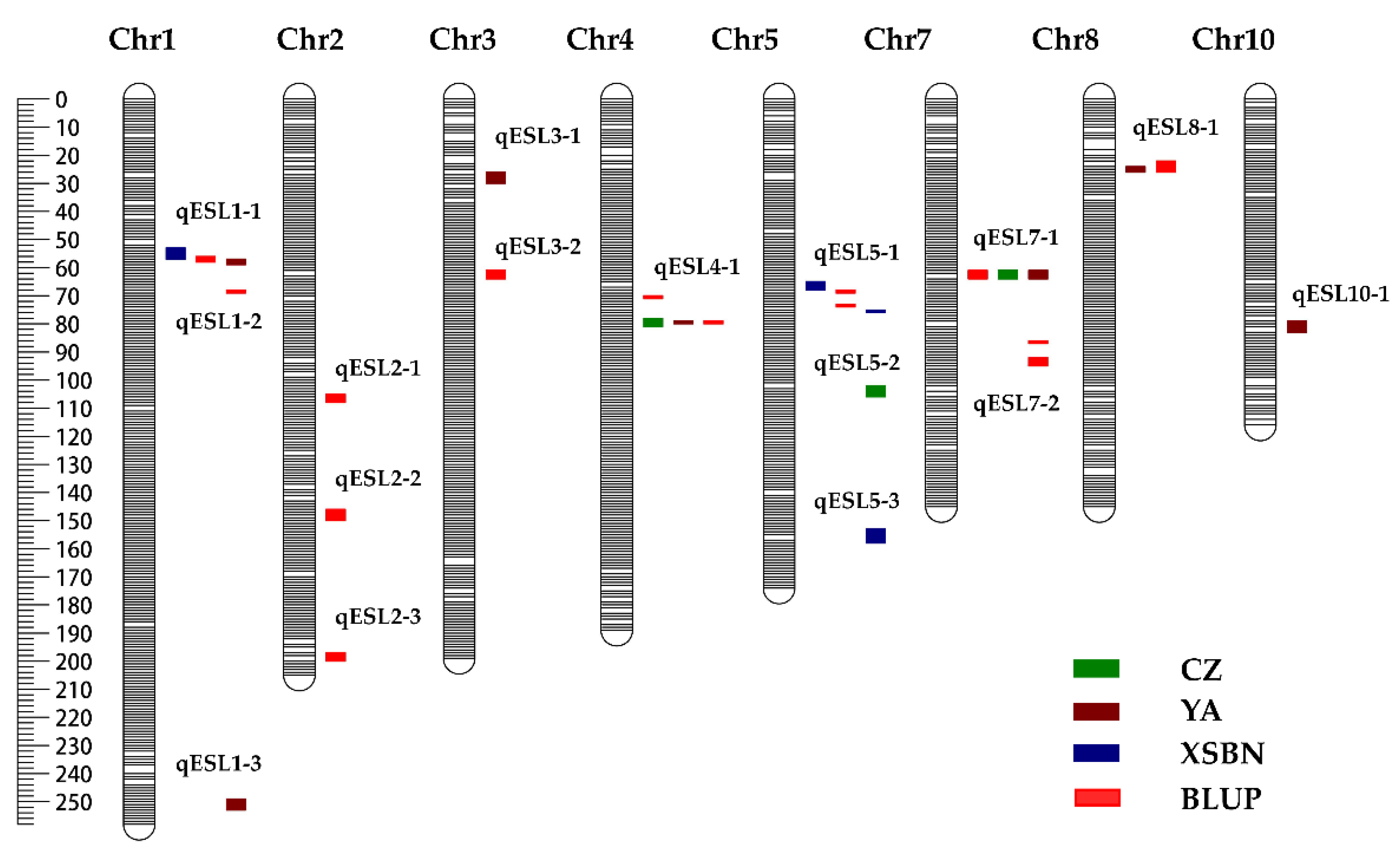
2.2. QTL Responsible for ESL
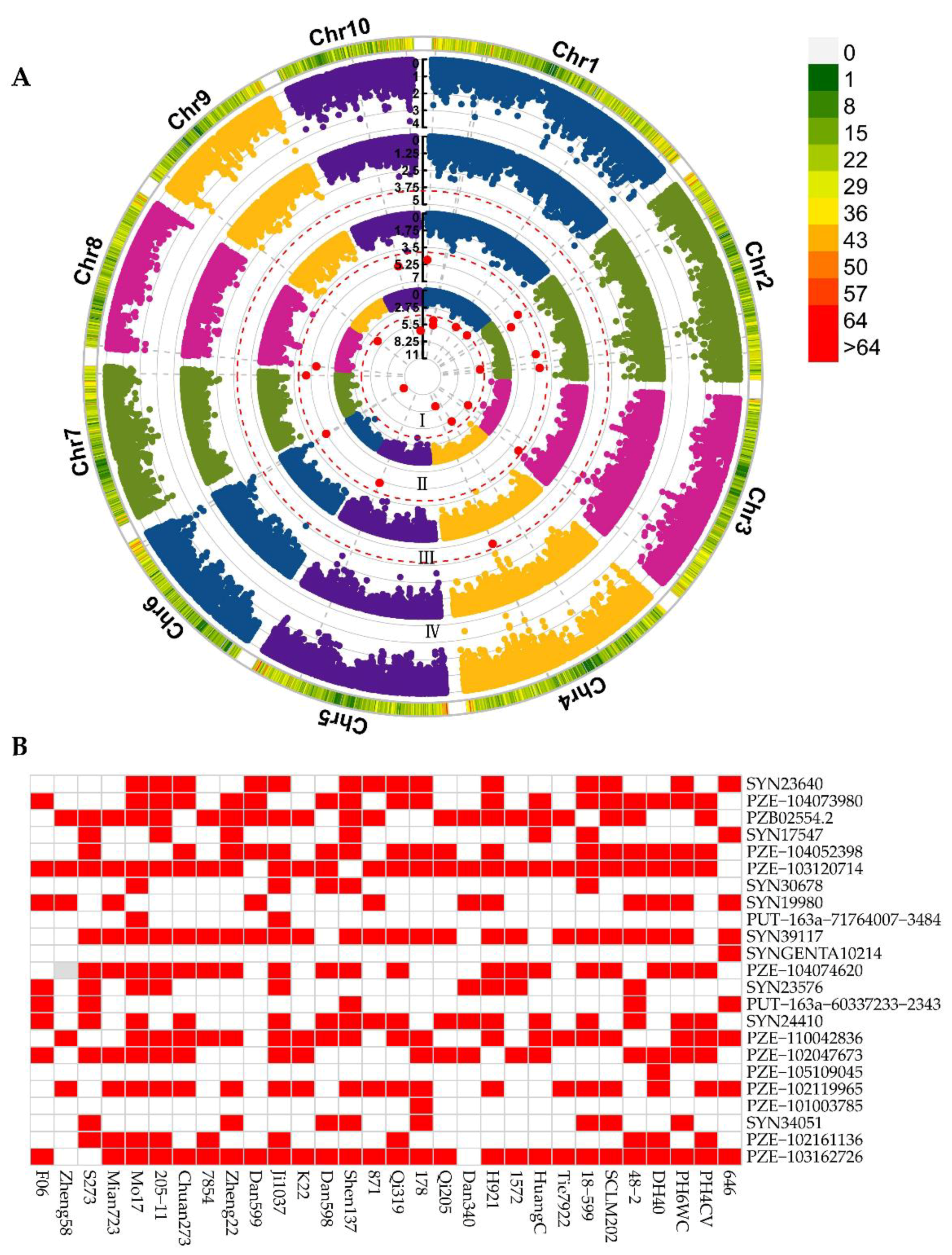
2.3. Significant Associations for ESL
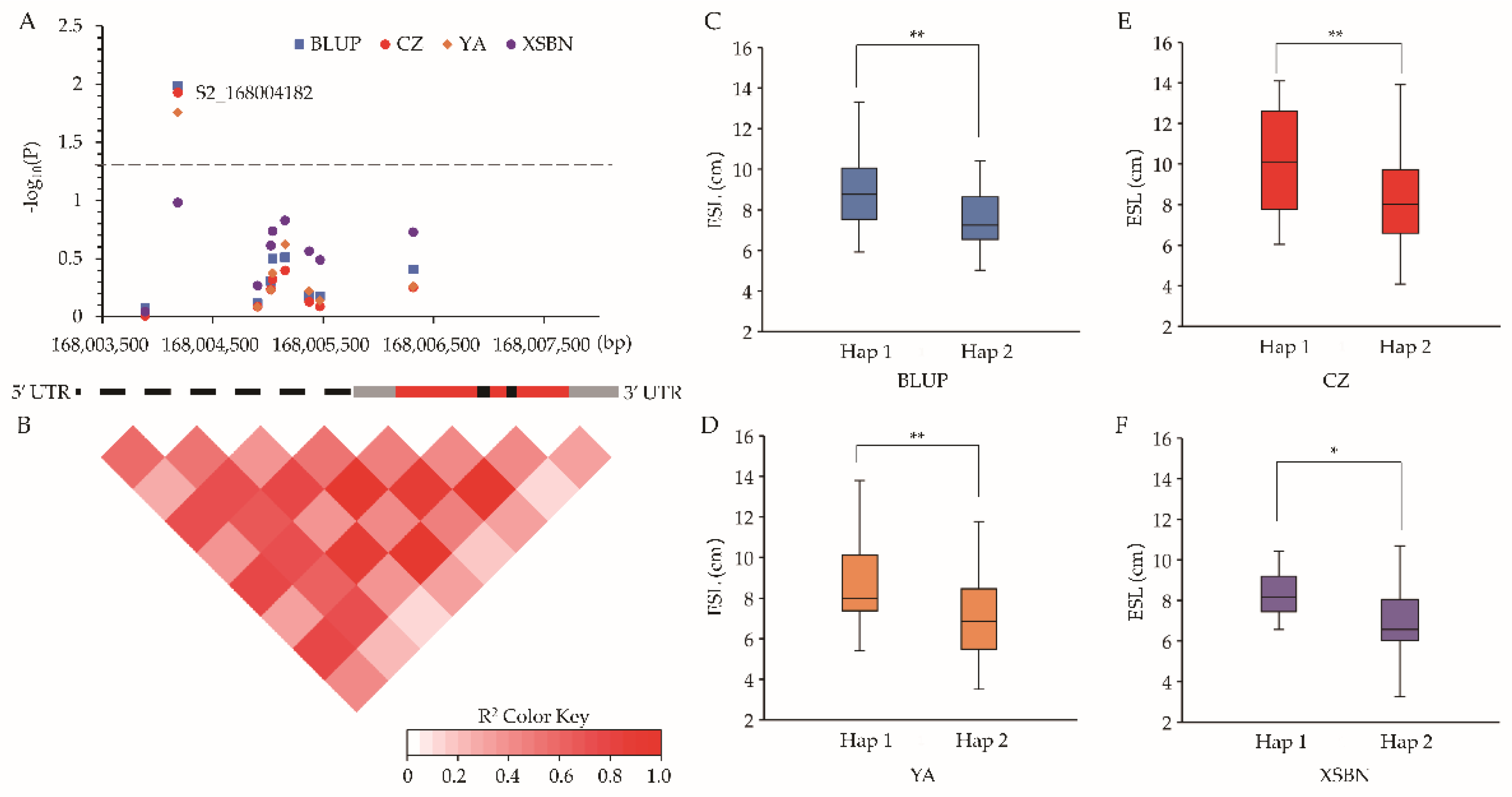
2.4. Candidate Genes Co-Localized by QTL Mapping and GWAS
2.5. Intragenic Variations Affecting ESL
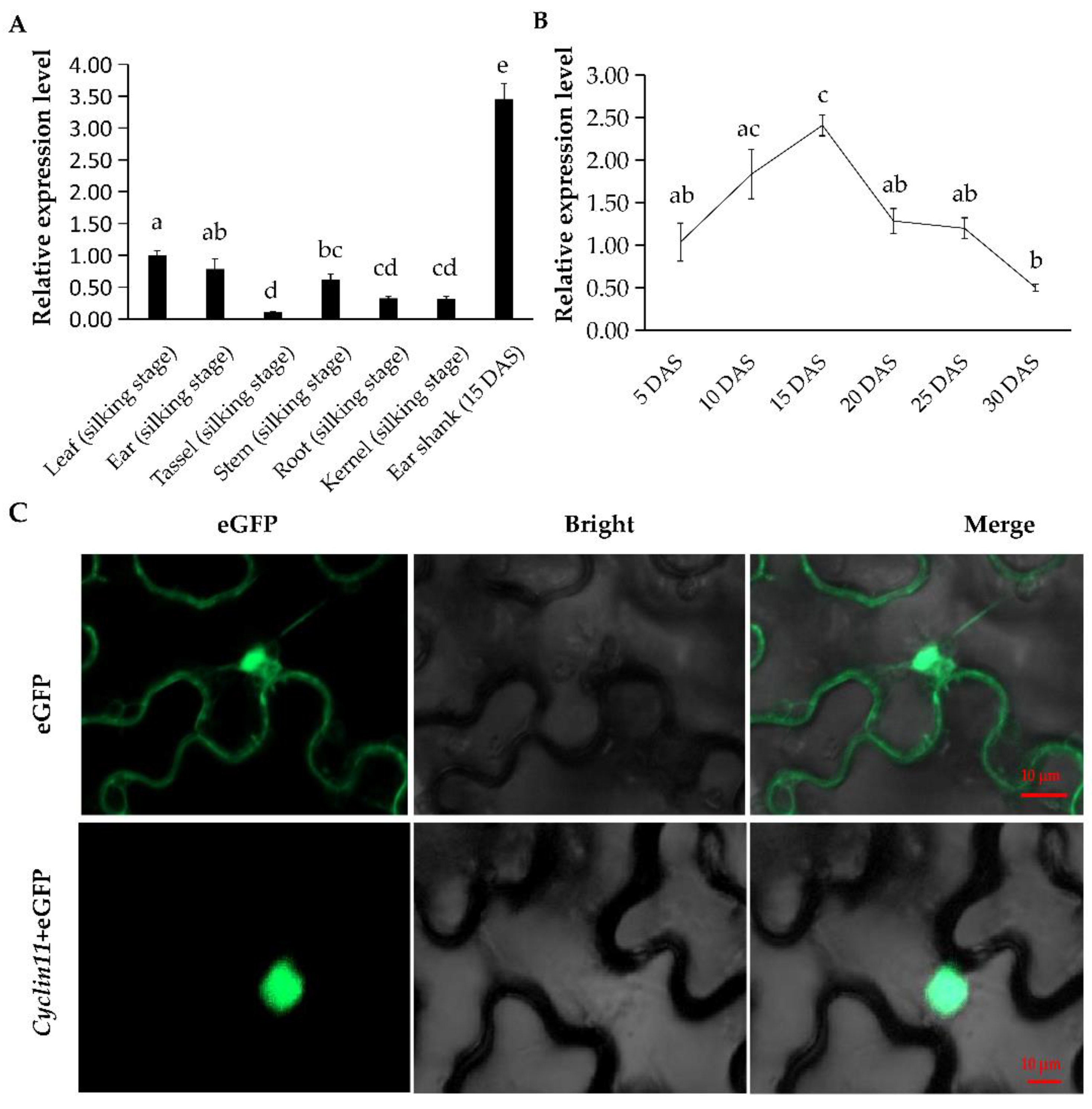
2.6. Temporal and Spatial Expression Profile of Cyclin11
3. Discussion
3.1. Combining the IBM Syn 10 DH Population and Association Panel to Detect the Genetic Basis of Maize ESL
3.2. Genetic Architecture of Maize ESL
3.3. Candidate Genes Involved in Maize ESL
4. Materials and Methods
4.1. Plant Materials and Field Trials
4.2. Sample Collection, Phenotyping and Statistical Analyses
4.3. QTL Mapping
4.4. Genome-Wide Association Study
4.5. DEG Identification
4.6. Gene-Based Association Analysis
4.7. RT-qPCR
4.8. Subcellular Localization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenge of Feeding 9 Billion People. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [Green Version]
- Evans, L.; Dunstone, R.; Rawson, H.M.; Williams, R. The Phloem of the Wheat Stem in Relation to Requirements for Assimilate by The Ear. Afr. J. Mar. Sci. 1970, 23, 743–752. [Google Scholar] [CrossRef]
- Nátrová, Z. Anatomical Characteristics of the Uppermost Internode of Winter Wheat Genoypes Differing in Stem Length. Biol. Plant 1991, 33, 491–494. [Google Scholar] [CrossRef]
- Borrás, L.; Slafer, G.A.; Otegui, M.E. Seed Dry Weight Response to Source–Sink Manipulations in Wheat, Maize and Soybean: A Quantitative Reappraisal. Field Crops Res. 2004, 86, 131–146. [Google Scholar] [CrossRef]
- Borrás, L.; Westgate, M.E.; Otegui, M.E. Control of Kernel Weight and Kernel Water Relations by Post-Flowering Source-Sink Ratio in Maize. Ann. Bot. 2003, 91, 857–867. [Google Scholar] [CrossRef]
- Ursache, R.; Miyashima, S.; Chen, Q.; Vatén, A.; Nakajima, K.; Carlsbecker, A.; Zhao, Y.; Helariutta, Y.; Dettmer, J. Tryptophan-Dependent Auxin Biosynthesis Is Required for HD-ZIP III-Mediated Xylem Patterning. Development 2014, 141, 1250–1259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donner, T.J.; Sherr, I.; Scarpella, E. Regulation of Preprocambial Cell State Acquisition by Auxin Signaling in Arabidopsis Leaves. Development 2009, 136, 3235–3246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramachandran, P.; Wang, G.; Augstein, F.; de Vries, J.; Carlsbecker, A. Continuous Root Xylem Formation and Vascular Acclimation to Water Deficit Involves Endodermal ABA Signalling via MiR165. Development 2018, 145, dev159202. [Google Scholar] [CrossRef] [Green Version]
- Etchells, J.P.; Provost, C.M.; Turner, S.R. Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene Signalling. PLoS Genet. 2012, 8, e1002997. [Google Scholar] [CrossRef] [Green Version]
- Hansen, M.; Kraft, T.; Ganestam, S.; Säll, T.; Nilsson, N.O. Linkage Disequilibrium Mapping of the Bolting Gene in Sea Beet Using AFLP Markers. Genet. Res. 2001, 77, 61–66. [Google Scholar] [CrossRef]
- Xiong, C.-Y.; Gong, Q.-Y.; Pei, H.; Liao, C.-J.; Yang, R.-C.; Li, G.-K.; Huang, J. Comparative Transcriptome Analysis Reveals Regulatory Networks during the Maize Ear Shank Elongation Process. Int. J. Mol. Sci. 2021, 22, 7029. [Google Scholar] [CrossRef]
- Liu, M.; He, W.; Zhang, A.; Zhang, L.; Sun, D.; Gao, Y.; Ni, P.; Ma, X.; Cui, Z.; Ruan, Y. Genetic Analysis of Maize Shank Length by QTL Mapping in Three Recombinant Inbred Line Populations. Plant Sci. 2021, 303, 110767. [Google Scholar] [CrossRef]
- Sun, G.; Zhang, X.; Duan, H.; Gao, J.; Li, N.; Su, P.; Xie, H.; Li, W.; Fu, Z.; Huang, Y.; et al. Dissection of the Genetic Architecture of Peduncle Vascular Bundle-Related Traits in Maize by a Genome-Wide Association Study. Plant Biotechnol. J. 2022, 20, 1042–1053. [Google Scholar] [CrossRef]
- Yan, Y.; Christensen, S.; Isakeit, T.; Engelberth, J.; Meeley, R.; Hayward, A.; Emery, R.J.N.; Kolomiets, M.V. Disruption of OPR7 and OPR8 Reveals the Versatile Functions of Jasmonic Acid in Maize Development and Defense. Plant Cell 2012, 24, 1420–1436. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Tan, X.; Yang, Y.; Liu, P.; Zhang, X.; Zhang, Y.; Wang, L.; Hu, Y.; Ma, L.; Li, Z.; et al. Analysis of the Genetic Architecture of Maize Kernel Size Traits by Combined Linkage and Association Mapping. Plant Biotechnol. J. 2020, 18, 207–221. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Zhang, S.; Shah, T.; Xie, C.; Hao, Z.; Li, X.; Farkhari, M.; Ribaut, J.-M.; Cao, M.; Rong, T.; et al. Joint Linkage-Linkage Disequilibrium Mapping Is a Powerful Approach to Detecting Quantitative Trait Loci Underlying Drought Tolerance in Maize. Proc. Natl. Acad. Sci. USA 2010, 107, 19585–19590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Guan, Z.; Li, Z.; Liu, P.; Ma, L.; Zhang, Y.; Pan, L.; He, S.; Zhang, Y.; Li, P.; et al. A Combination of Linkage Mapping and GWAS Brings New Elements on the Genetic Basis of Yield-Related Traits in Maize across Multiple Environments. Theor. Appl. Genet. 2020, 133, 2881–2895. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, S.; Zhou, Z.; Wang, S.; Dong, C.; Mu, C.; Song, Y.; Ma, P.; Li, C.; Wang, Z.; et al. Linkage Mapping and Genome-Wide Association Reveal Candidate Genes Conferring Thermotolerance of Seed-Set in Maize. J. Exp. Bot. 2019, 70, 4849–4864. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Warburton, M.; Crouch, J. Association Mapping for Enhancing Maize (Zea mays L.) Genetic Improvement. Crop Sci. 2011, 51, 433–449. [Google Scholar] [CrossRef]
- Tian, F.; Bradbury, P.J.; Brown, P.J.; Hung, H.; Sun, Q.; Flint-Garcia, S.; Rocheford, T.R.; McMullen, M.D.; Holland, J.B.; Buckler, E.S. Genome-Wide Association Study of Leaf Architecture in the Maize Nested Association Mapping Population. Nat. Genet. 2011, 43, 159–162. [Google Scholar] [CrossRef]
- Yang, N.; Lu, Y.; Yang, X.; Huang, J.; Zhou, Y.; Ali, F.; Wen, W.; Liu, J.; Li, J.; Yan, J. Genome Wide Association Studies Using a New Nonparametric Model Reveal the Genetic Architecture of 17 Agronomic Traits in an Enlarged Maize Association Panel. PLoS Genet. 2014, 10, e1004573. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh Bi, I.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A Unified Mixed-Model Method for Association Mapping That Accounts for Multiple Levels of Relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]
- Giraud, H.; Lehermeier, C.; Bauer, E.; Falque, M.; Segura, V.; Bauland, C.; Camisan, C.; Campo, L.; Meyer, N.; Ranc, N.; et al. Linkage Disequilibrium with Linkage Analysis of Multiline Crosses Reveals Different Multiallelic QTL for Hybrid Performance in the Flint and Dent Heterotic Groups of Maize. Genetics 2014, 198, 1717–1734. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Xia, A.; Zhang, A.; Luo, J.; Yang, X.; Zhang, L.; Ruan, Y.; He, Y. Linkage Mapping Combined with Association Analysis Reveals QTL and Candidate Genes for Three Husk Traits in Maize. Theor. Appl. Genet. 2018, 131, 2131–2144. [Google Scholar] [CrossRef]
- Zhang, C.; Zhou, Z.; Yong, H.; Zhang, X.; Hao, Z.; Zhang, F.; Li, M.; Zhang, D.; Li, X.; Wang, Z.; et al. Analysis of the Genetic Architecture of Maize Ear and Grain Morphological Traits by Combined Linkage and Association Mapping. Theor. Appl. Genet. 2017, 130, 1011–1029. [Google Scholar] [CrossRef]
- Vikram, P.; Swamy, B.M.; Dixit, S.; Ahmed, H.U.; Teresa Sta Cruz, M.; Singh, A.K.; Kumar, A. QDTY1.1, a Major QTL for Rice Grain Yield under Reproductive-Stage Drought Stress with a Consistent Effect in Multiple Elite Genetic Backgrounds. BMC Genet. 2011, 12, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Karki, S.; Biswal, A.K.; Lin, H.-C.; Dionora, M.J.; Rizal, G.; Yin, X.; Schuler, M.L.; Hughes, T.; Fouracre, J.P.; et al. Candidate Regulators of Early Leaf Development in Maize Perturb Hormone Signalling and Secondary Cell Wall Formation When Constitutively Expressed in Rice. Sci. Rep. 2017, 7, 4535. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, R.; Kumar, G.; Arya, P.; Jaswal, R.; Jain, P.; Singh, K.; Sharma, T.R. Genome-Wide Analysis and Expression Profiling of Rice Hybrid Proline-Rich Proteins in Response to Biotic and Abiotic Stresses, and Hormone Treatment. Plants 2019, 8, 343. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Liu, C.; Zhang, D.; He, C.; Zhang, J.; Li, Z. Effects of Maize Organ-Specific Drought Stress Response on Yields from Transcriptome Analysis. BMC Plant Biol. 2019, 19, 335. [Google Scholar] [CrossRef] [Green Version]
- Hou, F.; Zhou, X.; Liu, P.; Yuan, G.; Zou, C.; Lübberstedt, T.; Pan, G.; Ma, L.; Shen, Y. Genetic Dissection of Maize Seedling Traits in an IBM Syn10 DH Population under the Combined Stress of Lead and Cadmium. Mol. Genet. Genom. 2021, 296, 1057–1070. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, Y.; Guan, Z.; Liu, P.; He, Y.; Zou, C.; Li, P.; Gao, S.; Peng, H.; Yang, C.; et al. Combined Linkage Mapping and Association Analysis Reveals Genetic Control of Maize Kernel Moisture Content. Physiol. Plant 2020, 170, 508–518. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, N.; Vanhaeren, H.; Inzé, D. Leaf Size Control: Complex Coordination of Cell Division and Expansion. Trends Plant Sci. 2012, 17, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Meyerowitz, E.M. Control of Cell Division Patterns in Developing Shoots and Flowers of Arabidopsis Thaliana. Cold Spring Harb. Sym. 1997, 62, 369–375. [Google Scholar]
- Shiu, S.H.; Bleecker, A.B. Receptor-like Kinases from Arabidopsis Form a Monophyletic Gene Family Related to Animal Receptor Kinases. Proc. Natl. Acad. Sci. USA 2001, 98, 10763–10768. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, S.; Nguyen, B.; Wasti, S.D.; Xu, G. Plant Leucine-Rich Repeat Receptor Kinase (LRR-RK): Structure, Ligand Perception, and Activation Mechanism. Molecules 2019, 24, 3081. [Google Scholar] [CrossRef] [Green Version]
- Katsir, L.; Davies, K.A.; Bergmann, D.C.; Laux, T. Peptide Signaling in Plant Development. Curr. Biol. 2011, 21, R356–R364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boron, A.K.; van Orden, J.; Nektarios Markakis, M.; Mouille, G.; Adriaensen, D.; Verbelen, J.-P.; Höfte, H.; Vissenberg, K. Proline-Rich Protein-like PRPL1 Controls Elongation of Root Hairs in Arabidopsis Thaliana. J. Exp. Bot. 2014, 65, 5485–5495. [Google Scholar] [CrossRef] [Green Version]
- Zdanio, M.; Boron, A.K.; Balcerowicz, D.; Schoenaers, S.; Markakis, M.N.; Mouille, G.; Pintelon, I.; Suslov, D.; Gonneau, M.; Höfte, H.; et al. The Proline-Rich Family Protein EXTENSIN33 Is Required for Etiolated Arabidopsis Thaliana Hypocotyl Growth. Plant Cell Physiol. 2020, 61, 1191–1203. [Google Scholar] [CrossRef]
- Menges, M.; Pavesi, G.; Morandini, P.; Bögre, L.; Murray, J.A.H. Genomic Organization and Evolutionary Conservation of Plant D-Type Cyclins. Plant Physiol. 2007, 145, 1558–1576. [Google Scholar] [CrossRef] [Green Version]
- Collins, C.; Maruthi, N.M.; Jahn, C.E. CYCD3 D-Type Cyclins Regulate Cambial Cell Proliferation and Secondary Growth in Arabidopsis. J. Exp. Bot. 2015, 66, 4595–4606. [Google Scholar] [CrossRef] [Green Version]
- Forzani, C.; Aichinger, E.; Sornay, E.; Willemsen, V.; Laux, T.; Dewitte, W.; Murray, J.A.H. WOX5 Suppresses CYCLIN D Activity to Establish Quiescence at the Center of the Root Stem Cell Niche. Curr. Biol. 2014, 24, 1939–1944. [Google Scholar] [CrossRef] [PubMed]
- Hussain, T.; Tausend, P.; Graham, G.; Ho, J. Registration of IBM2 SYN10 Doubled Haploid Mapping Population of Maize. J. Plant Regist. 2007, 1, 81. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, H.; Li, L.; Lan, H.; Ren, Z.; Liu, D.; Wu, L.; Liu, H.; Jaqueth, J.; Li, B.; et al. Characterizing the Population Structure and Genetic Diversity of Maize Breeding Germplasm in Southwest China Using Genome-Wide SNP Markers. BMC Genom. 2016, 17, 697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Liu, P.; Zhang, X.; Zheng, Q.; Chen, M.; Ge, F.; Li, Z.; Sun, W.; Guan, Z.; Liang, T.; et al. Multi-Locus Genome-Wide Association Study Reveals the Genetic Architecture of Stalk Lodging Resistance-Related Traits in Maize. Front. Plant Sci. 2018, 9, 611. [Google Scholar] [CrossRef]
- Knapp, S.J.; Stroup, W.W.; Ross, W.M. Exact Confidence Intervals for Heritability on a Progeny Mean Basis1. Crop Sci. 1985, 25, 192–194. [Google Scholar] [CrossRef]
- Liu, H.; Niu, Y.; Gonzalez-Portilla, P.J.; Zhou, H.; Wang, L.; Zuo, T.; Qin, C.; Tai, S.; Jansen, C.; Shen, Y.; et al. An Ultra-High-Density Map as a Community Resource for Discerning the Genetic Basis of Quantitative Traits in Maize. BMC Genom. 2015, 16, 1078. [Google Scholar] [CrossRef] [Green Version]
- WinQTLCart. Available online: http://statgen.ncsu.edu/qtlcart/WQTLCart.htm (accessed on 20 July 2022).
- Churchill, G.A.; Doerge, R.W. Empirical Threshold Values for Quantitative Trait Mapping. Genetics 1994, 138, 963–971. [Google Scholar] [CrossRef]
- Ma, L.; Guan, Z.; Zhang, Z.; Zhang, X.; Zhang, Y.; Zou, C.; Peng, H.; Pan, G.; Lee, M.; Shen, Y.; et al. Identification of Quantitative Trait Loci for Leaf-Related Traits in an IBM Syn10 DH Maize Population across Three Environments. Plant Breed. 2018, 137, 127–138. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Xue, Y.; Guo, Z.; Li, W.; Tang, J. Genome-Wide Association Study Identifies Candidate Genes for Starch Content Regulation in Maize Kernels. Front. Plant Sci. 2016, 7, 1046. [Google Scholar] [CrossRef]
- Gawel, N.J.; Jarret, R.L. A Modified CTAB DNA Extraction Procedure ForMusa AndIpomoea. Plant Mol. Biol. Rep. 1991, 9, 262–266. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA Extraction Protocol for Plants Containing High Polysaccharide and Polyphenol Components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Hou, F.; Liu, K.; Zhang, N.; Zou, C.; Yuan, G.; Gao, S.; Zhang, M.; Pan, G.; Ma, L.; Shen, Y. Association Mapping Uncovers Maize ZmbZIP107 Regulating Root System Architecture and Lead Absorption under Lead Stress. Front. Plant Sci. 2022, 13, 1015151. [Google Scholar] [CrossRef] [PubMed]

| Population | |||||||
|---|---|---|---|---|---|---|---|
| IBM Syn 10 DH Population | Association Panel | ||||||
| Env. | CZ | YA | XSBN | CZ | YA | XSBN | |
| Mean (cm) | 8.88 | 7.12 | 7.28 | 8.93 | 7.77 | 7.69 | |
| SD | 2.55 | 2.09 | 2.26 | 2.45 | 2.16 | 2.02 | |
| Max (cm) | 19.93 | 14.3 | 14.61 | 17.3 | 14.77 | 15.19 | |
| Min (cm) | 3.37 | 3.10 | 2.28 | 3.74 | 3.50 | 3.27 | |
| CV (%) | 28.73 | 29.33 | 31.05 | 27.47 | 27.74 | 26.37 | |
| Mean ± SD (cm) | B73 | 6.63 ± 0.64 | 3.5 ± 0.62 | 10.18 ± 0.98 | - | - | - |
| Mo17 | 11.14 ± 1.03 | 9.16 ± 0.61 | 9.42 ± 1.37 | - | - | - | |
| p value | 0.006 ** | 0.0007 *** | 0.56 | - | - | - | |
| F value E * | 231.074 ** | 161.205 ** | |||||
| F value G * | 12.348 ** | 10.946 ** | |||||
| F value G × E * | 2.368 ** | 2.011 ** | |||||
| Heritability | 0.81 | 0.82 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, Z.; Xi, N.; Liu, H.; Liu, P.; Xiang, C.; Zhang, C.; Zou, C.; Cheng, X.; Yu, H.; Zhang, M.; et al. An Integration of Linkage Mapping and GWAS Reveals the Key Genes for Ear Shank Length in Maize. Int. J. Mol. Sci. 2022, 23, 15073. https://doi.org/10.3390/ijms232315073
Liang Z, Xi N, Liu H, Liu P, Xiang C, Zhang C, Zou C, Cheng X, Yu H, Zhang M, et al. An Integration of Linkage Mapping and GWAS Reveals the Key Genes for Ear Shank Length in Maize. International Journal of Molecular Sciences. 2022; 23(23):15073. https://doi.org/10.3390/ijms232315073
Chicago/Turabian StyleLiang, Zhenjuan, Na Xi, Hao Liu, Peng Liu, Chenchaoyang Xiang, Chen Zhang, Chaoying Zou, Xuyujuan Cheng, Hong Yu, Minyan Zhang, and et al. 2022. "An Integration of Linkage Mapping and GWAS Reveals the Key Genes for Ear Shank Length in Maize" International Journal of Molecular Sciences 23, no. 23: 15073. https://doi.org/10.3390/ijms232315073
APA StyleLiang, Z., Xi, N., Liu, H., Liu, P., Xiang, C., Zhang, C., Zou, C., Cheng, X., Yu, H., Zhang, M., Chen, Z., Pan, G., Yuan, G., Gao, S., Ma, L., & Shen, Y. (2022). An Integration of Linkage Mapping and GWAS Reveals the Key Genes for Ear Shank Length in Maize. International Journal of Molecular Sciences, 23(23), 15073. https://doi.org/10.3390/ijms232315073






