Pleiotropic Effects of APOB Variants on Lipid Profiles, Metabolic Syndrome, and the Risk of Diabetes Mellitus
Abstract
:1. Introduction
2. Results
2.1. Selection of Candidate Nonsynonymous Mutations of the APOB Gene
2.2. Genotype–Phenotype Association Analysis of APOB Nonsynonymous Mutations with Lipid Profiles and Metabolic Syndrome
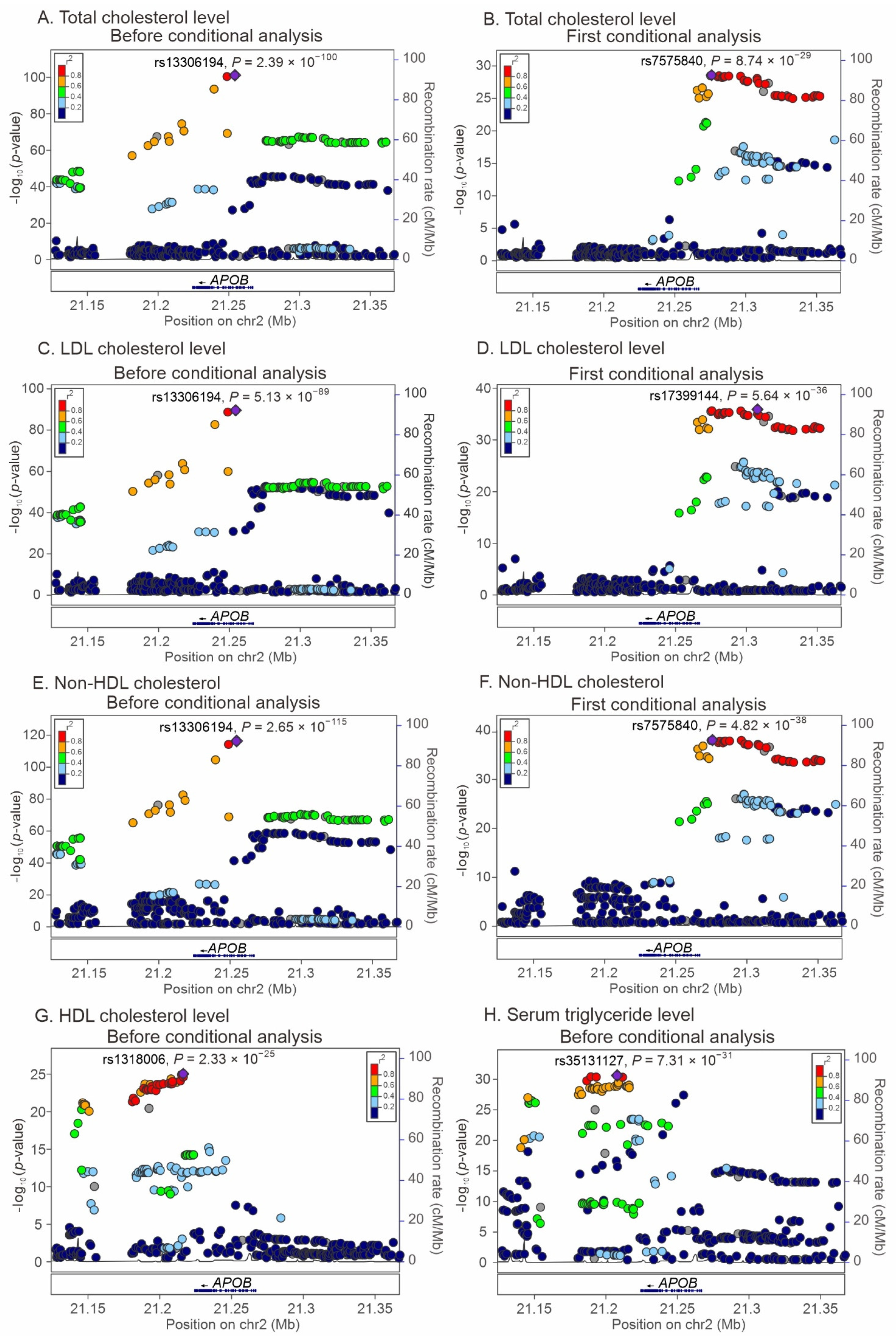
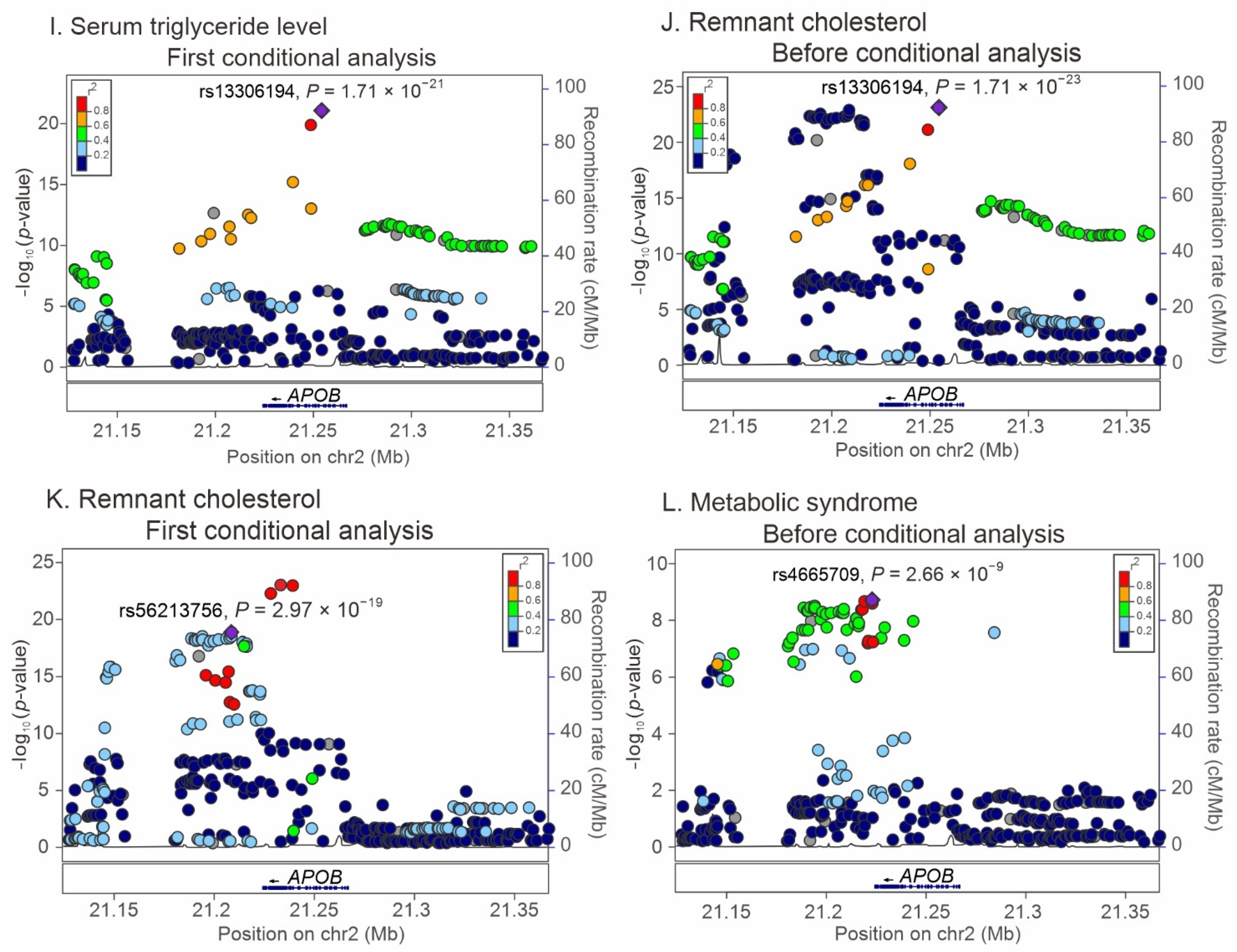
2.3. Regional Association Studies with Conditional Analyses for the Association of APOB Locus Variants with Lipid Profiles and Metabolic Syndrome
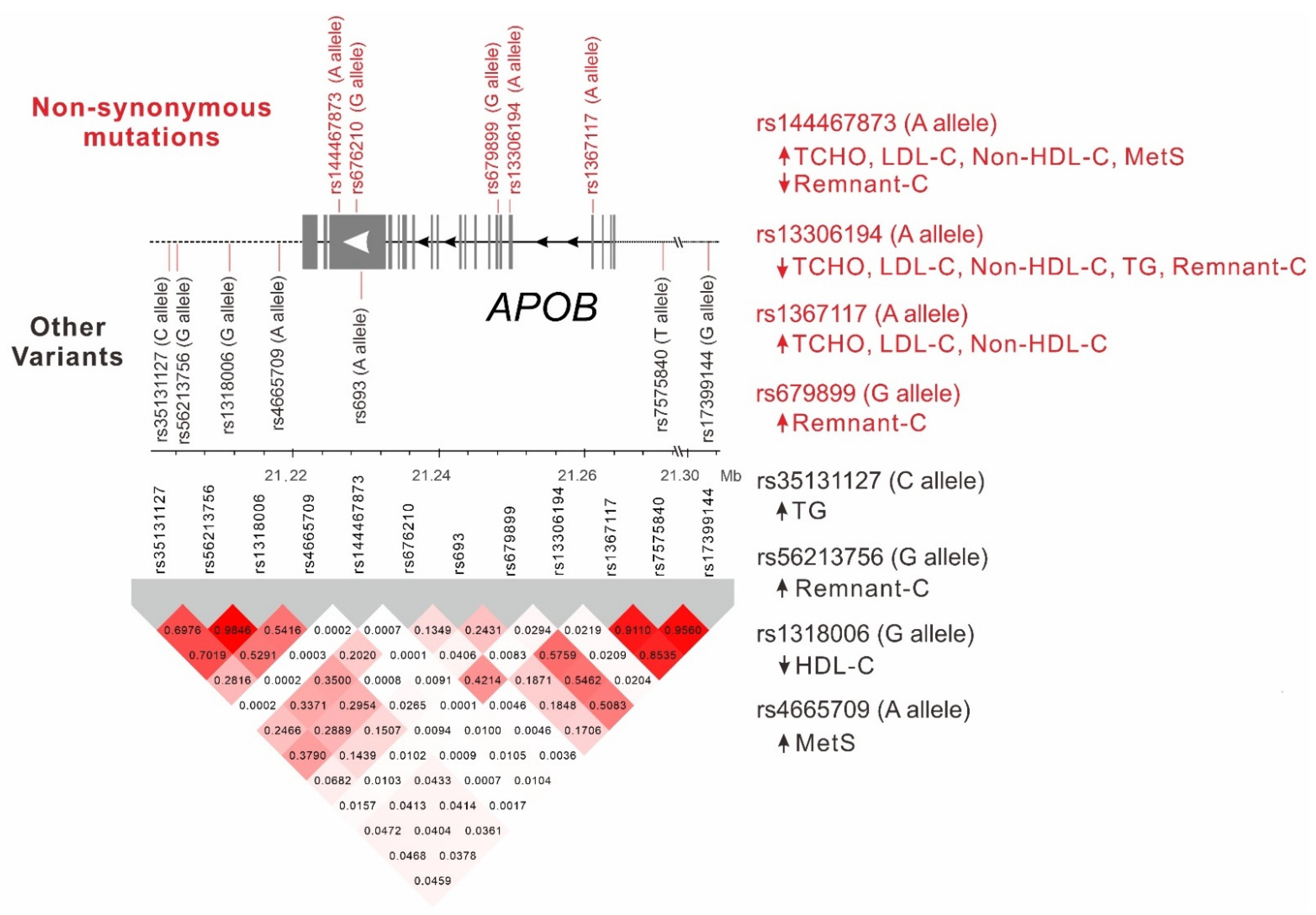
2.4. LD between APOB Nonsynonymous Mutations and Lead SNPs
2.5. Genotype–Phenotype Association Analysis of Lead SNPs Downstream of the APOB Gene with Lipid Profiles and Metabolic Syndrome
2.6. Stepwise Linear Regression Analysis for Lipid Profiles
2.7. Logistic Regression Analysis for Metabolic Syndrome
2.8. MR Analysis for the APOB Variants and WGRSs for Causal Relationship between LDL Cholesterol Levels and DM
3. Discussion
3.1. Ethnic-Specific APOB Variants for Total, LDL, and Non-HDL Cholesterol Levels in Taiwanese Individuals
3.2. Variants in the 3′ Intergenic Region of APOB as Genetic Determinants of Triglyceride and HDL and Remnant Cholesterol Levels
3.3. APOB Variants as Genetic Determinants of Remnant Cholesterol Levels
3.4. Novel APOB Variants Determining Metabolic Syndrom
3.5. MR Analyses
3.6. Limitations
4. Materials and Methods
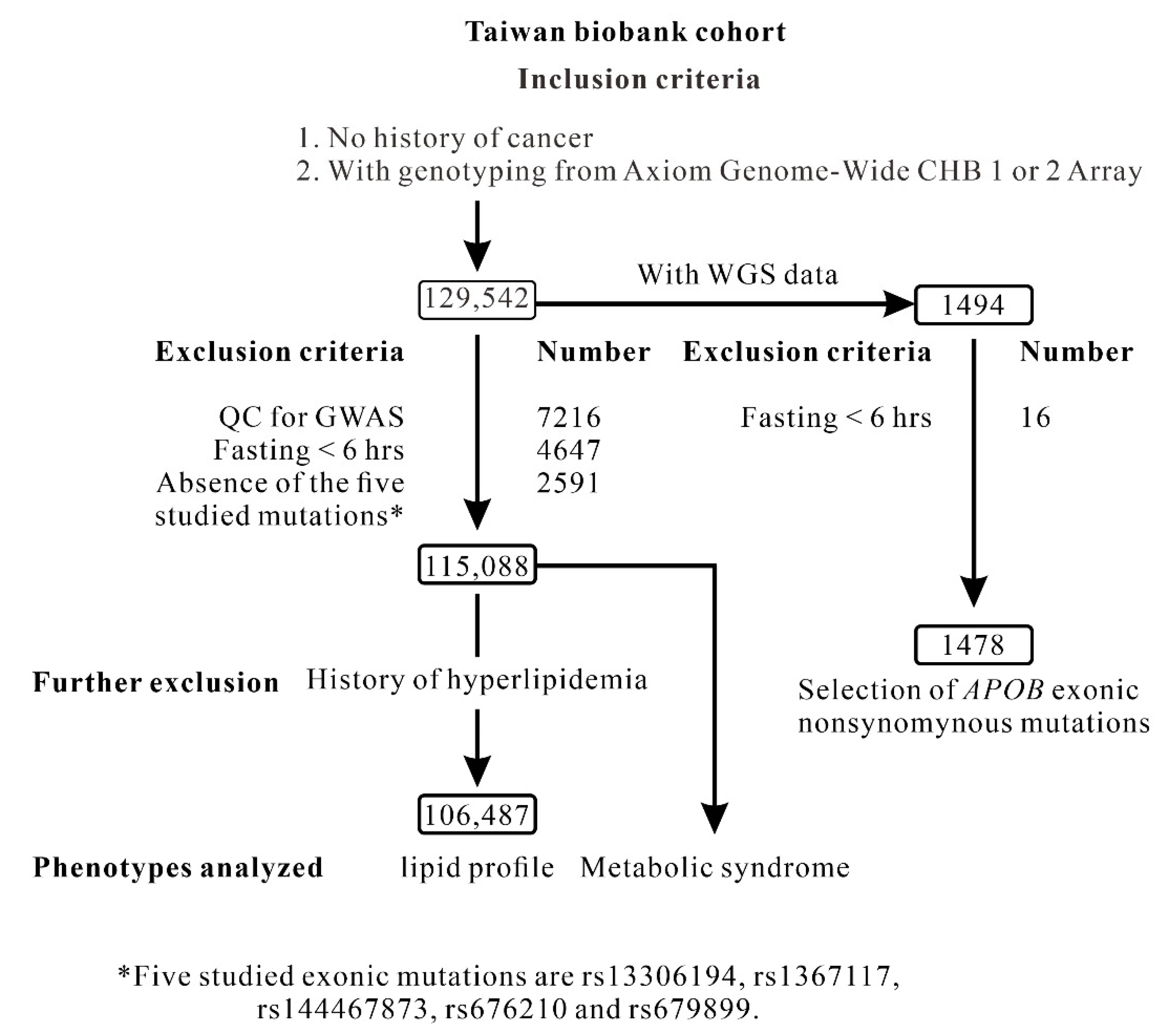
4.1. TWB Cohort
4.2. Clinical Phenotypes and Laboratory Examinations
4.3. Regional Association Analysis for the WGS Data
4.4. Regional Association Analysis for the GWAS Data
4.5. Statistical Analysis
4.6. MR Analysis for the APOB Variants and WGRSs with the Risk of DM through Their Associations with LDL-C Levels
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Behbodikhah, J.; Ahmed, S.; Elyasi, A.; Kasselman, L.J.; De Leon, J.; Glass, A.D.; Reiss, A.B. Apolipoprotein B and Cardiovascular Disease: Biomarker and Potential Therapeutic Target. Metabolites 2021, 11, 690. [Google Scholar] [CrossRef] [PubMed]
- Herscovitz, H.; Derksen, A.; Walsh, M.T.; McKnight, C.J.; Gantz, D.L.; Hadzopoulou-Cladaras, M.; Zannis, V.; Curry, C.; Small, D.M. The N-terminal 17% of apoB binds tightly and irreversibly to emulsions modeling nascent very low density lipoproteins. J. Lipid Res. 2001, 42, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Morita, S.Y. Metabolism and Modification of Apolipoprotein B-Containing Lipoproteins Involved in Dyslipidemia and Atherosclerosis. Biol. Pharm. Bull. 2016, 39, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Sniderman, A.D.; Thanassoulis, G.; Glavinovic, T.; Navar, A.M.; Pencina, M.; Catapano, A.; Ference, B.A. Apolipoprotein B Particles and Cardiovascular Disease: A Narrative Review. JAMA Cardiol. 2019, 4, 1287–1295. [Google Scholar] [CrossRef]
- Borén, J.; Williams, K.J. The central role of arterial retention of cholesterol-rich apolipoprotein-B-containing lipoproteins in the pathogenesis of atherosclerosis: A triumph of simplicity. Curr. Opin. Lipidol. 2016, 27, 473–483. [Google Scholar] [CrossRef]
- Benn, M.; Nordestgaard, B.G.; Jensen, G.B.; Tybjaerg-Hansen, A. Improving prediction of ischemic cardiovascular disease in the general population using apolipoprotein B: The Copenhagen City Heart Study. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 661–670. [Google Scholar] [CrossRef] [Green Version]
- Emerging Risk Factors, C.; Di Angelantonio, E.; Sarwar, N.; Perry, P.; Kaptoge, S.; Ray, K.K.; Thompson, A.; Wood, A.M.; Lewington, S.; Sattar, N.; et al. Major lipids, apolipoproteins, and risk of vascular disease. JAMA 2009, 302, 1993–2000. [Google Scholar] [CrossRef] [Green Version]
- De Graaf, J.; Couture, P.; Sniderman, A. ApoB in Clinical Care; Springer: Berlin, Germany, 2015. [Google Scholar]
- Blackhart, B.D.; Ludwig, E.M.; Pierotti, V.R.; Caiati, L.; Onasch, M.A.; Wallis, S.C.; Powell, L.; Pease, R.; Knott, T.J.; Chu, M.L.; et al. Structure of the human apolipoprotein B gene. J. Biol. Chem. 1986, 261, 15364–15367. [Google Scholar] [CrossRef]
- Andersen, L.H.; Miserez, A.R.; Ahmad, Z.; Andersen, R.L. Familial defective apolipoprotein B-100: A review. J. Clin. Lipidol. 2016, 10, 1297–1302. [Google Scholar] [CrossRef]
- Huang, C.C.; Niu, D.M.; Charng, M.J. Genetic Analysis in a Taiwanese Cohort of 750 Index Patients with Clinically Diagnosed Familial Hypercholesterolemia. J. Atheroscler. Thromb. 2022, 29, 639–653. [Google Scholar] [CrossRef]
- Teng, Y.N.; Pan, J.P.; Chou, S.C.; Tai, D.Y.; Lee-Chen, G.J. Familial defective apolipoprotein B-100: Detection and haplotype analysis of the Arg(3500)-->Gln mutation in hyperlipidemic Chinese. Atherosclerosis 2000, 152, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Tokgozoglu, L.; Kayikcioglu, M. Familial Hypercholesterolemia: Global Burden and Approaches. Curr. Cardiol. Rep. 2021, 23, 151. [Google Scholar] [CrossRef] [PubMed]
- Scheinfeldt, L.B.; Tishkoff, S.A. Recent human adaptation: Genomic approaches, interpretation and insights. Nat. Rev. Genet. 2013, 14, 692–702. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.H.; Yang, J.H.; Chiang, C.W.K.; Hsiung, C.N.; Wu, P.E.; Chang, L.C.; Chu, H.W.; Chang, J.; Song, I.W.; Yang, S.L.; et al. Population structure of Han Chinese in the modern Taiwanese population based on 10,000 participants in the Taiwan Biobank project. Hum. Mol. Genet. 2016, 25, 5321–5331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juang, J.J.; Lu, T.P.; Su, M.W.; Lin, C.W.; Yang, J.H.; Chu, H.W.; Chen, C.H.; Hsiao, Y.W.; Lee, C.Y.; Chiang, L.M.; et al. Rare variants discovery by extensive whole-genome sequencing of the Han Chinese population in Taiwan: Applications to cardiovascular medicine. J. Adv. Res. 2021, 30, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.J.; Lee, Y.; Park, S.; Kang, S.M.; Jang, Y.; Lee, J.H.; Lee, S.H. Rare and common variants of APOB and PCSK9 in Korean patients with extremely low low-density lipoprotein-cholesterol levels. PLoS ONE 2017, 12, e0186446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, S.; Kim, Y.J.; Han, S.; Hwang, M.Y.; Shin, D.M.; Park, M.Y.; Lu, Y.; Yoon, K.; Jang, H.M.; Kim, Y.K.; et al. The Korea Biobank Array: Design and Identification of Coding Variants Associated with Blood Biochemical Traits. Sci. Rep. 2019, 9, 1382. [Google Scholar] [CrossRef] [Green Version]
- Tang, C.S.; Zhang, H.; Cheung, C.Y.; Xu, M.; Ho, J.C.; Zhou, W.; Cherny, S.S.; Zhang, Y.; Holmen, O.; Au, K.W.; et al. Exome-wide association analysis reveals novel coding sequence variants associated with lipid traits in Chinese. Nat. Commun. 2015, 6, 10206. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.H.; Lee, Y.T.; Hsu, H.C.; Hsieh, L.L.; Wen, M.S.; Chern, M.S.; Wu, D. Further characterization of apolipoprotein B genetic variations in Taiwanese. Hum. Biol. 2001, 73, 451–460. [Google Scholar] [CrossRef]
- Yang, K.C.; Su, Y.N.; Shew, J.Y.; Yang, K.Y.; Tseng, W.K.; Wu, C.C.; Lee, Y.T. LDLR and ApoB are major genetic causes of autosomal dominant hypercholesterolemia in a Taiwanese population. J. Formos. Med. Assoc. Taiwan Yi Zhi 2007, 106, 799–807. [Google Scholar] [CrossRef]
- Chiou, K.R.; Charng, M.J. Genetic diagnosis of familial hypercholesterolemia in Han Chinese. J. Clin. Lipidol. 2016, 10, 490–496. [Google Scholar] [CrossRef] [PubMed]
- Miserez, A.R.; Keller, U. Differences in the phenotypic characteristics of subjects with familial defective apolipoprotein B-100 and familial hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 1995, 15, 1719–1729. [Google Scholar] [CrossRef] [PubMed]
- Pimstone, S.N.; Defesche, J.C.; Clee, S.M.; Bakker, H.D.; Hayden, M.R.; Kastelein, J.J. Differences in the phenotype between children with familial defective apolipoprotein B-100 and familial hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 1997, 17, 826–833. [Google Scholar] [CrossRef]
- Benn, M.; Watts, G.F.; Tybjærg-Hansen, A.; Nordestgaard, B.G. Mutations causative of familial hypercholesterolaemia: Screening of 98 098 individuals from the Copenhagen General Population Study estimated a prevalence of 1 in 217. Eur. Heart J. 2016, 37, 1384–1394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peloso, G.M.; Auer, P.L.; Bis, J.C.; Voorman, A.; Morrison, A.C.; Stitziel, N.O.; Brody, J.A.; Khetarpal, S.A.; Crosby, J.R.; Fornage, M.; et al. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am. J. Hum. Genet. 2014, 94, 223–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, H.; Damcott, C.M.; Rampersaud, E.; Pollin, T.I.; Horenstein, R.B.; McArdle, P.F.; Peyser, P.A.; Bielak, L.F.; Post, W.S.; Chang, Y.P.; et al. Familial defective apolipoprotein B-100 and increased low-density lipoprotein cholesterol and coronary artery calcification in the old order amish. Arch. Intern. Med. 2010, 170, 1850–1855. [Google Scholar] [CrossRef]
- Hoffmann, T.J.; Theusch, E.; Haldar, T.; Ranatunga, D.K.; Jorgenson, E.; Medina, M.W.; Kvale, M.N.; Kwok, P.Y.; Schaefer, C.; Krauss, R.M.; et al. A large electronic-health-record-based genome-wide study of serum lipids. Nat. Genet. 2018, 50, 401–413. [Google Scholar] [CrossRef]
- Sakaue, S.; Kanai, M.; Tanigawa, Y.; Karjalainen, J.; Kurki, M.; Koshiba, S.; Narita, A.; Konuma, T.; Yamamoto, K.; Akiyama, M.; et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 2021, 53, 1415–1424. [Google Scholar] [CrossRef]
- Spracklen, C.N.; Chen, P.; Kim, Y.J.; Wang, X.; Cai, H.; Li, S.; Long, J.; Wu, Y.; Wang, Y.X.; Takeuchi, F.; et al. Association analyses of East Asian individuals and trans-ancestry analyses with European individuals reveal new loci associated with cholesterol and triglyceride levels. Hum. Mol. Genet. 2017, 26, 1770–1784. [Google Scholar] [CrossRef] [Green Version]
- Guindo-Martínez, M.; Amela, R.; Bonàs-Guarch, S.; Puiggròs, M.; Salvoro, C.; Miguel-Escalada, I.; Carey, C.E.; Cole, J.B.; Rüeger, S.; Atkinson, E.; et al. The impact of non-additive genetic associations on age-related complex diseases. Nat. Commun. 2021, 12, 2436. [Google Scholar] [CrossRef]
- Gallois, A.; Mefford, J.; Ko, A.; Vaysse, A.; Julienne, H.; Ala-Korpela, M.; Laakso, M.; Zaitlen, N.; Pajukanta, P.; Aschard, H. A comprehensive study of metabolite genetics reveals strong pleiotropy and heterogeneity across time and context. Nat. Commun. 2019, 10, 4788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gudjonsson, A.; Gudmundsdottir, V.; Axelsson, G.T.; Gudmundsson, E.F.; Jonsson, B.G.; Launer, L.J.; Lamb, J.R.; Jennings, L.L.; Aspelund, T.; Emilsson, V.; et al. A genome-wide association study of serum proteins reveals shared loci with common diseases. Nat. Commun. 2022, 13, 480. [Google Scholar] [CrossRef] [PubMed]
- Klarin, D.; Damrauer, S.M.; Cho, K.; Sun, Y.V.; Teslovich, T.M.; Honerlaw, J.; Gagnon, D.R.; DuVall, S.L.; Li, J.; Peloso, G.M.; et al. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat. Genet. 2018, 50, 1514–1523. [Google Scholar] [CrossRef] [PubMed]
- Mäkelä, K.M.; Seppälä, I.; Hernesniemi, J.A.; Lyytikäinen, L.P.; Oksala, N.; Kleber, M.E.; Scharnagl, H.; Grammer, T.B.; Baumert, J.; Thorand, B.; et al. Genome-wide association study pinpoints a new functional apolipoprotein B variant influencing oxidized low-density lipoprotein levels but not cardiovascular events: AtheroRemo Consortium. Circulation. Cardiovasc. Genet. 2013, 6, 73–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mäkelä, K.M.; Traylor, M.; Oksala, N.; Kleber, M.E.; Seppälä, I.; Lyytikäinen, L.P.; Hernesniemi, J.A.; Kähönen, M.; Bevan, S.; Rothwell, P.M.; et al. Association of the novel single-nucleotide polymorphism which increases oxidized low-density lipoprotein levels with cerebrovascular disease events. Atherosclerosis 2014, 234, 214–217. [Google Scholar] [CrossRef] [PubMed]
- Richardson, T.G.; Leyden, G.M.; Wang, Q.; Bell, J.A.; Elsworth, B.; Davey Smith, G.; Holmes, M.V. Characterising metabolomic signatures of lipid-modifying therapies through drug target mendelian randomisation. PLoS Biol. 2022, 20, e3001547. [Google Scholar] [CrossRef]
- Wojcik, G.L.; Graff, M.; Nishimura, K.K.; Tao, R.; Haessler, J.; Gignoux, C.R.; Highland, H.M.; Patel, Y.M.; Sorokin, E.P.; Avery, C.L.; et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature 2019, 570, 514–518. [Google Scholar] [CrossRef] [PubMed]
- Hayat, M.; Kerr, R.; Bentley, A.R.; Rotimi, C.N.; Raal, F.J.; Ramsay, M. Genetic associations between serum low LDL-cholesterol levels and variants in LDLR, APOB, PCSK9 and LDLRAP1 in African populations. PLoS ONE 2020, 15, e0229098. [Google Scholar] [CrossRef] [Green Version]
- Long, T.; Lu, S.; Li, H.; Lin, R.; Qin, Y.; Li, L.; Chen, L.; Zhang, L.; Lv, Y.; Liang, D.; et al. Association of APOB and LIPC polymorphisms with type 2 diabetes in Chinese Han population. Gene 2018, 672, 150–155. [Google Scholar] [CrossRef]
- Yoshida, T.; Kato, K.; Yokoi, K.; Watanabe, S.; Metoki, N.; Satoh, K.; Aoyagi, Y.; Nishigaki, Y.; Nozawa, Y.; Yamada, Y. Association of candidate gene polymorphisms with chronic kidney disease in Japanese individuals with hypertension. Hypertens. Res. Off. J. Jpn. Soc. Hypertens. 2009, 32, 411–418. [Google Scholar] [CrossRef]
- Pérez-Campo, F.M.; De Castro-Orós, I.; Noriega, A.; Cofán, M.; Lamiquiz-Moneo, I.; Cenarro, A.; Ros, E.; Civeira, F.; Pocoví, M.; Rodríguez-Rey, J.C. Functional analysis of new 3’ untranslated regions genetic variants in genes associated with genetic hypercholesterolemias. J. Clin. Lipidol. 2017, 11, 532–542. [Google Scholar] [CrossRef] [PubMed]
- Chapman, M.J.; Ginsberg, H.N.; Amarenco, P.; Andreotti, F.; Borén, J.; Catapano, A.L.; Descamps, O.S.; Fisher, E.; Kovanen, P.T.; Kuivenhoven, J.A.; et al. Triglyceride-rich lipoproteins and high-density lipoprotein cholesterol in patients at high risk of cardiovascular disease: Evidence and guidance for management. Eur. Heart J. 2011, 32, 1345–1361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nordestgaard, B.G.; Benn, M.; Schnohr, P.; Tybjaerg-Hansen, A. Nonfasting triglycerides and risk of myocardial infarction, ischemic heart disease, and death in men and women. JAMA 2007, 298, 299–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varbo, A.; Benn, M.; Tybjærg-Hansen, A.; Jørgensen, A.B.; Frikke-Schmidt, R.; Nordestgaard, B.G. Remnant cholesterol as a causal risk factor for ischemic heart disease. J. Am. Coll. Cardiol. 2013, 61, 427–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quispe, R.; Martin, S.S.; Michos, E.D.; Lamba, I.; Blumenthal, R.S.; Saeed, A.; Lima, J.; Puri, R.; Nomura, S.; Tsai, M.; et al. Remnant cholesterol predicts cardiovascular disease beyond LDL and ApoB: A primary prevention study. Eur. Heart J. 2021, 42, 4324–4332. [Google Scholar] [CrossRef]
- Kaltoft, M.; Langsted, A.; Nordestgaard, B.G. Triglycerides and remnant cholesterol associated with risk of aortic valve stenosis: Mendelian randomization in the Copenhagen General Population Study. Eur. Heart J. 2020, 41, 2288–2299. [Google Scholar] [CrossRef] [PubMed]
- Ooi, E.M.; Afzal, S.; Nordestgaard, B.G. Elevated remnant cholesterol in 25-hydroxyvitamin D deficiency in the general population: Mendelian randomization study. Circulation. Cardiovasc. Genet. 2014, 7, 650–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kristiansson, K.; Perola, M.; Tikkanen, E.; Kettunen, J.; Surakka, I.; Havulinna, A.S.; Stancáková, A.; Barnes, C.; Widen, E.; Kajantie, E.; et al. Genome-wide screen for metabolic syndrome susceptibility Loci reveals strong lipid gene contribution but no evidence for common genetic basis for clustering of metabolic syndrome traits. Circulation. Cardiovasc. Genet. 2012, 5, 242–249. [Google Scholar] [CrossRef]
- Lind, L. Genome-Wide Association Study of the Metabolic Syndrome in UK Biobank. Metab. Syndr. Relat. Disord. 2019, 17, 505–511. [Google Scholar] [CrossRef]
- Hsu, L.A.; Chang, C.J.; Wu, S.; Teng, M.S.; Chou, H.H.; Chang, H.H.; Chang, P.Y.; Ko, Y.L. Association between functional variants of the ICAM1 and CRP genes and metabolic syndrome in Taiwanese subjects. Metab. Clin. Exp. 2010, 59, 1710–1716. [Google Scholar] [CrossRef]
- Bahrami, A.; Liberale, L.; Reiner, Ž.; Carbone, F.; Montecucco, F.; Sahebkar, A. Inflammatory Biomarkers for Cardiovascular Risk Stratification in Familial Hypercholesterolemia. Rev. Physiol. Biochem. Pharmacol. 2020, 177, 25–52. [Google Scholar] [CrossRef] [PubMed]
- Hsu, L.A.; Teng, M.S.; Wu, S.; Chou, H.H.; Ko, Y.L. Common and Rare PCSK9 Variants Associated with Low-Density Lipoprotein Cholesterol Levels and the Risk of Diabetes Mellitus: A Mendelian Randomization Study. Int. J. Mol. Sci. 2022, 23, 10418. [Google Scholar] [CrossRef] [PubMed]
- Besseling, J.; Kastelein, J.J.; Defesche, J.C.; Hutten, B.A.; Hovingh, G.K. Association between familial hypercholesterolemia and prevalence of type 2 diabetes mellitus. JAMA 2015, 313, 1029–1036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raczy, C.; Petrovski, R.; Saunders, C.T.; Chorny, I.; Kruglyak, S.; Margulies, E.H.; Chuang, H.Y.; Källberg, M.; Kumar, S.A.; Liao, A.; et al. Isaac: Ultra-fast whole-genome secondary analysis on Illumina sequencing platforms. Bioinformatics 2013, 29, 2041–2043. [Google Scholar] [CrossRef] [Green Version]
- Chait, A.; Ginsberg, H.N.; Vaisar, T.; Heinecke, J.W.; Goldberg, I.J.; Bornfeldt, K.E. Remnants of the Triglyceride-Rich Lipoproteins, Diabetes, and Cardiovascular Disease. Diabetes 2020, 69, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Teng, M.S.; Wu, S.; Hsu, L.A.; Chou, H.H.; Ko, Y.L. Pleiotropic Effects of Functional MUC1 Variants on Cardiometabolic, Renal, and Hematological Traits in the Taiwanese Population. Int. J. Mol. Sci. 2021, 22, 10641. [Google Scholar] [CrossRef]
- Yeh, K.H.; Hsu, L.A.; Teng, M.S.; Wu, S.; Chou, H.H.; Ko, Y.L. Pleiotropic Effects of Common and Rare GCKR Exonic Mutations on Cardiometabolic Traits. Genes 2022, 13, 491. [Google Scholar] [CrossRef]
- Hsu, L.A.; Chou, H.H.; Teng, M.S.; Wu, S.; Ko, Y.L. Circulating chemerin levels are determined through circulating platelet counts in nondiabetic Taiwanese people: A bidirectional Mendelian randomization study. Atherosclerosis 2021, 320, 61–69. [Google Scholar] [CrossRef]
- Palmer, T.M.; Lawlor, D.A.; Harbord, R.M.; Sheehan, N.A.; Tobias, J.H.; Timpson, N.J.; Davey Smith, G.; Sterne, J.A. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat. Methods Med. Res. 2012, 21, 223–242. [Google Scholar] [CrossRef] [Green Version]
- Grundy, S.M.; Cleeman, J.I.; Daniels, S.R.; Donato, K.A.; Eckel, R.H.; Franklin, B.A.; Gordon, D.J.; Krauss, R.M.; Savage, P.J.; Smith, S.C., Jr.; et al. Diagnosis and management of the metabolic syndrome: An American Heart Association/National Heart, Lung, and Blood Institute Scientific Statement. Circulation 2005, 11, 2735–2752. [Google Scholar] [CrossRef]
| Clinical and Laboratory Parameters * | With WGS Data (n = 1478) | With GWAS Data (n = 115,088) |
|---|---|---|
| Anthropology | ||
| Age (years) | 50.0 (40.0–59.0) # | 51.0 (40.0–59.0) # |
| Sex (male vs. female) | 739/739 | 41,467/73,621 |
| Body mass index (kg/m2) | 23.9 (21.9–26.4) | 23.8 (21.6–26.3) |
| Current smoking (%) | 10.08% (149) | 19.63% (22,590) |
| Lipid profile | ||
| Total cholesterol (mg/dL) | 191.0 (170.0–215.0) | 193.0 (171.0–217.0) |
| LDL cholesterol (mg/dL) | 119.0 (101.0–141.0) | 119.0 (99.0–141.0) |
| Non-HDL cholesterol (mg/dL) | 137.0 (116.0–160.0) | 138.0 (116.0–162.0) |
| HDL cholesterol (mg/dL) | 52.0 (44.0–63.0) | 53.0 (45.0–63.0) |
| Triglyceride (mg/dL) | 90.0 (64.0–130.0) | 91.0 (64.0–133.0) |
| Remnant cholesterol (mg/dL) | 16.0 (11.0–22.0) | 16.0 (11.0–23.0) |
| Metabolic syndrome (%) | 19.30% (285) | 25.53% (29,384) |
| Diabetes mellitus (%) | 9.07% (134) | 9.45% (10,879) |
| Genetic Variants | Genotypes | Beta | SE | p Value | ||
|---|---|---|---|---|---|---|
| APOB rs144467873 | GG (106,145) | GA (341) | AA (1) | |||
| Total cholesterol (mg/dL) | 193.0 (171.0–217.0) | 231.0 (206.0–255.5) | 278.0 | 0.0805 | 0.0041 | 2.29 × 10−85 |
| LDL cholesterol (mg/dL) | 119.0 (99.0–140.0) | 163.0 (138.0–188.0) | 207.0 | 0.1373 | 0.0061 | 1.96 × 10−110 |
| Non-HDL cholesterol (mg/dL) | 138.0 (116.0–162.0) | 176.0 (154.0–203.0) | 238.0 | 0.1148 | 0.0055 | 6.08 × 10−95 |
| HDL cholesterol (mg/dL) | 53.0 (45.0–63.0) | 53.0 (44.0–61.0) | 40.0 | −0.0124 | 0.0049 | 0.0114 |
| Triglyceride (mg/dL) | 91.0 (64.0–133.0) | 82.0 (60.0–127.0) | 150.0 | −0.0077 | 0.0115 | 0.5037 |
| Remnant cholesterol (mg/dL) | 16.0 (12.0–23.0) | 13.8 (8.0–19.0) | 31.0 | −0.1005 | 0.0142 | 1.71 × 10−12 |
| Metabolic syndrome (%) | 25.50% (29,235) | 33.94% (149) | 100% (1) | 0.6463 | 0.1161 | 2.58 × 10−8 |
| APOB rs676210 | AA (56,252) | AG (42,386) | GG (7849) | |||
| Total cholesterol (mg/dL) | 194.0 (172.0–218.0) | 192.0 (170.0–216.0) | 190.0 (168.0–213.0) | −0.0047 | 0.0004 | 1.53 × 10−36 |
| LDL cholesterol (mg/dL) | 120.0 (100.0–142.0) | 119.0 (99.0–139.0) | 117.0 (98.0–137.0) | −0.0063 | 0.0006 | 4.10 × 10−30 |
| Non-HDL cholesterol (mg/dL) | 139.0 (117.0–163.0) | 137.0 (116.0–161.0) | 135.0 (114.0–158.0) | −0.0053 | 0.0005 | 1.03 × 10−25 |
| HDL cholesterol (mg/dL) | 54.0 (45.0–63.0) | 53.0 (45.0–63.0) | 53.0 (44.0–62.0) | −0.0031 | 0.0004 | 1.75 × 10−12 |
| Triglyceride (mg/dL) | 90.0 (63.0–134.0) | 91.0 (64.0–133.0) | 91.0 (66.0–133.0) | 0.0021 | 0.0010 | 0.0424 |
| Remnant cholesterol (mg/dL) | 16.0 (12.0–23.0) | 16.0 (11.0–23.0) | 16.0 (12.0–23.0) | 0.0012 | 0.0013 | 0.3382 |
| Metabolic syndrome (%) | 25.42% (15,509) | 25.65% (11,727) | 25.66% (2148) | 0.0211 | 0.0124 | 0.0891 |
| APOB rs679899 | AA (77,368) | AG (26,778) | GG (2341) | |||
| Total cholesterol (mg/dL) | 192.0 (171.0–216.0) | 195.0 (172.0–218.0) | 198.0 (175.0–223.0) | 0.0052 | 0.0005 | 6.32 × 10−29 |
| LDL cholesterol (mg/dL) | 119.0 (99.0–140.0) | 121.0 (101.0–142.0) | 123.0 (103.0–145.0) | 0.0080 | 0.0007 | 2.50 × 10−30 |
| Non-HDL cholesterol (mg/dL) | 137.0 (115.0–161.0) | 140.0 (118.0–164.0) | 143.0 (120.0–168.0) | 0.0084 | 0.0006 | 1.93 × 10−40 |
| HDL cholesterol (mg/dL) | 53.0 (45.0–63.0) | 53.0 (45.0–63.0) | 53.0 (45.0–63.0) | −0.0030 | 0.0006 | 6.71 × 10−8 |
| Triglyceride (mg/dL) | 90.0 (64.0–133.0) | 92.0 (64.0–133.0) | 96.0 (65.0–138.0) | 0.0074 | 0.0013 | 1.23 × 10−8 |
| Remnant cholesterol (mg/dL) | 16.0 (11.0–23.0) | 17.0 (12.0–24.0) | 17.0 (12.0–25.0) | 0.0116 | 0.0016 | 2.47 × 10−13 |
| Metabolic syndrome (%) | 25.32% (21,139) | 26.08% (7579) | 26.03 (666) | 0.0488 | 0.0154 | 0.0016 |
| APOB rs13306194 | GG (77,614) | GA (26,682) | AA (2191) | |||
| Total cholesterol (mg/dL) | 194.0 (172.0–218.0) | 190.0 (169.0–213.0) | 185.0 (164.0–208.0) | −0.0100 | 0.0005 | 2.76 × 10−101 |
| LDL cholesterol (mg/dL) | 120.0 (100.0–142.0) | 117.0 (98.0–137.0) | 114.0 (94.0–134.0) | −0.0140 | 0.0007 | 5.13 × 10−89 |
| Non-HDL cholesterol (mg/dL) | 139.0 (117.0–163.0) | 135.0 (114.0–158.0) | 130.0 (110.0–151.0) | −0.0145 | 0.0006 | 2.65 × 10−115 |
| HDL cholesterol (mg/dL) | 53.0 (45.0–63.0) | 53.0 (45.0–63.0) | 54.0 (46.0–64.0) | 0.0014 | 0.0006 | 0.0137 |
| Triglyceride (mg/dL) | 91.0 (64.0–135.0) | 89.0 (63.0–130.0) | 85.0 (61.0–122.0) | −0.0143 | 0.0013 | 1.10 × 10−27 |
| Remnant cholesterol (mg/dL) | 17.0 (12.0–23.0) | 16.0 (11.0–23.0) | 15.0 (11.0–22.0) | −0.0158 | 0.0016 | 5.22 × 10−23 |
| Metabolic syndrome (%) | 25.80% (21,733) | 24.92% (7109) | 23.44% (542) | −0.0690 | 0.0159 | 1.40 × 10−5 |
| APOB rs1367117 | GG (80,541) | GA (24,135) | AA (1811) | |||
| Total cholesterol (mg/dL) | 192.0 (171.0–216.0) | 195.0 (173.0–219.0) | 200.0 (176.0–225.0) | 0.0070 | 0.0005 | 1.00 × 10−45 |
| LDL cholesterol (mg/dL) | 119.0 (99.0–140.0) | 122.0 (101.0–143.0) | 125.0 (103.0–148.0) | 0.0109 | 0.0007 | 5.52 × 10−50 |
| Non-HDL cholesterol (mg/dL) | 137.0 (115.0–161.0) | 140.0 (118.0–165.0) | 145.0 (121.0–171.0) | 0.0105 | 0.0007 | 4.12 × 10−56 |
| HDL cholesterol (mg/dL) | 53.0 (45.0–63.0) | 53.0 (45.0–63.0) | 53.0 (45.0–63.0) | −0.0020 | 0.0006 | 0.0006 |
| Triglyceride (mg/dL) | 90.0 (64.0–133.0) | 91.0 (64.0–134.0) | 95.0 (65.0–140.0) | 0.0048 | 0.0014 | 0.0004 |
| Remnant cholesterol (mg/dL) | 16.0 (11.0–23.0) | 17.0 (12.0–23.0) | 17.0 (12.0–25.0) | 0.0070 | 0.0017 | 2.76 × 10−5 |
| Metabolic syndrome (%) | 25.42% (22,094) | 25.75% (6742) | 27.41% (548) | 0.0362 | 0.0163 | 0.0258 |
| Total Cholesterol | LDL Cholesterol | Non-HDL Cholesterol | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | R2 | p Value | Beta | SE | R2 | p Value | Beta | SE | R2 | p Value | |
| Age (years) | 0.0013 | 0.00002 | 0.034 | <10−307 | 0.0014 | 0.00003 | 0.0168 | <10−307 | 0.0019 | 0.00003 | 0.0352 | <10−307 |
| Sex (male vs. female) | 0.014 | 0.0005 | 0.0047 | 5.87 × 10−171 | - | - | - | - | −0.0052 | 0.0008 | 0.0007 | 7.93 × 10−12 |
| Body mass index (kg/m2) | 0.0016 | 0.0001 | 0.0053 | 1.49 × 10−133 | 0.005 | 0.0001 | 0.0257 | <10−307 | 0.0061 | 0.0001 | 0.0485 | <10−307 |
| Current smoking (%) | - | - | - | - | −0.0029 | 0.0009 | 0.0001 | 0.0014 | 0.0031 | 0.0009 | 0.0001 | 0.0005 |
| APOB rs144467873 (GG vs. GA vs. AA) | 0.0759 | 0.0041 | 0.0033 | 8.24 × 10−76 | 0.1302 | 0.0062 | 0.0045 | 4.36 × 10−99 | 0.1079 | 0.0056 | 0.0036 | 9.82 × 10−84 |
| APOB rs13306194 (GG vs. GA vs. AA) | −0.0093 | 0.0005 | 0.0041 | 2.11 × 10−85 | −0.0128 | 0.0007 | 0.0035 | 2.04 × 10−73 | −0.0133 | 0.0006 | 0.0045 | 2.07 × 10−96 |
| APOB rs1367117 (GG vs. GA vs. AA) | 0.0047 | 0.0005 | 0.0008 | 3.37 × 10−21 | 0.0075 | 0.0007 | 0.0009 | 6.84 × 10−24 | 0.0072 | 0.0007 | 0.0010 | 5.60 × 10−27 |
| HDL Cholesterol | Triglyceride | Remnant Cholesterol | ||||||||||
| Age (years) | 0.0001 | 0.00003 | 0.0002 | 7.03 × 10−9 | 0.003 | 0.0001 | 0.0194 | <10−307 | 0.0050 | 0.0001 | 0.0418 | <10−307 |
| Sex (male vs. female) | 0.0623 | 0.0007 | 0.0863 | <10−307 | −0.0566 | 0.0016 | 0.0199 | 4.78 × 10−285 | −0.0149 | 0.0019 | 0.0005 | 5.84 × 10−15 |
| Body mass index (kg/m2) | −0.0094 | 0.0001 | 0.1645 | <10−307 | 0.0221 | 0.0002 | 0.1505 | <10−307 | 0.0100 | 0.0002 | 0.0231 | <10−307 |
| Current smoking (%) | −0.0112 | 0.0008 | 0.0014 | 5.00 × 10−44 | 0.0411 | 0.0019 | 0.0036 | 5.95 × 10−107 | 0.0319 | 0.0023 | 0.0033 | 3.01 × 10−44 |
| APOB rs1318006 (AA vs. AG vs. GG) | −0.0064 | 0.0006 | 0.0008 | 2.33 × 10−25 | ||||||||
| APOB rs35131127 (TT vs. TC vs. CC) | 0.017 | 0.0017 | 0.0010 | 1.09 × 10−24 | ||||||||
| APOB rs13306194 (GG vs. GA vs. AA) | −0.0125 | 0.0013 | 0.0007 | 2.66 × 10−21 | −0.0138 | 0.0016 | 0.0009 | 2.82 × 10−17 | ||||
| APOB rs144467873 (GG vs. GA vs. AA) | −0.1048 | 0.0143 | 0.0004 | 2.32 × 10−13 | ||||||||
| APOB rs56213756 (CC vs. CG vs. GG) | 0.0124 | 0.0019 | 0.0007 | 3.70 × 10−11 | ||||||||
| APOB rs679899 (AA vs. AG vs. GG) | 0.0060 | 0.0017 | 0.0001 | 0.0006 | ||||||||
| Beta | SE | OR (95% CI) | p Value | |
|---|---|---|---|---|
| Age (years) | 0.0870 | 0.0185 | 1.0909 (1.0519–1.1312) | 2.70 × 10−6 |
| Sex (male vs. female) | 0.0664 | 0.0008 | 1.0687 (1.0670–1.0704) | <10−307 |
| Body mass index (kg/m2) | 0.3127 | 0.0025 | 1.3672 (1.3605–1.3738) | <10−307 |
| Current smoking (%) | 0.2767 | 0.0214 | 1.3188 (1.2646–1.3753) | 3.14 × 10−38 |
| APOB rs144467873 (GG vs. GA vs. AA) | 0.6564 | 0.1162 | 1.9279 (1.5354–2.4209) | 1.60 × 10−8 |
| APOB rs4665709 (GG vs. GA vs. AA) | 0.1189 | 0.0212 | 1.1263 (1.0804–1.1742) | 2.16 × 10−8 |
| TA | TB | GA | TA-TB | GA-TA | GA-TB | IVA-TB | IVA-TB-adjTA | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | P a | Beta | SE | P a | Beta | SE | P a | Beta | SE | P | Beta | SE | P b | |||
| LDL-C level | DM | APOB rs144467873 | −2.3131 | 0.0988 | 3.25 × 10−121 | 0.1373 | 0.0061 | 1.96 × 10−110 | 0.1182 | 0.2095 | 0.5726 | 0.8691 | 1.5404 | 0.5726 a (0.7473 c) | 2.8639 | 1.5556 | 0.0656 |
| APOB rs13306194 | −2.3131 | 0.0988 | 3.25 × 10−121 | −0.0140 | 0.0007 | 5.13 × 10−89 | 0.0561 | 0.0233 | 0.0158 | −4.0072 | 1.6611 | 0.0158 a (0.0002 c) | −1.8901 | 1.6720 | 0.2583 | ||
| APOB rs1367117 | −2.3131 | 0.0988 | 3.25 × 10−121 | 0.0109 | 0.0007 | 5.52 × 10−50 | −0.0534 | 0.0256 | 0.0368 | −4.8537 | 2.3245 | 0.0368 a (0.0272 c) | −2.6217 | 2.3389 | 0.2623 | ||
| WGRS_APOB_3SNPs | −2.3131 | 0.0988 | 3.25 × 10−121 | 0.8796 | 0.0277 | 5.81 × 10−221 | −2.1620 | 1.0200 | 0.0340 | −2.4568 | 1.1591 | 0.0340 a (0.0029 c) | −0.3710 | 1.1688 | 0.7509 | ||
| WGRS_APOB_2SNPs | −2.3131 | 0.0988 | 3.25 × 10−121 | 0.8805 | 0.0375 | 9.08 × 10−122 | −3.6711 | 1.2769 | 0.0040 | −4.1717 | 1.4511 | 0.0040 a (8.2 × 10−5 c) | −2.0614 | 1.4608 | 0.1582 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, S.-J.; Tuan, W.-L.; Hsu, L.-A.; Er, L.-K.; Teng, M.-S.; Wu, S.; Ko, Y.-L. Pleiotropic Effects of APOB Variants on Lipid Profiles, Metabolic Syndrome, and the Risk of Diabetes Mellitus. Int. J. Mol. Sci. 2022, 23, 14963. https://doi.org/10.3390/ijms232314963
Jang S-J, Tuan W-L, Hsu L-A, Er L-K, Teng M-S, Wu S, Ko Y-L. Pleiotropic Effects of APOB Variants on Lipid Profiles, Metabolic Syndrome, and the Risk of Diabetes Mellitus. International Journal of Molecular Sciences. 2022; 23(23):14963. https://doi.org/10.3390/ijms232314963
Chicago/Turabian StyleJang, Shih-Jung, Wei-Lun Tuan, Lung-An Hsu, Leay-Kiaw Er, Ming-Sheng Teng, Semon Wu, and Yu-Lin Ko. 2022. "Pleiotropic Effects of APOB Variants on Lipid Profiles, Metabolic Syndrome, and the Risk of Diabetes Mellitus" International Journal of Molecular Sciences 23, no. 23: 14963. https://doi.org/10.3390/ijms232314963
APA StyleJang, S.-J., Tuan, W.-L., Hsu, L.-A., Er, L.-K., Teng, M.-S., Wu, S., & Ko, Y.-L. (2022). Pleiotropic Effects of APOB Variants on Lipid Profiles, Metabolic Syndrome, and the Risk of Diabetes Mellitus. International Journal of Molecular Sciences, 23(23), 14963. https://doi.org/10.3390/ijms232314963





