DGCR8 Microprocessor Subunit Mutation and Expression Deregulation in Thyroid Lesions
Abstract
1. Introduction
2. Results
2.1. c.1552G>A p.(E518K) in a Poorly Differentiated Thyroid Carcinoma
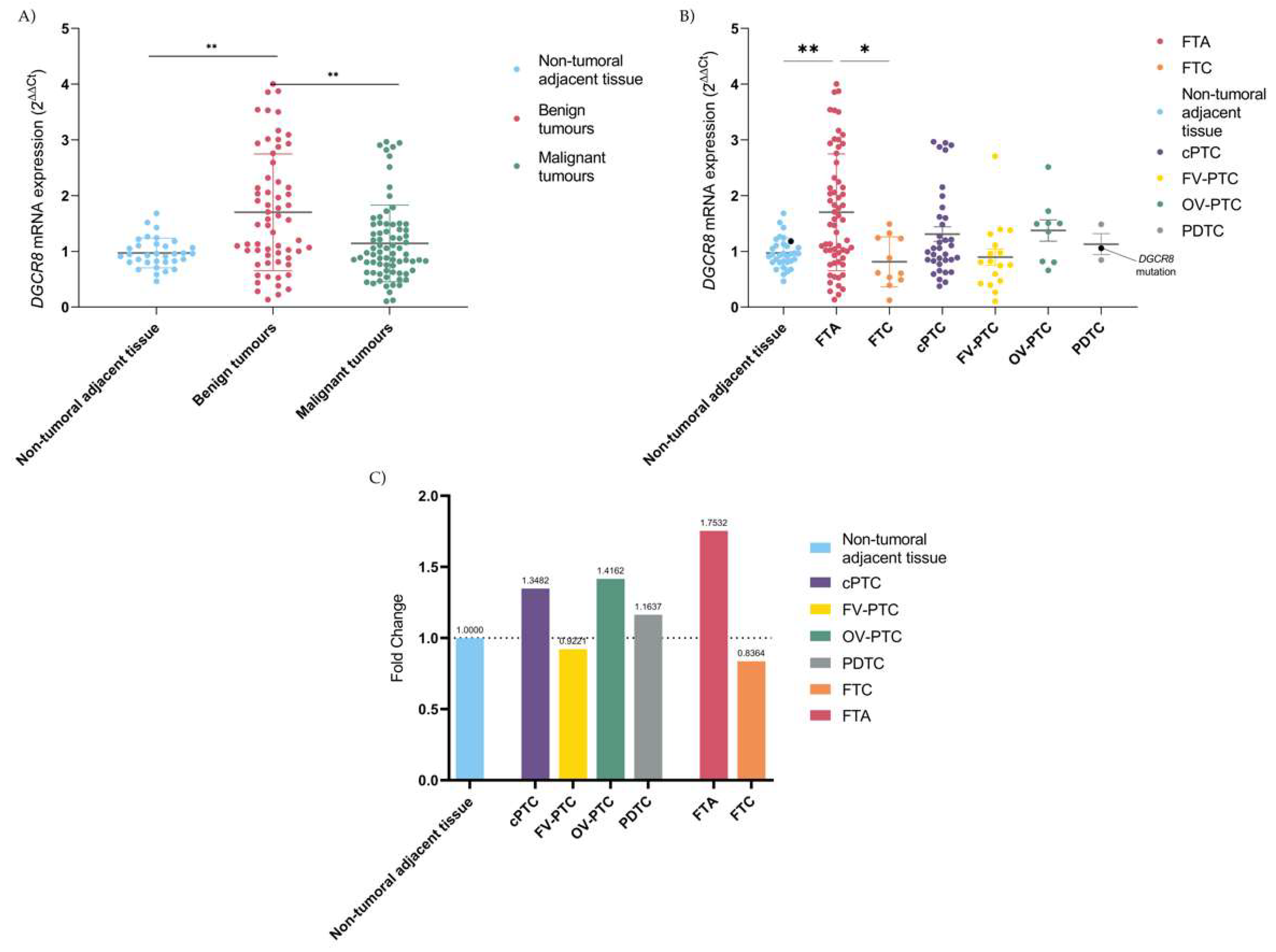
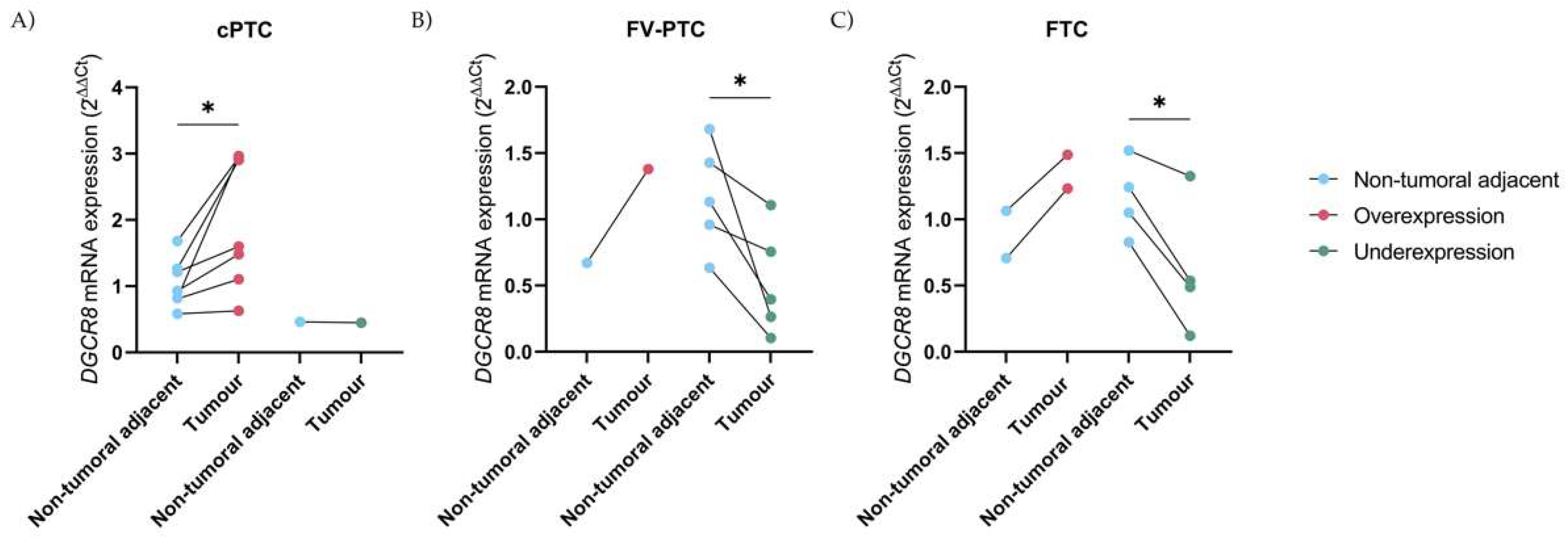
2.2. Deregulation of DGCR8 mRNA Expression in Follicular-Patterned Tumours
2.3. DGCR8 Immunoexpression in Thyroid Cancer
2.4. DGCR8 Expression, and Clinicopathological Associations
3. Discussion
4. Materials and Methods
4.1. Samples
4.2. DGCR8 Amplification and Genotyping
4.3. Quantitative PCR Analysis
4.4. Immunohistochemistry
4.5. Statistical Methods
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, W.T.; Wang, Y. Dgcr8 knockout approaches to understand microRNA functions in vitro and in vivo. Cell. Mol. Life Sci. 2019, 76, 1697–1711. [Google Scholar] [CrossRef] [PubMed]
- Nikiforova, M.N.; Chiosea, S.I.; Nikiforov, Y.E. MicroRNA expression profiles in thyroid tumors. Endocr. Pathol. 2009, 20, 85–91. [Google Scholar] [CrossRef]
- Ouellet, D.L.; Perron, M.P.; Gobeil, L.A.; Plante, P.; Provost, P. MicroRNAs in gene regulation: When the smallest governs it all. J. Biomed. Biotechnol. 2006, 69616. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Arroyo, V.M.; Nam, Y. Dynamic Protein-RNA recognition in primary MicroRNA processing. Curr Opin Struct Biol 2022, 76, 102442. [Google Scholar] [CrossRef] [PubMed]
- Marini, F.; Luzi, E.; Brandi, M.L. MicroRNA Role in Thyroid Cancer Development. J. Thyroid Res. 2011, 2011, 407123. [Google Scholar] [CrossRef] [PubMed]
- Macias, S.; Cordiner, R.A.; Caceres, J.F. Cellular functions of the microprocessor. Biochem. Soc. Trans. 2013, 41, 838–843. [Google Scholar] [CrossRef] [PubMed]
- Rivera, B.; Nadaf, J.; Fahiminiya, S.; Apellaniz-Ruiz, M.; Saskin, A.; Chong, A.S.; Sharma, S.; Wagener, R.; Revil, T.; Condello, V.; et al. DGCR8 microprocessor defect characterizes familial multinodular goiter with schwannomatosis. J. Clin. Investig. 2020, 130, 1479–1490. [Google Scholar] [CrossRef]
- Robertson, J.C.; Jorcyk, C.L.; Oxford, J.T. DICER1 Syndrome: DICER1 Mutations in Rare Cancers. Cancers 2018, 10, 143. [Google Scholar] [CrossRef] [PubMed]
- Slade, I.; Bacchelli, C.; Davies, H.; Murray, A.; Abbaszadeh, F.; Hanks, S.; Barfoot, R.; Burke, A.; Chisholm, J.; Hewitt, M.; et al. DICER1 syndrome: Clarifying the diagnosis, clinical features and management implications of a pleiotropic tumour predisposition syndrome. J. Med. Genet. 2011, 48, 273–278. [Google Scholar] [CrossRef]
- Fuziwara, C.S.; Kimura, E.T. MicroRNAs in thyroid development, function and tumorigenesis. Mol. Cell. Endocrinol. 2017, 456, 44–50. [Google Scholar] [CrossRef]
- Bartram, M.P.; Amendola, E.; Benzing, T.; Schermer, B.; de Vita, G.; Muller, R.U. Mice lacking microRNAs in Pax8-expressing cells develop hypothyroidism and end-stage renal failure. BMC Mol. Biol. 2016, 17, 11. [Google Scholar] [CrossRef] [PubMed]
- Markopoulos, G.S.; Roupakia, E.; Tokamani, M.; Chavdoula, E.; Hatziapostolou, M.; Polytarchou, C.; Marcu, K.B.; Papavassiliou, A.G.; Sandaltzopoulos, R.; Kolettas, E. A step-by-step microRNA guide to cancer development and metastasis. Cell. Oncol. 2017, 40, 303–339. [Google Scholar] [CrossRef]
- Li, X.; Abdel-Mageed, A.B.; Mondal, D.; Kandil, E. MicroRNA expression profiles in differentiated thyroid cancer, a review. Int. J. Clin. Exp. Med. 2013, 6, 74–80. [Google Scholar] [PubMed]
- Poma, A.M.; Condello, V.; Denaro, M.; Torregrossa, L.; Elisei, R.; Vitti, P.; Basolo, F. DICER1 somatic mutations strongly impair miRNA processing even in benign thyroid lesions. Oncotarget 2019, 10, 1785–1797. [Google Scholar] [CrossRef] [PubMed]
- Schultz, K.A.P.; Rednam, S.P.; Kamihara, J.; Doros, L.; Achatz, M.I.; Wasserman, J.D.; Diller, L.R.; Brugières, L.; Druker, H.; Schneider, K.A. PTEN, DICER1, FH, and their associated tumor susceptibility syndromes: Clinical features, genetics, and surveillance recommendations in childhood. Clin. Cancer Res. 2017, 23, e76–e82. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.A.; Jo, M.H.; Choi, Y.G.; Park, J.; Kwon, S.C.; Hohng, S.; Kim, V.N.; Woo, J.S. Functional Anatomy of the Human Microprocessor. Cell 2015, 161, 1374–1387. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Lv, Z.; Ding, H.; Fang, X.; Sun, M. Association of miRNA biosynthesis genes DROSHA and DGCR8 polymorphisms with cancer susceptibility: A systematic review and meta-analysis. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef]
- Kim, J.; Park, W.J.; Jeong, K.J.; Kang, S.H.; Kwon, S.Y.; Kim, S.; Park, J.W. Racial Differences in Expression Levels of miRNA Machinery-Related Genes, Dicer, Drosha, DGCR8, and AGO2, in Asian Korean Papillary Thyroid Carcinoma and Comparative Validation Using the Cancer Genome Atlas. Int. J. Genom. 2017, 2017, 5789769. [Google Scholar] [CrossRef] [PubMed]
- Guo, F. Drosha and DGCR8 in MicroRNA Biogenesis. In Eukaryotic RNases and Their Partners in RNA Degradation and Biogenesis, Part B; Academic Press Elsevier: Waltham, MA, USA, 2012; pp. 101–121. [Google Scholar] [CrossRef]
- Sellier, C.; Hwang, V.J.; Dandekar, R.; Durbin-Johnson, B.; Charlet-Berguerand, N.; Ander, B.P.; Sharp, F.R.; Angkustsiri, K.; Simon, T.J.; Tassone, F. Decreased DGCR8 expression and miRNA dysregulation in individuals with 22q11.2 deletion syndrome. PLoS ONE 2014, 9, e103884. [Google Scholar] [CrossRef] [PubMed]
- Paulsson, J.O.; Rafati, N.; DiLorenzo, S.; Chen, Y.; Haglund, F.; Zedenius, J.; Juhlin, C.C. Whole-genome sequencing of follicular thyroid carcinomas reveal recurrent mutations in microRNA processing subunit DGCR8. J. Clin. Endocrinol. Metab. 2021, 106, 3265–3282. [Google Scholar] [CrossRef]
- Castro, P.; Eknæs, M.; Teixeira, M.R.; Danielsen, H.E.; Soares, P.; Lothe, R.A.; Sobrinho-Simões, M. Adenomas and follicular carcinomas of the thyroid display two major patterns of chromosomal changes. J. Pathol. A J. Pathol. Soc. Great Br. Irel. 2005, 206, 305–311. [Google Scholar] [CrossRef]
- Vardapour, R.; Kehl, T.; Kneitz, S.; Ludwig, N.; Meese, E.; Lenhof, H.P.; Gessler, M. The DGCR8 E518K mutation found in Wilms tumors leads to a partial miRNA processing defect that alters gene expression patterns and biological processes. Carcinogenesis 2022, 43, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Pestana, A.; Batista, R.; Celestino, R.; Canberk, S.; Sobrinho-Simoes, M.; Soares, P. Comprehensive Assessment of TERT mRNA Expression across a Large Cohort of Benign and Malignant Thyroid Tumours. Cancers 2020, 12, 1846. [Google Scholar] [CrossRef] [PubMed]
- Soares, P.; Povoa, A.A.; Melo, M.; Vinagre, J.; Maximo, V.; Eloy, C.; Cameselle-Teijeiro, J.M.; Sobrinho-Simoes, M. Molecular Pathology of Non-familial Follicular Epithelial-Derived Thyroid Cancer in Adults: From RAS/BRAF-like Tumor Designations to Molecular Risk Stratification. Endocr. Pathol. 2021, 32, 44–62. [Google Scholar] [CrossRef] [PubMed]
- Chong, A.S.; Nikiforov, Y.E.; Condello, V.; Wald, A.I.; Nikiforova, M.N.; Foulkes, W.D.; Rivera, B. Prevalence and Spectrum of DICER1 Mutations in Adult-onset Thyroid Nodules with Indeterminate Cytology. J. Clin. Endocrinol. Metab. 2021, 106, 968–977. [Google Scholar] [CrossRef] [PubMed]
- Paulsson, J.O.; Zedenius, J.; Juhlin, C.C. TERT Promoter Mutated Follicular Thyroid Carcinomas Exhibit a Distinct microRNA Expressional Profile with Potential Implications for Tumor Progression. Endocr. Pathol. 2021, 32, 513–516. [Google Scholar] [CrossRef]
- Wasserman, J.D.; Sabbaghian, N.; Fahiminiya, S.; Chami, R.; Mete, O.; Acker, M.; Wu, M.K.; Shlien, A.; de Kock, L.; Foulkes, W.D. DICER1 Mutations Are Frequent in Adolescent-Onset Papillary Thyroid Carcinoma. J. Clin. Endocrinol. Metab. 2018, 103, 2009–2015. [Google Scholar] [CrossRef]
- Canberk, S.; Ferreira, J.C.; Pereira, L.; Batista, R.; Vieira, A.F.; Soares, P.; Sobrinho Simoes, M.; Maximo, V. Analyzing the Role of DICER1 Germline Variations in Papillary Thyroid Carcinoma. Eur. Thyroid J. 2021, 9, 296–303. [Google Scholar] [CrossRef]
- Puppin, C.; Durante, C.; Sponziello, M.; Verrienti, A.; Pecce, V.; Lavarone, E.; Baldan, F.; Campese, A.F.; Boichard, A.; Lacroix, L.; et al. Overexpression of genes involved in miRNA biogenesis in medullary thyroid carcinomas with RET mutation. Endocrine 2014, 47, 528–536. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Swede, H.; Cassarino, D.; Fleming, E.; Fire, A.; Dadras, S.S. Up-regulated Dicer expression in patients with cutaneous melanoma. PLoS ONE 2011, 6, e20494. [Google Scholar] [CrossRef]
- Fardmanesh, H.; Shekari, M.; Movafagh, A.; Alizadeh Shargh, S.; Poursadegh Zonouzi, A.A.; Shakerizadeh, S.; Poursadegh Zonouzi, A.; Hosseinzadeh, A. Upregulation of the double-stranded RNA binding protein DGCR8 in invasive ductal breast carcinoma. Gene 2016, 581, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Triboulet, R.; Chang, H.M.; Lapierre, R.J.; Gregory, R.I. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA 2009, 15, 1005–1011. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, R.; Osamura, R.; Klöppel, G.; Rosai, J. WHO Classification of Tumours of Endocrine Organs, 4th ed.; IARC Press: Lyon, France, 2017; Volume 10. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

| Histological Subtypes | ||||||||
|---|---|---|---|---|---|---|---|---|
| Clinicopathological Variables * | FTA (n = 86) | MNG (n = 15) | FTC (n = 22) | cPTC (n = 44) | FV-PTC (n = 22) | OV-PTC (n = 14) | PDTC (n = 5) | ATC (n = 2) |
| Age (mean, y/o) | 43.2 | 45.3 | 47.8 | 40.2 | 42.3 | 45 | 65.6 | N.D. |
| Gender (female), n (%) | 69 (80.2) | 15 (100.0) | 16 (72.7) | 34 (77.3) | 22 (100.0) | 10 (71.4) | 4 (66.7) | N.D. |
| Tumour size (mean, mm) | 35 | 23 | 40 | 25 | 33 | 38 | 51 | N.D. |
| Lymphocytic infiltrate, n (%) | 20/67 (29.9) | N.D. | 3/14 (21.4) | 17/37 (45.9) | 7/17 (41.2) | 4/13 (30.8) | 0/5 (0.0) | N.D. |
| Vascular invasion, n (%) | 0/70 (0.0) | N.D. | 8/17 (47.1) | 19/37 (51.4) | 5/18 (27.7) | 6/13 (46.2) | 2/3 (66.7) | N.D. |
| Lymph node metastasis, n (%) | 0/1 (0.0) | N.D. | 0/7 (0.0) | 14/22 (63.6) | 5/6 (83.3) | 4/4 (100.0) | 2/3 (66.7) | N.D. |
| Minimal extrathyroidal extension, n (%) | 0/32 (0.0) | N.D. | 1/14 (7.14) | 15/37 (40.5) | 5/18 (27.7) | 7/12 (58.3) | 2/5 (40.0) | 1/1 (100.0) |
| Molecular characterization † | FTA | MNG | FTC | cPTC | FV-PTC | OV-PTC | PDTC | ATC |
| BRAF, nm/nt (%) | 0/23 (0.0) | 0/2 (0.0) | 0/18 (0.0) | 18/42 (42.9) | 4/20 (20.0) | 4/13 (30.8) | 0/4 (0.0) | 0/1 (0.0) |
| NRAS, nm/nt (%) | 4/86 (4.7) | 0/2 (0.0) | 4/21 (19.0) | 2/42 (4.8) | 3/21 (14.3) | 2/12 (16.7) | 0/4 (0.0) | 0/1 (0.0) |
| RET/PTC, nm/nt (%) | 0/74 (0.0) | 0/2 (0.0) | 1/13 (7.7) | 5/36 (13.9) | 0/20 (0.0) | 2/14 (14.3) | 0/4 (0.0) | N.D. |
| PAX8/PPARg, nm/nt (%) | 3/76 (3.9) | 0/2 (0.0) | 3/16 (18.8) | 0/41 (0.0) | 1/21 (4.8) | 0/14 (0.0) | 0/4 (0.0) | N.D. |
| TERTp, nm/nt (%) | 0/84 (0.0) | 0/2 (0.0) | 0/17 (0.0) | 0/41 (0.0) | 0/20 (0.0) | 2/14 (14.3) | 1/4 (25.0) | 0/1 (0.0) |
| DGCR8, nm/nt (%) | 0/86 (0.0) | 0/15 (0.0) | 0/22 (0.0) | 0/44 (0.0) | 0/22 (0.0) | 0/14 (0.0) | 1/5 (20.0) | 0/2 (0.0) |
| Score | |||||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 6 | 9 | |
| Histotype n, (%) | |||||||
| FTA n = 30 | 6 (20.0) | 5 (16.7) | 4 (13.3) | 2 (6.7) | 2 (6.7) | 6 (20.0) | 5 (16.7) |
| FTC n = 15 | 3 (20.0) | 0 (0) | 2 (13.3) | 1 (6.7) | 2 (13.3) | 5 (33.3) | 2 (13.3) |
| PTC n = 50 | 13 (26.0) | 3 (6.0) | 7 (14.0) | 2 (4.0) | 4 (8.0) | 14 (28.0) | 7 (14.0) |
| PDTC n = 4 | 0 (0) | 1 (25.0) | 2 (50.0) | 0 (0) | 0 (0) | 0 (0) | 1 (25.0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodrigues, L.; Canberk, S.; Macedo, S.; Soares, P.; Vinagre, J. DGCR8 Microprocessor Subunit Mutation and Expression Deregulation in Thyroid Lesions. Int. J. Mol. Sci. 2022, 23, 14812. https://doi.org/10.3390/ijms232314812
Rodrigues L, Canberk S, Macedo S, Soares P, Vinagre J. DGCR8 Microprocessor Subunit Mutation and Expression Deregulation in Thyroid Lesions. International Journal of Molecular Sciences. 2022; 23(23):14812. https://doi.org/10.3390/ijms232314812
Chicago/Turabian StyleRodrigues, Lia, Sule Canberk, Sofia Macedo, Paula Soares, and João Vinagre. 2022. "DGCR8 Microprocessor Subunit Mutation and Expression Deregulation in Thyroid Lesions" International Journal of Molecular Sciences 23, no. 23: 14812. https://doi.org/10.3390/ijms232314812
APA StyleRodrigues, L., Canberk, S., Macedo, S., Soares, P., & Vinagre, J. (2022). DGCR8 Microprocessor Subunit Mutation and Expression Deregulation in Thyroid Lesions. International Journal of Molecular Sciences, 23(23), 14812. https://doi.org/10.3390/ijms232314812








