Comparative Genomics and Physiological Characterization of Two Aerobic Spore Formers Isolated from Human Ileal Samples
Abstract
:1. Introduction
2. Results
2.1. Genome Sequence and Phylogenetic Analysis
Functional Annotation of SF106 and SF174 Genomes
2.2. Genome Analyses
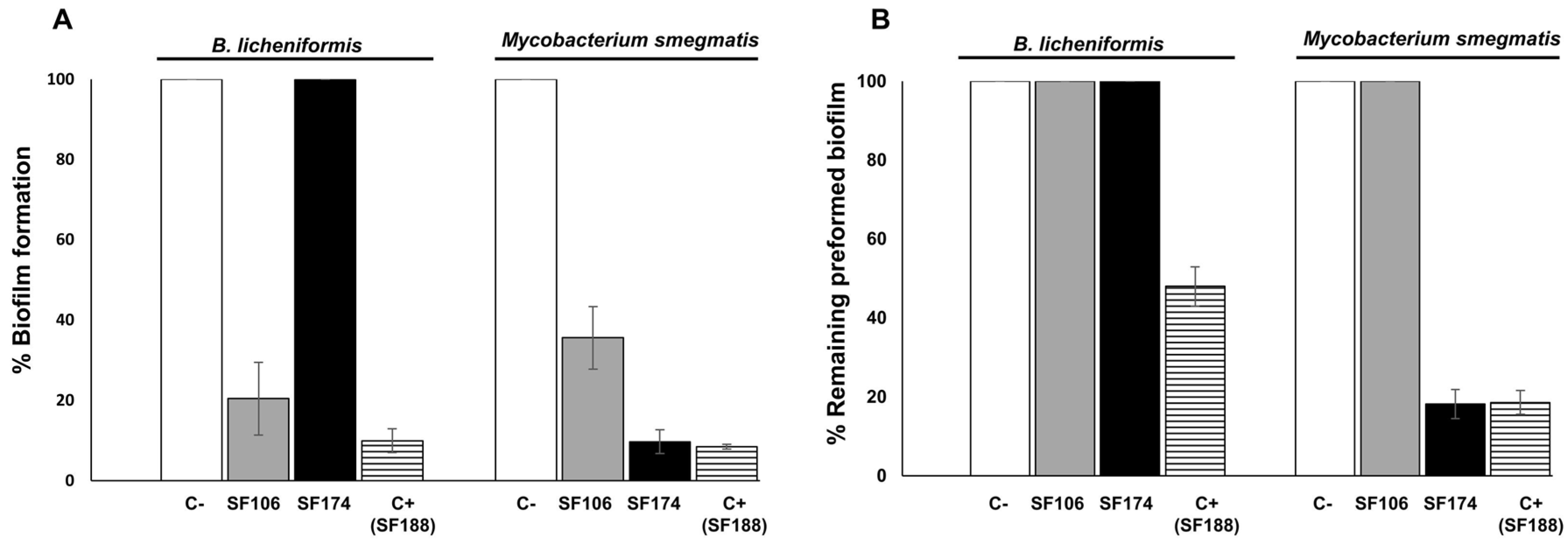
2.3. Antimicrobial and Antibiofilm Activities
2.4. Antibiotic-Resistance
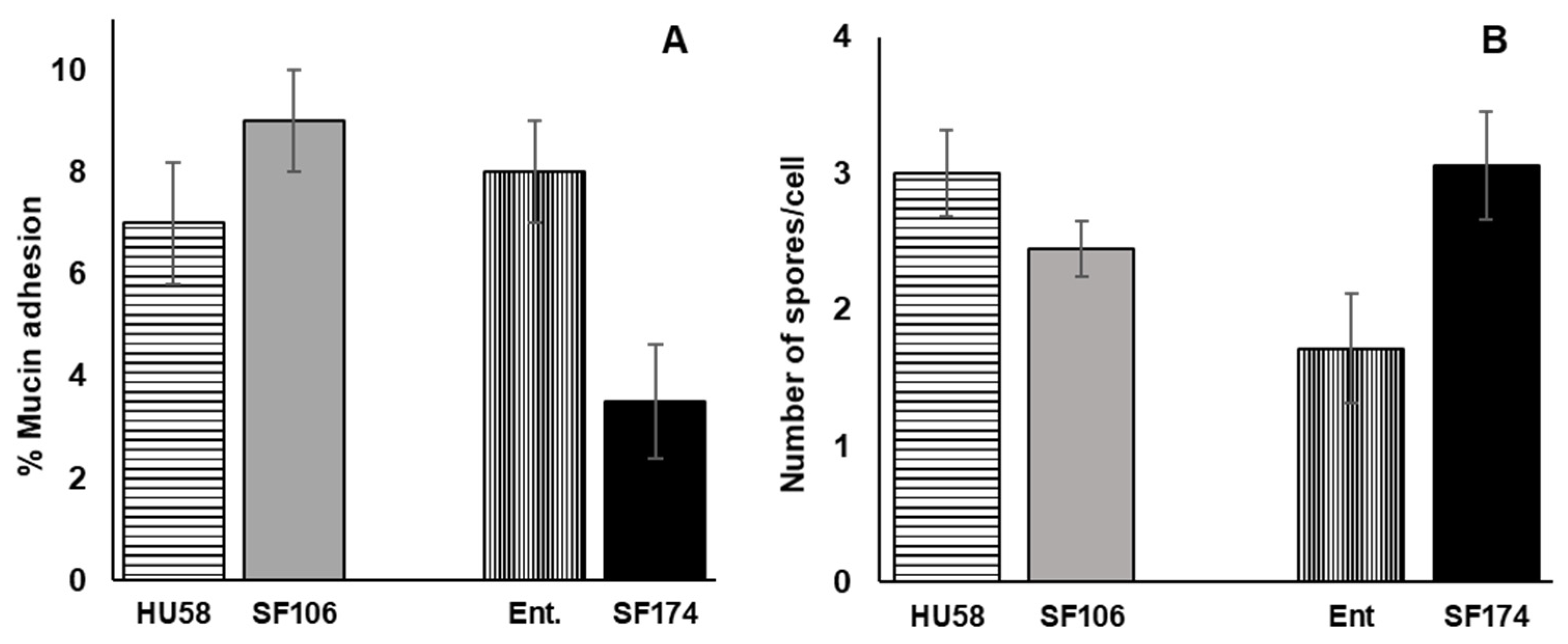
2.5. Interactions of SF106 and SF174 with Mucin and Human Epithelial Cells In Vitro
3. Discussion
4. Materials and Methods
4.1. Bacterial Growth, Spore Production and Purification
4.2. Whole-Genome Sequencing and Bioinformatic Analysis of SF106 and SF174 Genomes
4.3. The Genome Annotations and Genome and Pangenome Analysis
4.4. Antimicrobial and Antibiofilm Activities
4.5. Antibiofilm Assay
4.6. Minimum Inhibitory Concentrations
4.7. MTT Assay
4.8. Mucin Adherence Assay
4.9. Adherence to HCT116 Cells
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Christie, G.; Setlow, P. Bacillus spore germination: Knowns, unknowns and what we need to learn. Cell. Signal. 2020, 74, 109729. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, W.L.; Munakata, N.; Horneck, G.; Melosh, H.J.; Setlow, P. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 2000, 64, 548–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wörmer, L.; Hoshino, T.; Bowles, M.W.; Viehweger, B.; Adhikari, R.R.; Xiao, N.; Uramoto, G.; Konneke, M.; Lazar, C.; Inagaki, F.; et al. Microbial dormancy in the marine subsurface: Global endospore abundance and response to burial. Sci. Adv. 2019, 5, eaav1024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cutting, S.M. Bacillus probiotics. Food Microbiol. 2011, 28, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Egan, M.; Dempsey, E.; Ryan, C.A.; Ross, R.P.; Stanton, C. The sporobiota of the human gut. Gut Microbes 2021, 13, e1863134. [Google Scholar] [CrossRef]
- Saggese, A.; Baccigalupi, L.; Ricca, E. Spore formers as beneficial microbes for humans and animals. Appl. Microbiol. 2021, 1, 498–509. [Google Scholar] [CrossRef]
- Casula, G.; Cutting, S.M. Bacillus probiotics: Spore germination in the gastrointestinal tract. Appl. Environ. Microbiol. 2002, 68, 2344–2352. [Google Scholar] [CrossRef] [Green Version]
- Hoa, T.T.; Duc, L.H.; Isticato, R.; Baccigalupi, L.; Ricca, E.; Van, P.H.; Cutting, S.M. Fate and dissemination of Bacillus subtilis spores in a murine model. Appl. Environ. Microbiol. 2001, 67, 3819–3823. [Google Scholar] [CrossRef] [Green Version]
- Tam, N.K.; Uyen, N.Q.; Hong, H.A.; Duc, L.H.; Hoa, T.T.; Serra, C.R.; Henriques, A.O.; Cutting, S.M. The intestinal life cycle of Bacillus subtilis and close relatives. J. Bacteriol. 2006, 188, 2692–2700. [Google Scholar] [CrossRef] [Green Version]
- Rhee, K.-J.; Sethupathi, P.; Driks, A.; Lanning, D.K.; Knight, K.L. Role of commensal bacteria in development of gut-associated lymphoid tissues and preimmune antibody repertoire. J. Immunol. 2004, 172, 1118. [Google Scholar] [CrossRef]
- Mazzoli, A.; Donadio, G.; Lanzilli, M.; Saggese, A.; Guarino, A.M.; Rivetti, M.; Crescenzo, R.; Ricca, E.; Ferrandino, I.; Iossa, S.; et al. Bacillus megaterium SF185 spores exert protective effects against oxidative stress in vivo and in vitro. Sci. Rep. 2014, 9, 12082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Arienzo, R.; Maurano, F.; Mazzarella, G.; Luongo, D.; Stefanile, R.; Ricca, E.; Rossi, M. Bacillus subtilis spores reduce susceptibility to Citrobacter rodentium-mediated enteropathy in a mouse model. Res. Microbiol. 2006, 157, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Paparo, L.; Tripodi, L.; Bruno, C.; Pisapia, L.; Damiano, C.; Pastore, L.; Berni Canani, R. Protective action of Bacillus clausii probiotic strains in an invitro model of Rotavirus infection. Sci. Rep. 2020, 10, 12636. [Google Scholar] [CrossRef] [PubMed]
- Kotowicz, N.; Bhardwaj, R.K.; Ferreira, W.T.; Hong, H.A.; Olender, A.; Ramirez, J.; Cutting, S.M. Safety and probiotic evaluation of two Bacillus strains producing antioxidant compounds. Benef. Microbes 2019, 10, 759–771. [Google Scholar] [CrossRef]
- Marzorati, M.; Van den Abbeele, P.; Bubeck, S.; Bayne, T.; Krishnan, K.; Young, A. Treatment with a spore-based probiotic containing five strains of Bacillus induced changes in the metabolic activity and community composition of the gut microbiota in a SHIME® model of the human gastrointestinal system. Food Res. Intern. 2021, 149, 110676. [Google Scholar] [CrossRef]
- Huang, J.M.; La Ragione, R.; Nunez, A.; Cutting, S.M. Immunostimulatory activity of Bacillus spores. FEMS Immunol. Med. Microbiol. 2008, 53, 195–203. [Google Scholar] [CrossRef] [Green Version]
- Petruk, G.; Donadio, G.; Lanzilli, M.; Isticato, R.; Monti, D.M. Alternative use of Bacillus subtilis spores: Protection against environmental oxidative stress in human normal keratinocytes. Sci. Rep. 2018, 8, 1745. [Google Scholar] [CrossRef] [Green Version]
- Saggese, A.; Culurciello, R.; Casillo, A.; Corsaro, M.M.; Ricca, E.; Baccigalupi, L. A marine isolate of Bacillus pumilus secretes a pumilacidin active against Staphylococcus aureus. Mar. Drugs 2018, 16, 180. [Google Scholar] [CrossRef] [Green Version]
- Saggese, A.; De Luca, Y.; Baccigalupi, L.; Ricca, E. An antimicrobial peptide specifically active against Listeria monocytogenes is secreted by Bacillus pumilus SF214. BMC Microbiol. 2022, 22, 3. [Google Scholar] [CrossRef]
- Fujita, M.; Musch, M.W.; Nakagawa, Y.; Hu, S.; Alverdy, J.; Kohgo, Y.; Schneewind, O.; Jabri, B.; Chang, E.B. The Bacillus subtilis quorum-sensing molecule CSF contribute to intestinal homoestasis via OCTN2, a host cell membrane transporter. Cell Host Microbe 2007, 1, 299–308. [Google Scholar] [CrossRef]
- Di Luccia, B.; D’Apuzzo, E.; Varriale, F.; Baccigalupi, L.; Ricca, E.; Pollice, A. Bacillus megaterium SF185 induces stress pathways and affects the cell cycle distribution of human intestinal epithelial cells. Benef. Microbes 2016, 7, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Fakhry, S.; Sorrentini, I.; Ricca, E.; De Felice, M.; Baccigalupi, L. Characterization of spore forming Bacilli isolated from the human gastrointestinal tract. J. Appl. Microbiol. 2008, 105, 2178–2186. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Gupta, R.S. A phylogenomic and comparative genomic framework for resolving the polyphyly of the genus Bacillus: Proposal for six new genera of Bacillus species, Peribacillus gen nov., Cytobacillus gen. nov., Mesobacillus gen. nov., Neobacillus gen. nov., Metabacillus gen. nov. and Alkalihalobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 2020, 70, 406–438. [Google Scholar]
- Manzo, N.; D’Apuzzo, E.; Coutinho, P.M.; Cutting, S.M.; Henrissat, B.; Ricca, E. Carbohydrate-active enzymes from pigmented Bacilli: A genomic approach to assess carbohydrate utilization and degradation. BMC Microbiol. 2011, 11, 198. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Mahowald, M.A.; Ley, R.E.; Lozupone, C.A.; Hamady, M.; Martens, E.C.; Henrissat, B.; Coutinho, P.M.; Minx, P.; Latreille, P.; et al. Evolution of Symbiotic Bacteria in the Distal Human Intestine. PLoS Biol. 2007, 5, e156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Kaoutari, A.; Armougom, F.; Gordon, J.I.; Raoult, D.; Henrissat, B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 2013, 11, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Ozer, E.A. ClustaAGE: A tool for clustering and distribution analysis of bacterial accessory genomic elements. BMC Bioinform. 2018, 19, 150. [Google Scholar] [CrossRef] [PubMed]
- Guinebretiere, M.H.; Broussolle, V.; Nguyen-The, C. Enterotoxigenic profiles of food-poisoning and food-borne Bacillus cereus strains. J. Clin. Microbiol. 2002, 40, 3053–3056. [Google Scholar] [CrossRef] [Green Version]
- Phelps, R.J.; McKillip, J.L. Enterotoxin production in natural isolates of Bacillaceae outside the Bacillus cereus group. Appl. Environ. Microbiol. 2002, 68, 3147–3151. [Google Scholar] [CrossRef] [Green Version]
- Babasaki, K.; Takao, T.; Shimonishi, Y.; Kurahashi, K. Subtilosin A, a new antibiotic peptide produced by Bacillus subtilis 168: Isolation, structural analysis, and biogenesis. J. Biochem. 1985, 98, 585–603. [Google Scholar] [CrossRef]
- Shelburne, C.E.; An, F.Y.; Dholpe, V.; Ramamoorthy, A.; Lopatin, D.E.; Lantz, M.S. The spectrum of antimicrobial activity of the bacteriocin subtilosin A. J. Antimicrob. Chemother. 2007, 59, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Algburi, A.; Zehm, S.; Netrebov, V.; Bren, A.B.; Chistykov, V.; Chikindas, M.L. Subtilosin Prevents Biofilm Formation by Inhibiting Bacterial Quorum Sensing. Probiotics Antimicrob Proteins 2017, 9, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Nannan, C.; Vu, H.G.; Gillis, A.; Caulier, S.; Nguyen, T.T.; Mahillon, J. Bacilysin within the Bacillus subtilis group: Gene prevalence versus antagonistic activity against Gram-negative foodborne pathogens. J. Biotechnol. 2021, 327, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.B.; Walsh, C.T. Action and Timing of BacC and BacD in the Late Stages of Biosynthesis of the Dipeptide Antibiotic Bacilysin. Biochemistry 2013, 52, 889–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vetter, N.D.; Palmer, D.R.J. Substrate substitution in Kanosamine biosynthesis using phosphonates and phosphite rescue. Biochemistry 2021, 60, 1926–1932. [Google Scholar] [CrossRef] [PubMed]
- Prasertanan, T.; Palmer, D.R.J. The kanosamine biosynthetic pathway in Bacillus cereus UW85: Functional and kinetic characterization of KabA, KabB, and KabC. Arch. Biochem. Biophys. 2019, 676, 108139. [Google Scholar] [CrossRef]
- Gotz, F. Epidermin and gallidermin: Staphylococcal lantibiotics. Int. J. Med. Microbiol. 2014, 304, 63–71. [Google Scholar] [CrossRef]
- Schrempf, H.; Merling, P. Extracellular Streptomyces lividans vesicles: Composition, biogenesis and antimicrobial activity. Microb. Biotechnol. 2015, 8, 644–658. [Google Scholar] [CrossRef]
- Baidya, A.K.; Bhattacharya, S.; Dubey, G.P.; Mamou, G.; Ben-Yehuda, S. Bacterial nanotubes: A conduit for intercellular molecular trade. Curr. Opin. Microbiol. 2017, 42, 1–6. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority. Guidance on the Assessment of Bacterial Susceptibility to Antimicrobials of Human and Veterinary Importance. 2012. Available online: https://www.efsa.europa.eu/sites/default/files/consultation/120323.pdf (accessed on 1 February 2021).
- Gueimonde, M.; Snachez, B.; Reyes-Gavilan, C.G.; Margoles, A. Antibiotic resistance in probiotic bacteria. Front. Microbiol. 2013, 4, 202. [Google Scholar] [CrossRef] [Green Version]
- Salvetti, E.; Orrù, L.; Capozzi, V.; Martina, A.; Lamontana, A.; Kellers, D.; Cash, H.; Felis, G.E.; Cattivelli, L.; Torriani, S.; et al. Integrate genome-based assessment of safety for probiotic strains: Bacillus coagulans GBI-30, 6086 as a case study. Appl. Microbiol. Biotechnol. 2016, 100, 4595–4605. [Google Scholar] [CrossRef] [PubMed]
- Cox, G.; Wright, G.D. Intrinsic antibiotic resistance: Mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol. 2013, 303, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Rajput, A.; Dominguez San Martin, I.; Rose, R.; Beko, A.; Levea, C.; Sharratt, E.; Mazurchuk, R.; Hoffman, R.M.; Brattain, M.G.; Wang, J. Characterization of HCT116 human colon cancer cells in an orthotopic model. J. Surg. Res. 2008, 147, 276–281. [Google Scholar] [CrossRef]
- Marzorati, M.; Van den Abbeele, P.; Bubeck, S.S.; Bayne, T.; Krishnan, K.; Young, A.; Mehta, D.; DeSouza, A. Bacillus subtilis HU58 and Bacillus coagulans SC208 Probiotics Reduced the Effects of Antibiotic-Induced Gut Microbiome Dysbiosis in An M-SHIME ® Model. Microorganisms 2020, 8, 1028. [Google Scholar] [CrossRef]
- Rooney, A.P.; Price, N.P.J.; Ehrhardt, C.; Swezey, J.L.; Bannan, J.D. Phylogeny and molecular taxonomy of the Bacillus subtilis species complex and description of Bacillus subtilis subsp. inaquosorum subsp. nov. Int. J. Syst. Evol. Microbiol. 2009, 59, 2429–2436. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Chain, F.; Martín, R.; Bermúdez-Humarán, L.G.; Courau, S.; Langella, P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb. Cell Fact. 2013, 16, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossi, M.; Amaretti, A.; Raimondi, S. Folate production by probiotic bacteria. Nutrients 2011, 3, 118–134. [Google Scholar] [CrossRef] [Green Version]
- Ren, L.; Peng, C.; Hu, X.; Han, Y.; Huang, H. Microbial production of vitamin K2: Current status and future prospects. Biotechnol. Adv. 2020, 39, 107453. [Google Scholar] [CrossRef]
- Mayr, J.A.; Zimmermnn, F.A.; Fauth, C.; Bergheim, C.; Meierhofer, D.; Radmayr, D.; Zschocke, J.; Koch, J.; Sperl, W. Lipoic Acid Synthetase Deficiency Causes Neonatal-Onset Epilepsy, Defective Mitochondrial Energy Metabolism, and Glycine Elevation. Am. J. Hum. Genet. 2011, 89, 792–797. [Google Scholar] [CrossRef] [Green Version]
- Nicholson, W.L.; Setlow, P. Sporulation, Germination, and Outgrowth. In Molecular Biological Methods for Bacillus; Harwood, C.R., Cutting, S.M., Eds.; John Wiley and Sons: New York, NY, USA, 1990; pp. 391–450. [Google Scholar]
- Saggese, A.; Isticato, R.; Cangiano, G.; Ricca, E.; Baccigalupi, L. CotG-Like Modular Proteins Are Common among Spore-Forming Bacilli. J. Bacteriol. 2016, 198, 1513–1520. [Google Scholar] [CrossRef] [Green Version]
- Lugli, G.A.; Milani, C.; Mancabelli, L.; Van Sinderen, D.; Ventura, M. MEGAnnotator: A user-friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiol. Lett. 2016, 363, fnw049. [Google Scholar] [CrossRef]
- Shillinger, U.; Lucke, F.K. Antibacterial activity of Lactobacillus sake isolated from meat. Appl. Environ. Microbiol. 1989, 55, 1901–1906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zanfardino, A.; Migliardi, A.; D’Alonzo, D.; Lombardi, A.; Varcamonti, M.; Cordone, A. Inactivation of MSMEG_0412 gene drastically affects surface related properties of Mycobacterium smegmatis. BMC Microbiol. 2016, 16, 267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vittoria, M.; Saggese, A.; Di Gregorio Barletta, G.; Castaldi, S.; Isticato, R.; Baccigalupi, L.; Ricca, E. Sporulation efficiency and spore quality in a human intestinal isolate of Bacillus cereus. bioRxiv 2022. [Google Scholar] [CrossRef]
- Huang, J.Y.; Lee, S.M.; Mazmanian, S.K. The human commensal Bacteroides fragilis binds intestinal mucin. Anaerobe 2011, 17, 137–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maia, A.R.; Reyes-Ramírez, R.; Pizarro-Guajardo, M.; Saggese, A.; Castro-Córdova, P.; Isticato, R.; Ricca, E.; Paredes-Sabja, D.; Baccigalupi, L. Induction of a Specific Humoral Immune Response by Nasal Delivery of Bcla2ctd of Clostridioides difficile. Int. J. Mol. Sci. 2020, 21, 1277. [Google Scholar] [CrossRef]
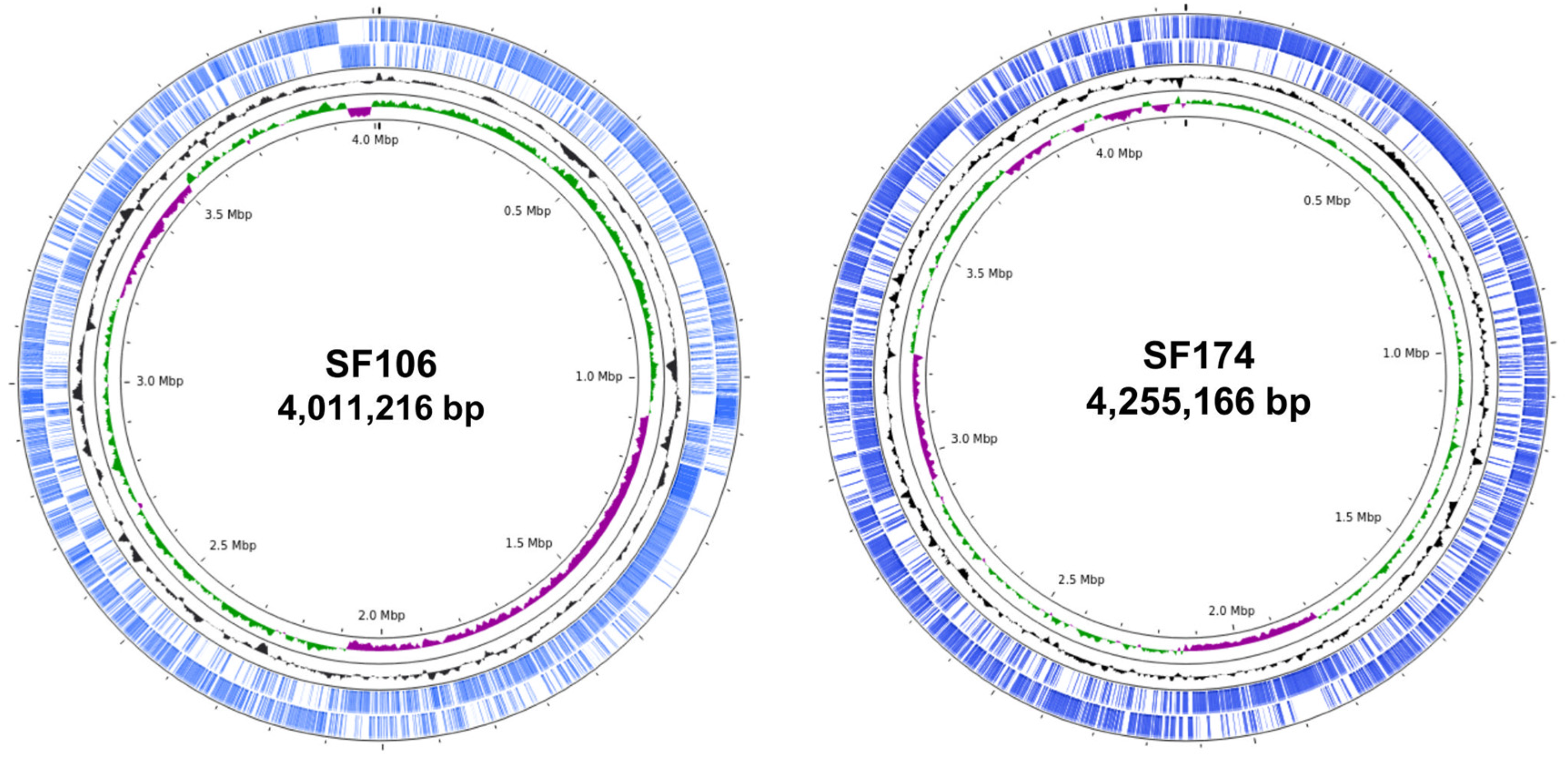
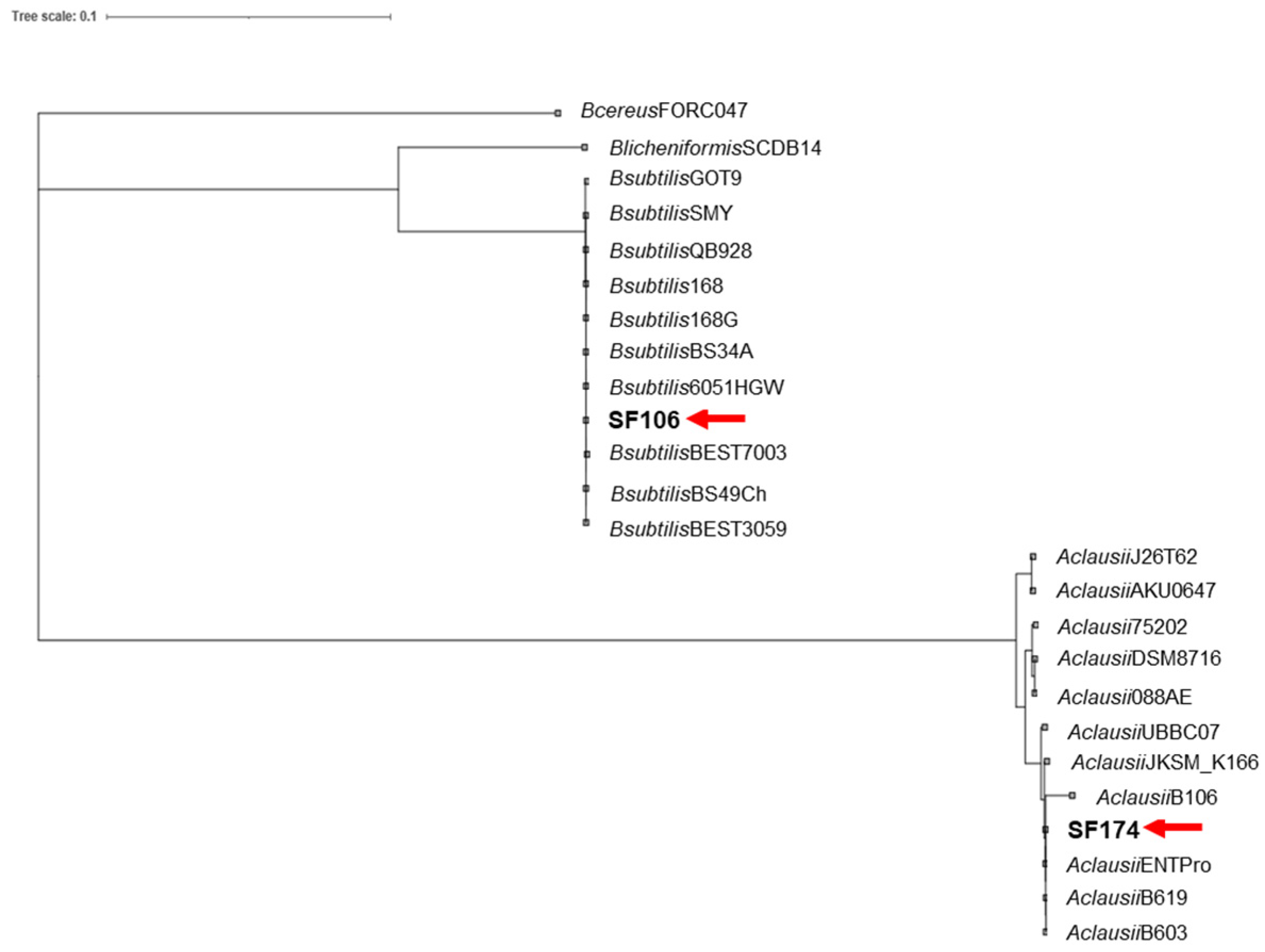
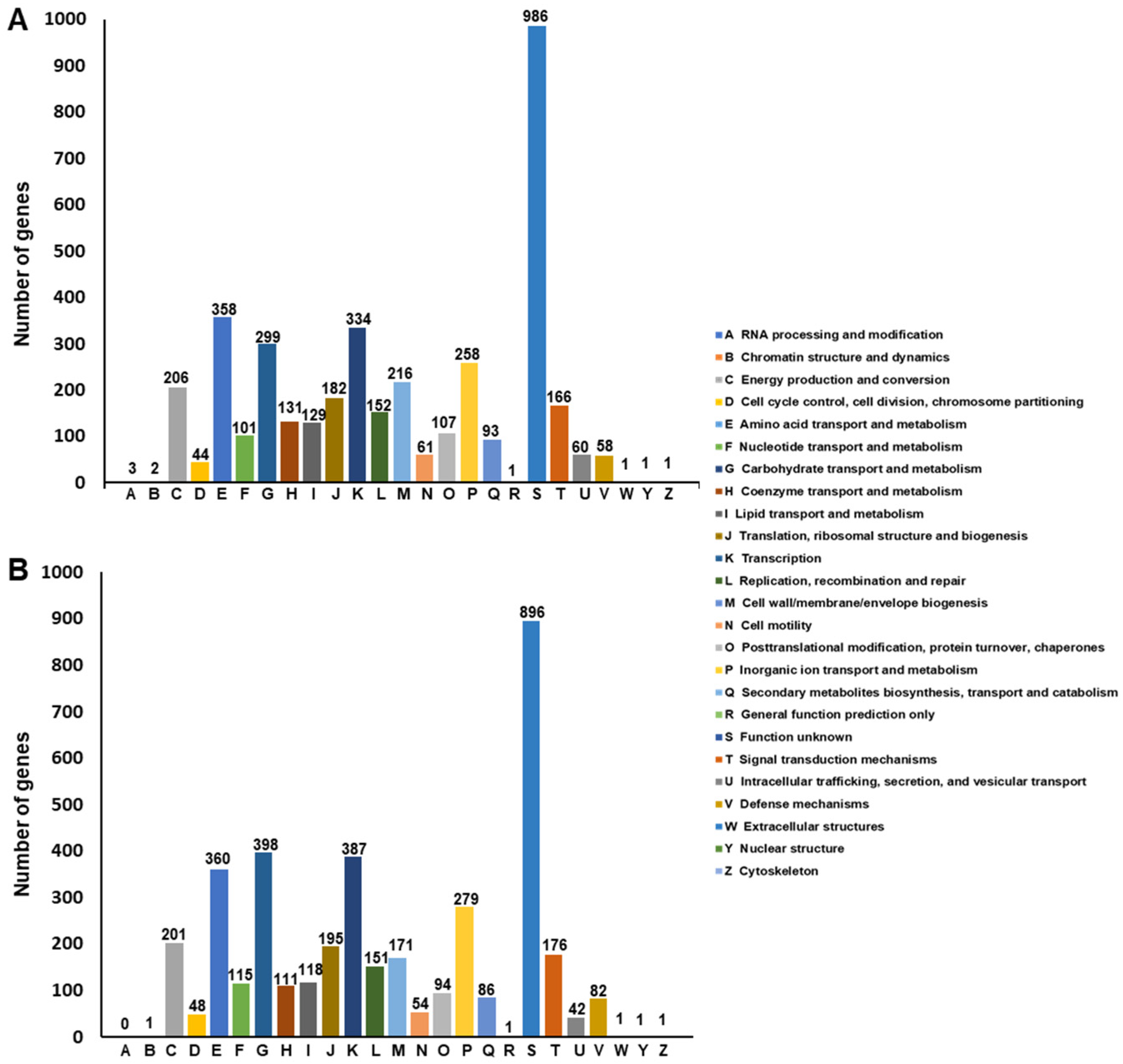


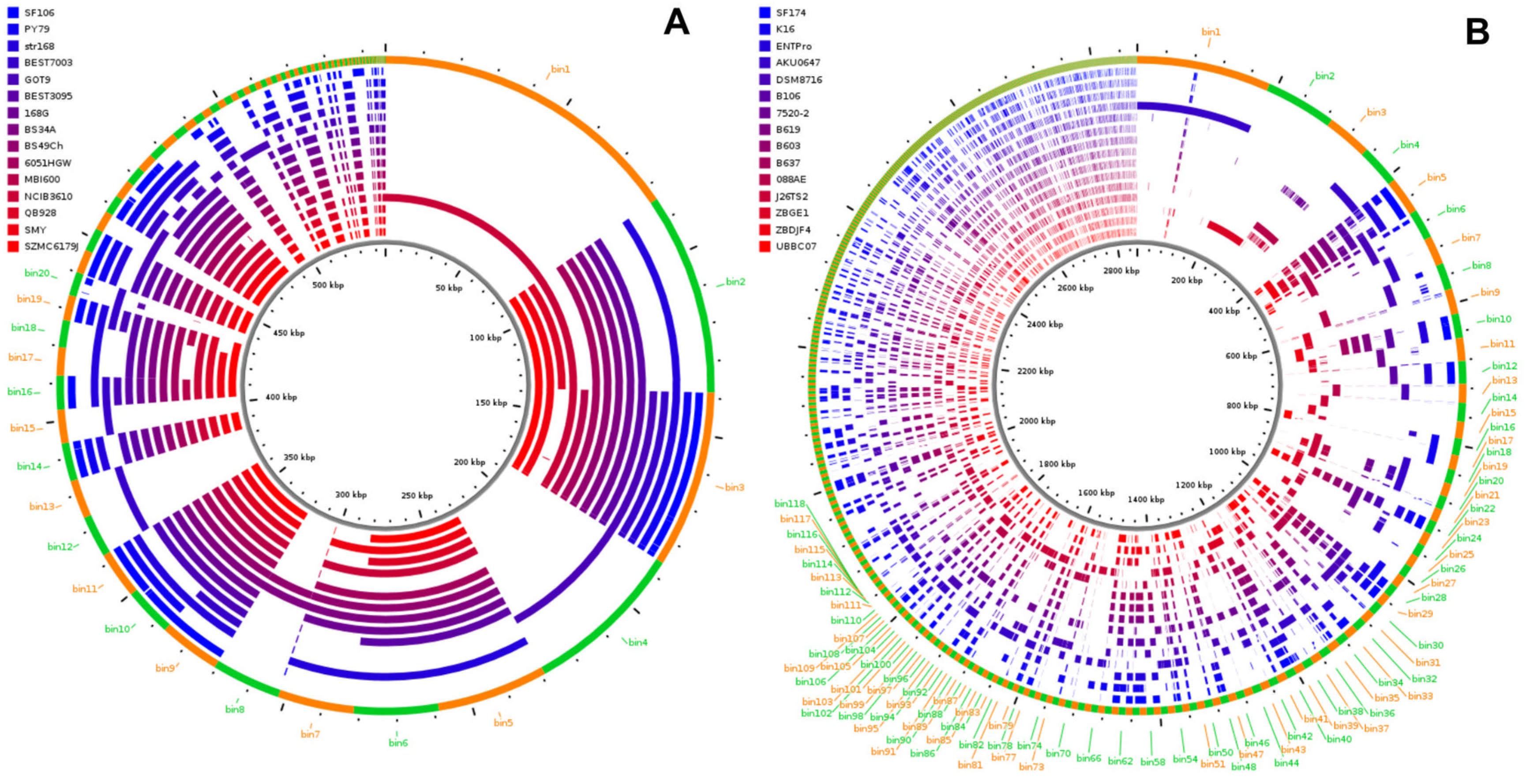
| SF106 | SF174 | |
|---|---|---|
| Size (bp) | 4,011,216 | 4,255,166 |
| Number of contigs | 11 | 40 |
| Average GC percentage | 43.75 | 44.67 |
| Number of predicted ORFs | 3992 | 4225 |
| Number of predicted rRNA genes | 8 | 12 |
| Number of predicted tRNA genes | 63 | 75 |
| Strain | GH | GT | PL | CE | CBM | Total |
|---|---|---|---|---|---|---|
| SF106 | 53 | 39 | 7 | 14 | 22 | 135 |
| B. subtilis Natto BEST195 | 55 | 38 | 5 | 13 | 34 | 145 |
| SF174 | 54 | 28 | 4 | 12 | 13 | 111 |
| A. clausii EntPro | 54 | 30 | 4 | 12 | 13 | 113 |
| Strain | GH2 | GH3 | GH18 | CE4 | CE9 |
|---|---|---|---|---|---|
| SF106 | 0 | 1 | 4 | 6 | 1 |
| SF174 | 1 | 2 | 2 | 6 | 1 |
| Target Bacteria | SF106 | SF174 |
|---|---|---|
| Pseudomonas fluorescens ATCC 13525 | - | - |
| Salmonella enterica typhi ATCC 14028 | - | - |
| Escherichia coli ATCC 25922 | - | - |
| Shigella sonnei ATCC 25931 | - | - |
| Bacillus subtilis NCBI 3610 | - | - |
| Bacillus cereus GC105 | +s/c | - |
| Bacillus licheniformis SG277 | - | - |
| Listeria monocytogenes ATCC7644 | +s/c | +c |
| Lactobacillus rhamnosus GG | - | - |
| Mycobacterium smegmatis mc2 155 | - | - |
| Staphylococcus aureus ATCC 6538 | +c | - |
| Staphylococcus aureus ATCC 25923 | +c | - |
| Staphylococcus aureus ATCC 43300 MRSA | +c | - |
| Staphylococcus aureus ST398 MDR | +c | - |
| Staphylococcus aureus 392 MDR | +c | - |
| Candida albicans | + | + |
| Antibiotic | B. subtilis SF106 a | A. clausi SF174 a | Bacillus EFSA Breakpoint a |
|---|---|---|---|
| Tetracycline | 4.00 | 7.50 | 8 |
| Vancomycin | 2.00 | 0.50 | 4 |
| Streptomycin | 32.00 | 16.00 | 8 |
| Clindamycin | 1.19 | 16.00 | 4 |
| Gentamycin | 0.44 | 0.19 | 4 |
| Erythromycin | 0.07 | 16.00 | 4 |
| Kanamycin | 0.75 | 7.50 | 8 |
| Chloramphenicol | 1.00 | 24.00 | 8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saggese, A.; Giglio, R.; D’Anzi, N.; Baccigalupi, L.; Ricca, E. Comparative Genomics and Physiological Characterization of Two Aerobic Spore Formers Isolated from Human Ileal Samples. Int. J. Mol. Sci. 2022, 23, 14946. https://doi.org/10.3390/ijms232314946
Saggese A, Giglio R, D’Anzi N, Baccigalupi L, Ricca E. Comparative Genomics and Physiological Characterization of Two Aerobic Spore Formers Isolated from Human Ileal Samples. International Journal of Molecular Sciences. 2022; 23(23):14946. https://doi.org/10.3390/ijms232314946
Chicago/Turabian StyleSaggese, Anella, Rosa Giglio, Nicola D’Anzi, Loredana Baccigalupi, and Ezio Ricca. 2022. "Comparative Genomics and Physiological Characterization of Two Aerobic Spore Formers Isolated from Human Ileal Samples" International Journal of Molecular Sciences 23, no. 23: 14946. https://doi.org/10.3390/ijms232314946
APA StyleSaggese, A., Giglio, R., D’Anzi, N., Baccigalupi, L., & Ricca, E. (2022). Comparative Genomics and Physiological Characterization of Two Aerobic Spore Formers Isolated from Human Ileal Samples. International Journal of Molecular Sciences, 23(23), 14946. https://doi.org/10.3390/ijms232314946








