Different Effects of Different Lactobacillus acidophilus Strains on DSS-Induced Colitis
Abstract
1. Introduction
2. Results
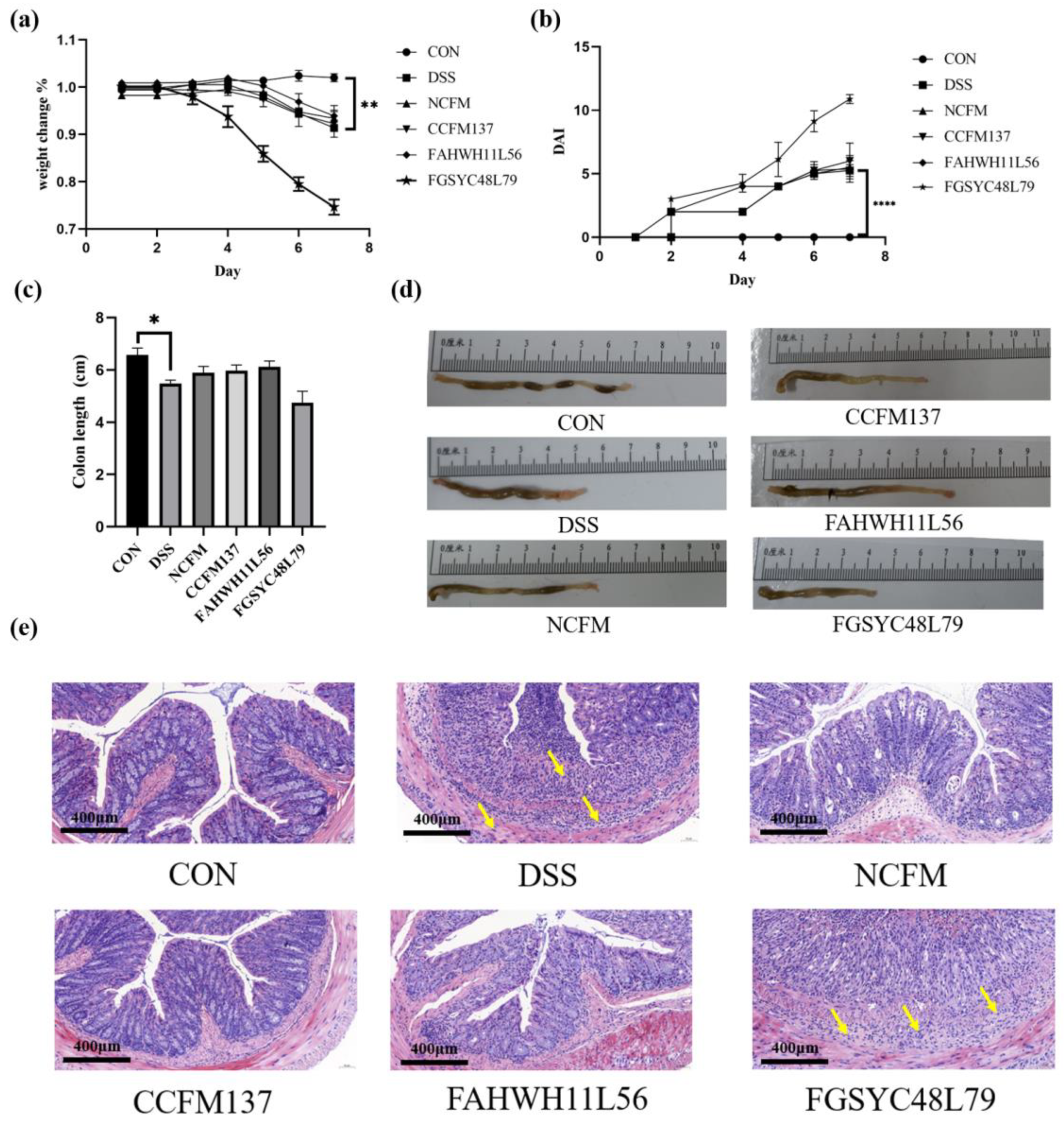
2.1. L. acidophilus Improved the Symptoms of Mice with Colitis
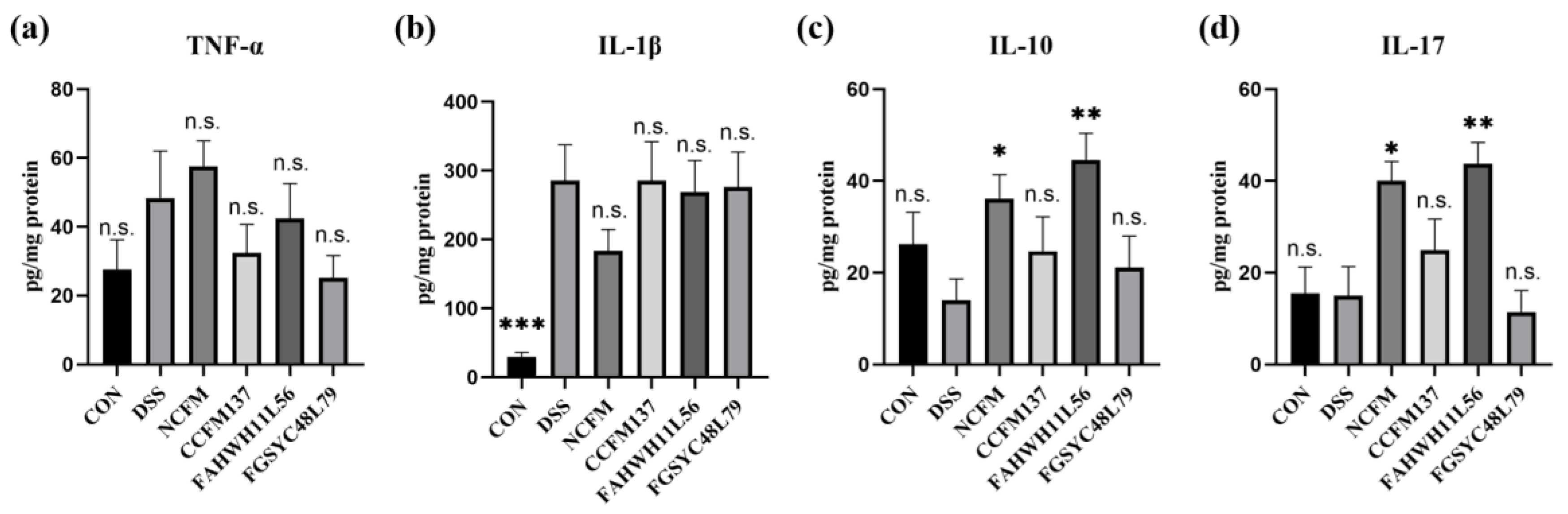
2.2. L. acidophilus Affected the Cytokines in the Colon of Mice
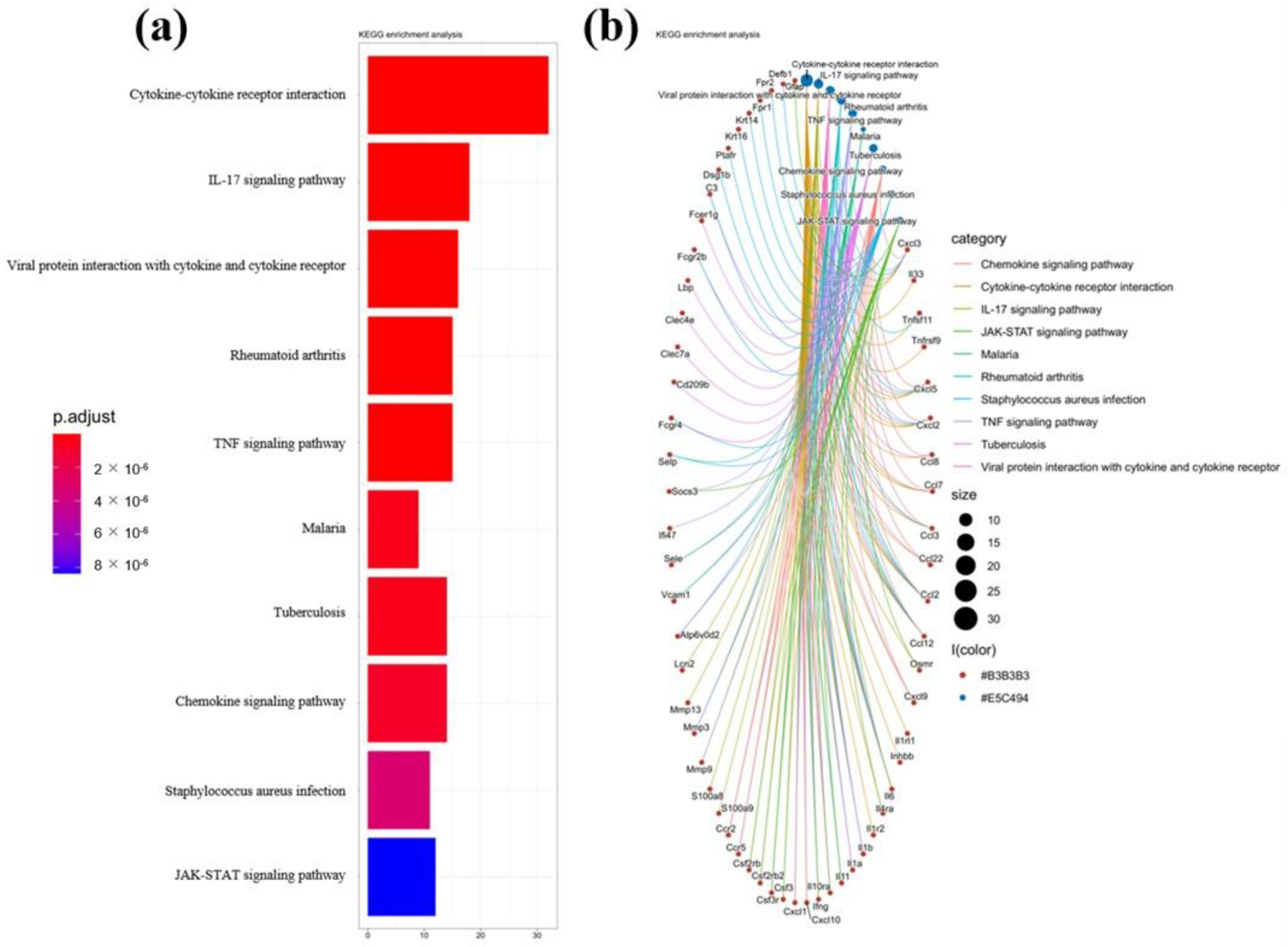
2.3. L. acidophilus Affected the CCL2/CCR2 Axis and CCL3/CCR1 Axis Downstream IL-17 Signal Pathway in the Colon of Mice
2.4. L. acidophilus Modified the Diversity of Gut Microbiota in DSS-Induced Colitis Mice
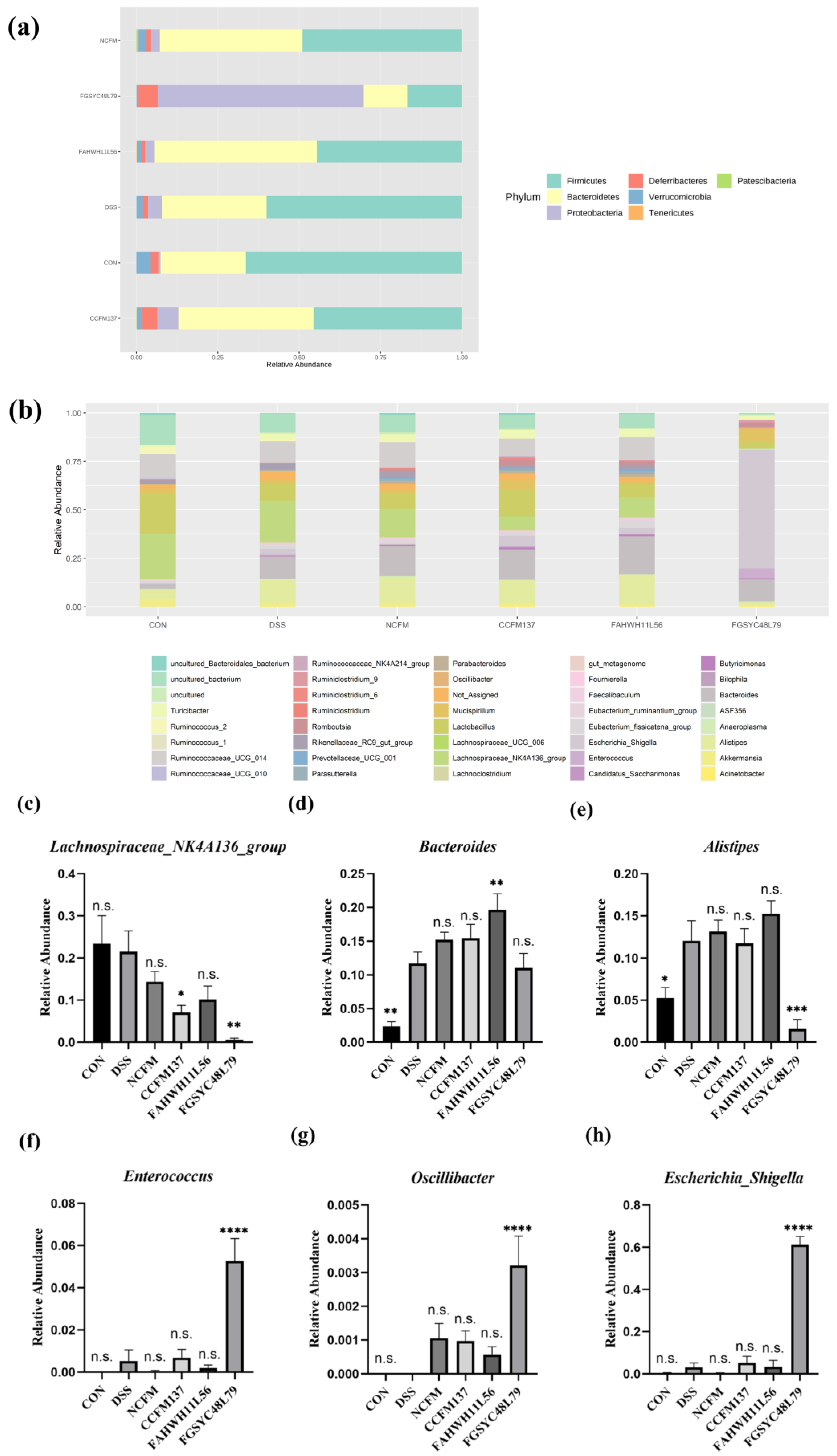
2.5. L. acidophilus Modified the Composition of Gut Microbiota in DSS-Induced Colitis Mice
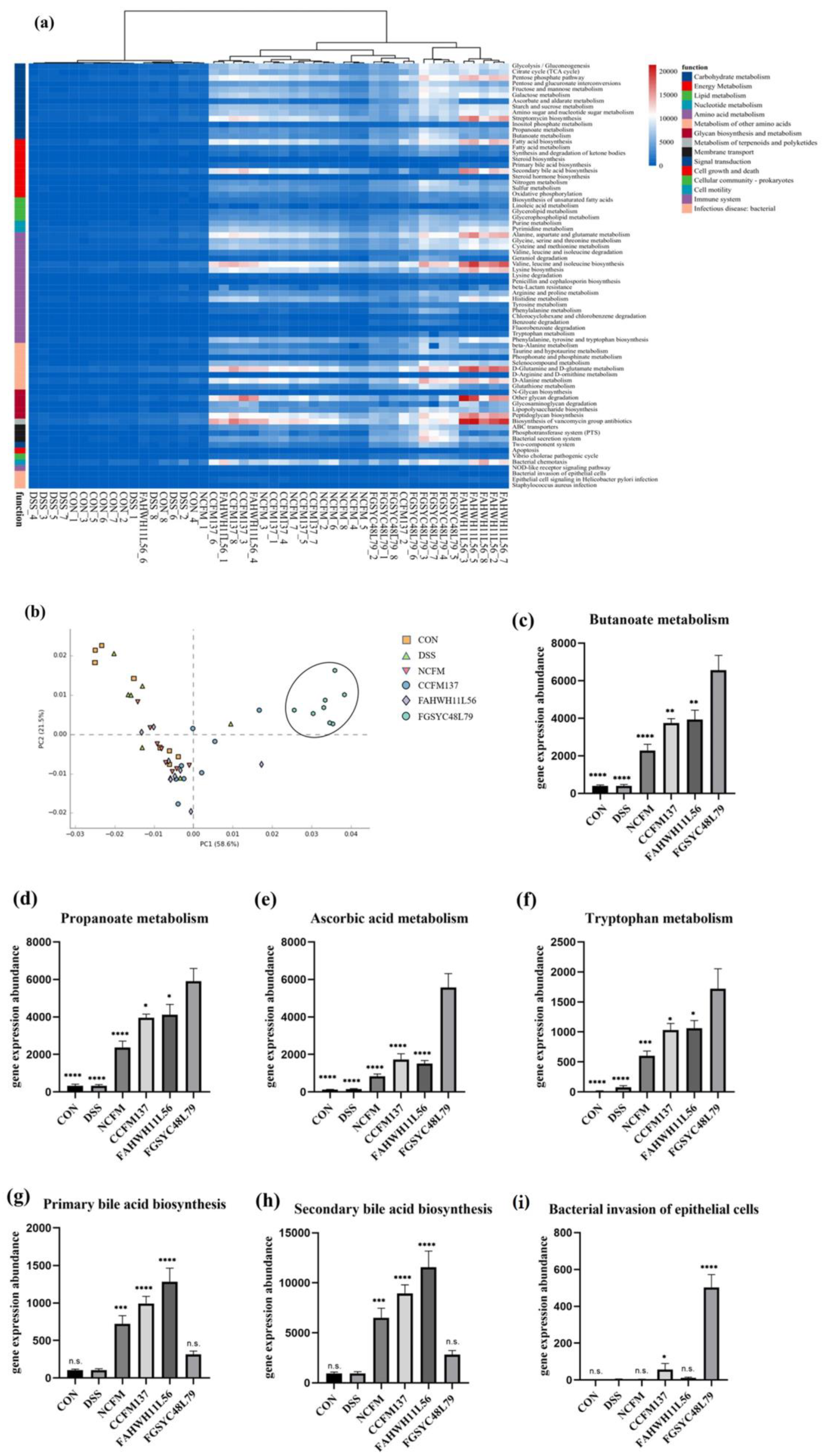
2.6. PICRUSt Analysis of Gut Microbiota in DSS-Induced Colitis Mice Modified by L. acidophilus
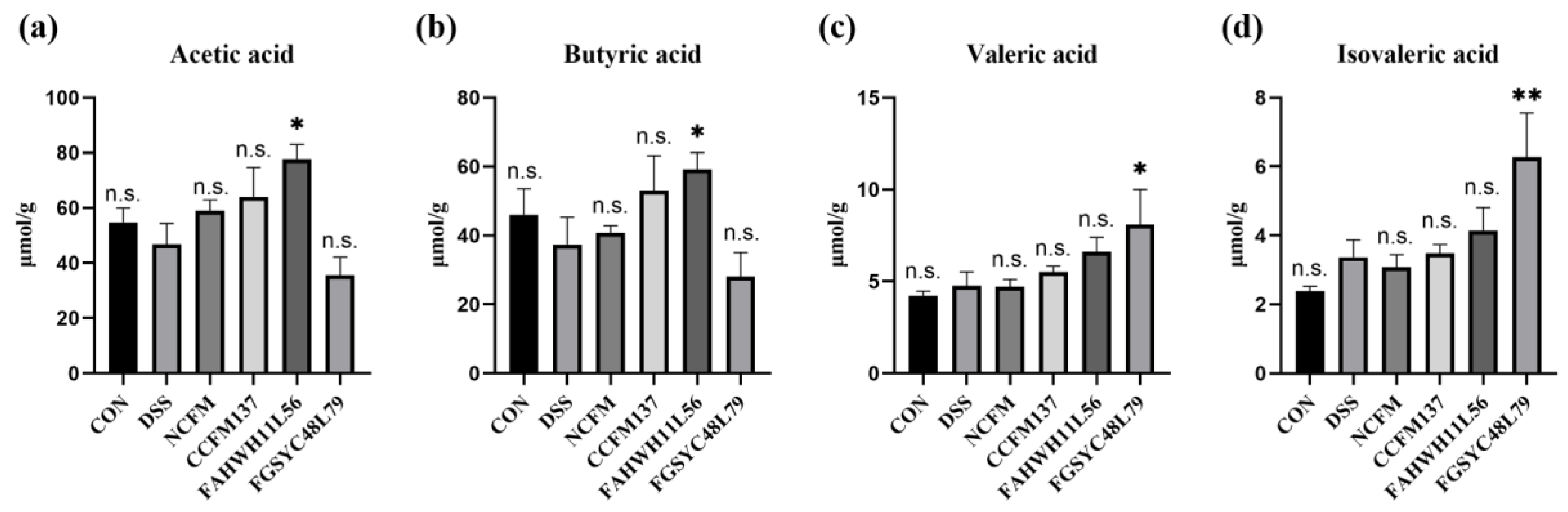
2.7. L. acidophilus Modified the Fecal SCFA in DSS-Induced Colitis Mice
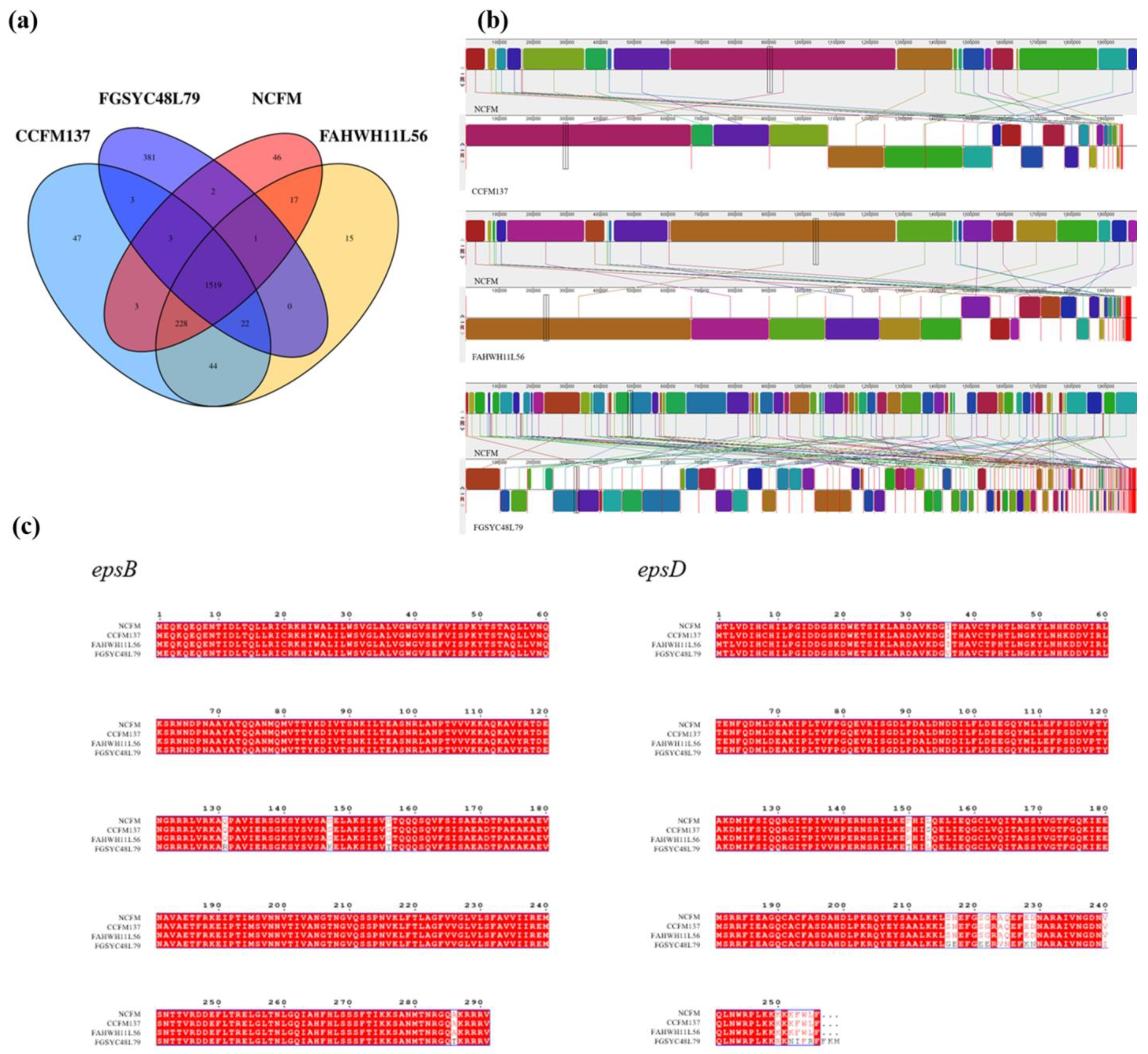
2.8. Genome Analysis of L. acidophilus
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Preparation
4.2. Animal Trial Design
4.3. Assessment of Colitis Symptoms
4.4. Biochemical Assays
4.5. RNA Isolation and Real-Time Quantitative-PCR
4.6. DNA Extraction and Pyrosequencing
4.7. Short Chain Fatty Acid Determination
4.8. Genome Analysis
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, Y.Z.; Li, Y.Y. Inflammatory bowel disease: Pathogenesis. World J. Gastroenterol. 2014, 20, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.X.; Chen, Y.X.; Wang, Y.Y.; Li, Y.; Zhang, X.J.; Zheng, H.H.; Ma, F.L.; Ma, C.W.; Lu, B.Y.; Xie, Z.Y.; et al. Beneficial changes of gut microbiota and metabolism in weaned rats with Lactobacillus acidophilus NCFM and Bifidobacterium lactis Bi-07 supplementation. J. Funct. Foods 2018, 48, 252–265. [Google Scholar] [CrossRef]
- Gao, X.W.; Mubasher, M.; Fang, C.Y.; Reifer, C.; Miller, L.E. Dose-response efficacy of a proprietary probiotic formula of Lactobacillus acidophilus CL1285 and Lactobacillus casei LBC80R for antibiotic-associated diarrhea and clostridium difficile-Associated diarrhea prophylaxis in adult patients. Am. J. Gastroenterol. 2010, 105, 1636–1641. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Xu, P.; Wu, Z.; Pan, D. Anti-inflammatory activity of surface layer protein SlpA of Lactobacillus acidophilus CICC 6074 in LPS-induced RAW 264.7 cells and DSS-induced mice colitis. J. Funct. Foods 2018, 51, 16–27. [Google Scholar] [CrossRef]
- Kim, W.K.; Han, D.H.; Jang, Y.J.; Park, S.; Jang, S.J.; Lee, G.; Han, H.S.; Ko, G. Alleviation of DSS-induced colitis via Lactobacillus acidophilus treatment in mice. Food Funct. 2021, 12, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.T.; Wang, H.X.; Xiang, C.Z.; Mu, J.F.; Zhao, X. Preventive Effect of Lactobacillus acidophilus XY27 on DSS-Induced Ulcerative Colitis in Mice. Drug Des. Dev. Ther. 2020, 14, 5645–5657. [Google Scholar] [CrossRef]
- Park, J.S.; Choi, J.; Jhun, J.; Kwon, J.Y.; Lee, B.I.; Yang, C.W.; Park, S.H.; Cho, M.L. Lactobacillus acidophilus Improves Intestinal Inflammation in an Acute Colitis Mouse Model by Regulation of Th17 and Treg Cell Balance and Fibrosis Development. J. Med. Food 2018, 21, 215–224. [Google Scholar] [CrossRef]
- Mohamadzadeh, M.; Pfeiler, E.A.; Brown, J.B.; Zadeh, M.; Gramarossa, M.; Managlia, E.; Bere, P.; Sarraj, B.; Khan, M.W.; Pakanati, K.C.; et al. Regulation of induced colonic inflammation by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc. Natl. Acad. Sci. USA 2011, 108, 4623–4630. [Google Scholar] [CrossRef]
- Huang, Z.; Zhou, X.Y.; Stanton, C.; Ross, R.P.; Zhao, J.X.; Zhang, H.; Yang, B.; Chen, W. Comparative Genomics and Specific Functional Characteristics Analysis of Lactobacillus acidophilus. Microorganisms 2021, 9, 1992. [Google Scholar] [CrossRef]
- Cui, Y.; Wei, H.Y.; Lu, F.G.; Liu, X.W.; Liu, D.L.; Gu, L.; Ouyang, C.H. Different Effects of Three Selected Lactobacillus Strains in Dextran Sulfate Sodium-Induced Colitis in BALB/c Mice. PLoS ONE 2016, 11, e0148241. [Google Scholar] [CrossRef]
- Zhang, Q.P.; Wu, Y.Q.; Wang, J.; Wu, G.J.; Long, W.M.; Xue, Z.S.; Wang, L.H.; Zhang, X.J.; Pang, X.Y.; Zhao, Y.F.; et al. Accelerated dysbiosis of gut microbiota during aggravation of DSS-induced colitis by a butyrate-producing bacterium. Sci. Rep. 2016, 6, 27572. [Google Scholar] [CrossRef] [PubMed]
- Arai, Y.; Takanashi, H.; Kitagawa, H.; Okayasu, I. Involvement of interleukin-1 in the development of ulcerative colitis induced by dextran sulfate sodium in mice. Cytokine 1998, 10, 890–896. [Google Scholar] [CrossRef] [PubMed]
- Popivanova, B.K.; Kitamura, K.; Wu, Y.; Kondo, T.; Kagaya, T.; Kaneko, S.; Oshima, M.; Fujii, C.; Mukaida, N. Blocking TNF-alpha in mice reduces colorectal carcinogenesis associated with chronic colitis. J. Clin. Investig. 2008, 118, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Schottelius, A.J.G.; Mayo, M.W.; Sartor, R.B.; Badwin, A.S. Interleukin-10 signaling blocks inhibitor of kappa B kinase activity and nuclear factor kappa B DNA binding. J. Biol. Chem. 1999, 274, 31868–31874. [Google Scholar] [CrossRef]
- Veldhoen, M. Interleukin 17 is a chief orchestrator of immunity. Nat. Immunol. 2017, 18, 612–621. [Google Scholar] [CrossRef]
- Chen, L.; Zou, Y.; Peng, J.; Lu, F.; Yin, Y.; Li, F.; Yang, J. Lactobacillus acidophilus Suppresses Colitis-Associated Activation of the IL-23/Th17 Axis. J. Immunol. Res. 2015, 2015, 909514. [Google Scholar] [CrossRef]
- Sarra, M.; Pallone, F.; MacDonald, T.T.; Monteleone, G. IL-23/IL-17 Axis in IBD. Inflamm. Bowel Dis. 2010, 16, 1808–1813. [Google Scholar] [CrossRef]
- Zhang, H.-j.; Xu, B.; Wang, H.; Xu, B.; Wang, G.-d.; Jiang, M.-z.; Lei, C.; Ding, M.-l.; Yu, P.-f.; Nie, Y.-z.; et al. IL-17 is a protection effector against the adherent-invasive Escherichia coli in murine colitis. Mol. Immunol. 2018, 93, 166–172. [Google Scholar] [CrossRef]
- McClellan, J.L.; Davis, J.M.; Steiner, J.L.; Enos, R.T.; Jung, S.H.; Carson, J.A.; Pena, M.M.; Carnevale, K.A.; Berger, F.G.; Murphy, E.A. Linking tumor-associated macrophages, inflammation, and intestinal tumorigenesis: Role of MCP-1. Am. J. Physiol. -Gastrointest. Liver Physiol. 2012, 303, G1087–G1095. [Google Scholar] [CrossRef]
- Chun, E.; Lavoie, S.; Michaud, M.; Gallini, C.A.; Kim, J.; Soucy, G.; Odze, R.; Glickman, J.N.; Garrett, W.S. CCL2 Promotes Colorectal Carcinogenesis by Enhancing Polymorphonuclear Myeloid-Derived Suppressor Cell Population and Function. Cell Rep. 2015, 12, 244–257. [Google Scholar] [CrossRef]
- Sanford, D.E.; Belt, B.A.; Panni, R.Z.; Mayer, A.; Deshpande, A.D.; Carpenter, D.; Mitchem, J.B.; Plambeck-Suess, S.M.; Worley, L.A.; Goetz, B.D.; et al. Inflammatory Monocyte Mobilization Decreases Patient Survival in Pancreatic Cancer: A Role for Targeting the CCL2/CCR2 Axis. Clin. Cancer Res. 2013, 19, 3404–3415. [Google Scholar] [CrossRef] [PubMed]
- Li, X.G.; Yao, W.B.; Yuan, Y.; Chen, P.Z.; Li, B.; Li, J.Q.; Chu, R.A.; Song, H.Y.; Xie, D.; Jiang, X.Q.; et al. Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut 2017, 66, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Izumi, K.; Fang, L.Y.; Mizokami, A.; Namiki, M.; Li, L.; Lin, W.J.; Chang, C. Targeting the androgen receptor with siRNA promotes prostate cancer metastasis through enhanced macrophage recruitment via CCL2/CCR2-induced STAT3 activation. Embo Mol. Med. 2013, 5, 1383–1401. [Google Scholar] [CrossRef] [PubMed]
- Ajuebor, M.N.; Hogaboam, C.M.; Le, T.; Proudfoot, A.E.I.; Swain, M.G. CCL3/MIP-1 alpha is pro-inflammatory in murine T cell-mediated hepatitis by recruiting CCR1-expressing CD4(+) T cells to the liver. Eur. J. Immunol. 2004, 34, 2907–2918. [Google Scholar] [CrossRef]
- Yang, X.Q.; Lu, P.R.; Fujii, C.; Nakamoto, Y.; Gao, J.L.; Kaneko, S.; Murphy, P.M.; Mukaida, N. Essential contribution of a chemokine, CCL3, and its receptor, CCR1, to hepatocellular carcinoma progression. Int. J. Cancer 2006, 118, 1869–1876. [Google Scholar] [CrossRef] [PubMed]
- Silva, T.A.; Ribeiro, F.L.L.; De Oliveira-Neto, H.H.; Watanabe, S.; Alencar, R.; Fukada, S.Y.; Cunha, F.Q.; Leles, C.R.; Mendonca, E.F.; Batista, A.C. Dual role of CCL3/CCR1 in oral squamous cell carcinoma: Implications in tumor metastasis and local host defense. Oncol. Rep. 2007, 18, 1107–1113. [Google Scholar]
- Lavelle, A.; Lennon, G.; O’Sullivan, O.; Docherty, N.; Balfe, A.; Maguire, A.; Mulcahy, H.E.; Doherty, G.; O’Donoghue, D.; Hyland, J.; et al. Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut 2015, 64, 1553–1561. [Google Scholar] [CrossRef]
- Shin, N.-R.; Whon, T.W.; Bae, J.-W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Shao, X.; Sun, C.Z.; Tang, X.; Zhang, X.S.; Han, D.; Liang, S.; Qu, R.; Hui, X.D.; Shan, Y.W.; Hu, L.H.; et al. Anti-Inflammatory and Intestinal Microbiota Modulation Properties of Jinxiang Garlic (Allium sativum L.) Polysaccharides toward Dextran Sodium Sulfate-Induced Colitis. J. Agric. Food Chem. 2020, 68, 12295–12309. [Google Scholar] [CrossRef]
- Alam, M.T.; Amos, G.C.A.; Murphy, A.R.J.; Murch, S.; Wellington, E.M.H.; Arasaradnam, R.P. Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathogens 2020, 12, 1. [Google Scholar] [CrossRef]
- Berry, D.; Reinisch, W. Intestinal microbiota: A source of novel biomarkers in inflammatory bowel diseases? Best Pract. Res. Clin. Gastroenterol. 2013, 27, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Parker, B.J.; Wearsch, P.A.; Veloo, A.C.M.; Rodriguez-Palacios, A. The GenusAlistipes: Gut Bacteria with Emerging Implications to Inflammation, Cancer, and Mental Health. Front. Immunol. 2020, 11, 906. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.J.; Goeser, L.; Lu, K.; Faith, J.J.; Hansen, J.J. Enterococcus faecalis Glucosamine Metabolism Exacerbates Experimental Colitis. Cell. Mol. Gastroenterol. Hepatol. 2021, 12, 1373–1389. [Google Scholar] [CrossRef]
- Li, M.; Wu, Y.Q.; Hu, Y.X.; Zhao, L.P.; Zhang, C.H. Initial gut microbiota structure affects sensitivity to DSS-induced colitis in a mouse model. Sci. China-Life Sci. 2018, 61, 762–769. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.P.; Wang, W.; Zhou, R.; Ng, S.C.; Li, J.; Huang, M.F.; Zhou, F.; Wang, X.; Shen, B.; Kamm, M.A.; et al. Characteristics of Fecal and Mucosa-Associated Microbiota in Chinese Patients with Inflammatory Bowel Disease. Medicine 2014, 93, e51. [Google Scholar] [CrossRef]
- Yan, H.Y.; Wang, H.J.; Zhang, X.L.; Li, X.Q.; Yu, J. Ascorbic acid ameliorates oxidative stress and inflammation in dextran sulfate sodium-induced ulcerative colitis in mice. Int. J. Clin. Exp. Med. 2015, 8, 20245–20253. [Google Scholar]
- Islam, J.; Sato, S.; Watanabe, K.; Watanabe, T.; Ardiansyah; Hirahara, K.; Aoyama, Y.; Tomita, S.; Aso, H.; Komai, M.; et al. Dietary tryptophan alleviates dextran sodium sulfate-induced colitis through aryl hydrocarbon receptor in mice. J. Nutr. Biochem. 2017, 42, 43–50. [Google Scholar] [CrossRef]
- Duboc, H.; Rajca, S.; Rainteau, D.; Benarous, D.; Maubert, M.A.; Quervain, E.; Thomas, G.; Barbu, V.; Humbert, L.; Despras, G.; et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2013, 62, 531–539. [Google Scholar] [CrossRef]
- Martinez-Augustin, O.; de Medina, F.S. Intestinal bile acid physiology and pathophysiology. World J. Gastroenterol. 2008, 14, 5630–5640. [Google Scholar] [CrossRef]
- Roager, H.M.; Sulek, K.; Skov, K.; Frandsen, H.L.; Smedsgaard, J.; Wilcks, A.; Skov, T.H.; Villas-Boas, S.G.; Licht, T.R. Lactobacillus acidophilus NCFM affects vitamin E acetate metabolism and intestinal bile acid signature in monocolonized mice. Gut Microbes 2014, 5, 296–303. [Google Scholar] [CrossRef]
- Gill, P.A.; van Zelm, M.C.; Muir, J.G.; Gibson, P.R. short chain fatty acids as potential therapeutic agents in human gastrointestinal and inflammatory disorders. Aliment. Pharmacol. Ther. 2018, 48, 15–34. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, J.; Redondo-Blanco, S.; Gutierrez-del-Rio, I.; Miguelez, E.M.; Villar, C.J.; Lombo, F. Colon microbiota fermentation of dietary prebiotics towards short-chain fatty acids and their roles as anti-inflammatory and antitumour agents: A review. J. Funct. Foods 2016, 25, 511–522. [Google Scholar] [CrossRef]
- Hallert, C.; Bjorck, I.; Nyman, M.; Pousette, A.; Granno, C.; Svensson, H. Increasing fecal butyrate in ulcerative colitis patients by diet: Controlled pilot study. Inflamm. Bowel Dis. 2003, 9, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Daïen, C.I.; Tan, J.; Audo, R.; Mielle, J.; Quek, L.E.; Krycer, J.R.; Angelatos, A.; Duraes, M.; Pinget, G.; Ni, D.; et al. Gut-derived acetate promotes B10 cells with antiinflammatory effects. JCI Insight 2021, 6, e144156. [Google Scholar] [CrossRef]
- Sengul, N.; Isik, S.; Aslim, B.; Ucar, G.; Demirbag, A.E. The Effect of Exopolysaccharide-Producing Probiotic Strains on Gut Oxidative Damage in Experimental Colitis. Dig. Dis. Sci. 2011, 56, 707–714. [Google Scholar] [CrossRef] [PubMed]
- El-Deeb, N.M.; Yassin, A.M.; Al-Madboly, L.A.; El-Hawiet, A. A novel purified Lactobacillus acidophilus 20079 exopolysaccharide, LA-EPS-20079, molecularly regulates both apoptotic and NF-kappa B inflammatory pathways in human colon cancer. Microb. Cell Factories 2018, 17, 29. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Waliullah, S.; Godfrey, V.; Khan, M.A.W.; Ramachandran, R.A.; Cantarel, B.L.; Behrendt, C.; Peng, L.; Hooper, L.V.; Zaki, H. Dietary simple sugars alter microbial ecology in the gut and promote colitis in mice. Sci. Transl. Med. 2020, 12, eaay6218. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, B.; Ross, R.P.; Jin, Y.; Stanton, C.; Zhao, J.X.; Zhang, H.; Chen, W. Orally Administered CLA Ameliorates DSS-Induced Colitis in Mice via Intestinal Barrier Improvement, Oxidative Stress Reduction, and Inflammatory Cytokine and Gut Microbiota Modulation. J. Agric. Food Chem. 2019, 67, 13282–13298. [Google Scholar] [CrossRef]
- Yan, S.; Yang, B.; Zhao, J.C.; Zhao, J.X.; Stanton, C.; Ross, R.P.; Zhang, H.; Chen, W. A ropy exopolysaccharide producing strain Bifidobacterium longum subsp. longum YS108R alleviates DSS-induced colitis by maintenance of the mucosal barrier and gut microbiota modulation. Food Funct. 2019, 10, 1595–1608. [Google Scholar] [CrossRef]
- Sun, X.W.; Gao, Y.; Wang, X.; Hu, G.; Wang, Y.; Feng, B.; Hu, Y.X.; Mu, X.; Zhang, Y.; Dong, H. Escherichia coli O-101-induced diarrhea develops gut microbial dysbiosis in rats. Exp. Ther. Med. 2019, 17, 824–834. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; He, Z.J.; Zhou, Y.H.; Ross, R.P.; Stanton, C.; Zhao, J.X.; Zhang, H.; Yang, B.; Chen, W. Lactobacillus plantarum relieves diarrhea caused by enterotoxin-producing Escherichia coli through inflammation modulation and gut microbiota regulation. Food Funct. 2020, 11, 10362–10374. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Stoeckert, C.J.; Roos, D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef]
- Xavier, R.; Patrice, G. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]


| Primer | Sequence |
|---|---|
| CCL2-F | 5′-TTAAAAACCTGGATCGGAACCAA-3′ |
| CCL2-R | 5′-GCATTAGCTTCAGATTTACGGGT-3′ |
| CCL3-F | 5′-TTCTCTGTACCATGACACTCTGC-3′ |
| CCL3-R | 5′-CGTGGAATCTTCCGGCTGTAG-3′ |
| CCR2-F | 5′-ATCCACGGCATACTATCAACATC-3′ |
| CCR2-R | 5′-CAAGGCTCACCATCATCGTAG-3′ |
| CCR1-F | 5′-CTCATGCAGCATAGGAGGCTT-3′ |
| CCR1-R | 5′-ACATGGCATCACCAAAAATCCA-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Z.; Gong, L.; Jin, Y.; Stanton, C.; Ross, R.P.; Zhao, J.; Yang, B.; Chen, W. Different Effects of Different Lactobacillus acidophilus Strains on DSS-Induced Colitis. Int. J. Mol. Sci. 2022, 23, 14841. https://doi.org/10.3390/ijms232314841
Huang Z, Gong L, Jin Y, Stanton C, Ross RP, Zhao J, Yang B, Chen W. Different Effects of Different Lactobacillus acidophilus Strains on DSS-Induced Colitis. International Journal of Molecular Sciences. 2022; 23(23):14841. https://doi.org/10.3390/ijms232314841
Chicago/Turabian StyleHuang, Zheng, Lei Gong, Yan Jin, Catherine Stanton, Reynolds Paul Ross, Jianxin Zhao, Bo Yang, and Wei Chen. 2022. "Different Effects of Different Lactobacillus acidophilus Strains on DSS-Induced Colitis" International Journal of Molecular Sciences 23, no. 23: 14841. https://doi.org/10.3390/ijms232314841
APA StyleHuang, Z., Gong, L., Jin, Y., Stanton, C., Ross, R. P., Zhao, J., Yang, B., & Chen, W. (2022). Different Effects of Different Lactobacillus acidophilus Strains on DSS-Induced Colitis. International Journal of Molecular Sciences, 23(23), 14841. https://doi.org/10.3390/ijms232314841






