Decoding Gene Expression Signatures Underlying Vegetative to Inflorescence Meristem Transition in the Common Bean
Abstract
:1. Introduction
2. Results
2.1. Morphological Developmental Changes Occurring during Inflorescence Meristem Differentiation
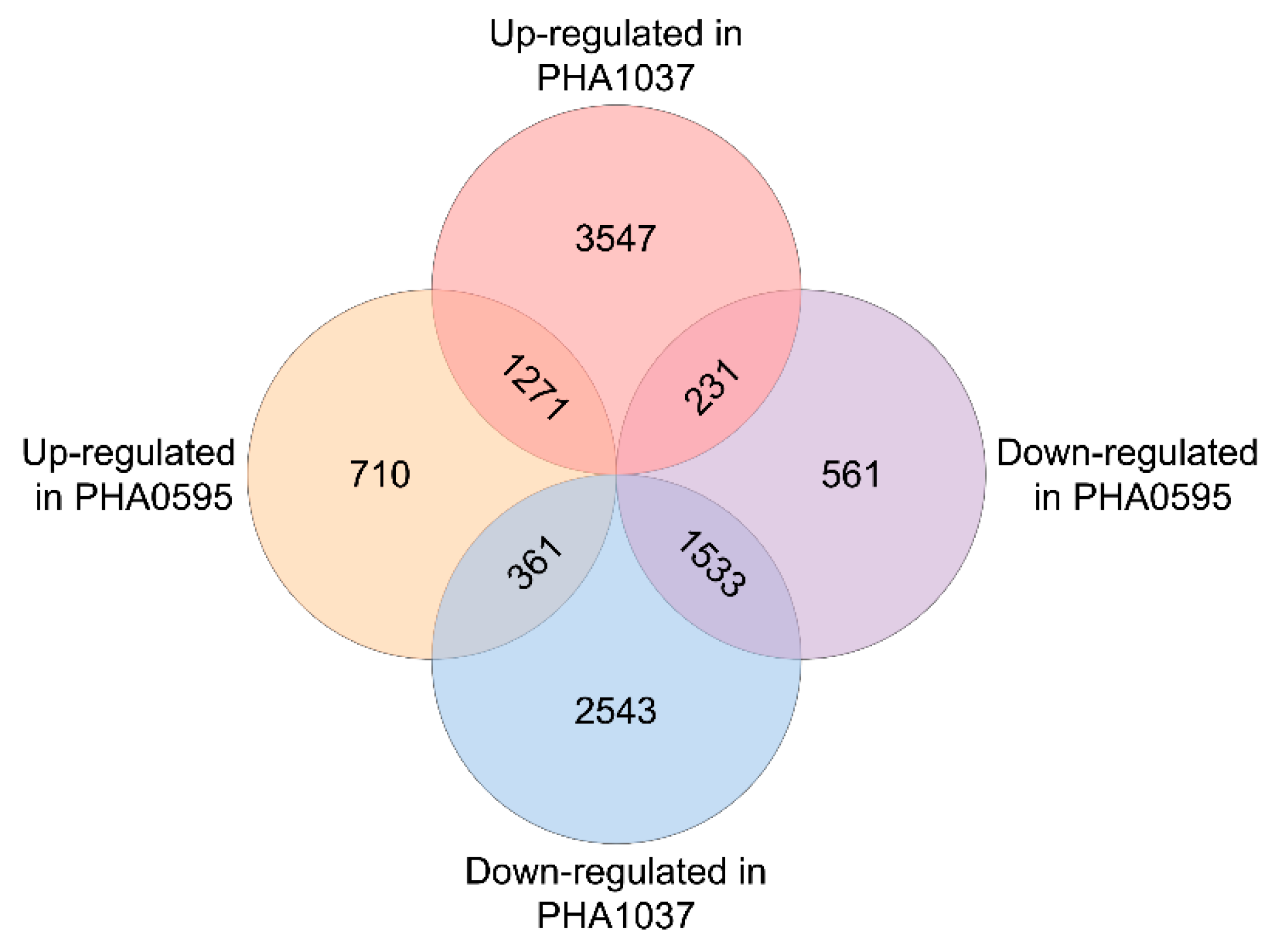
2.2. Gene Expression Changes upon Differentiation to Inflorescence Meristem
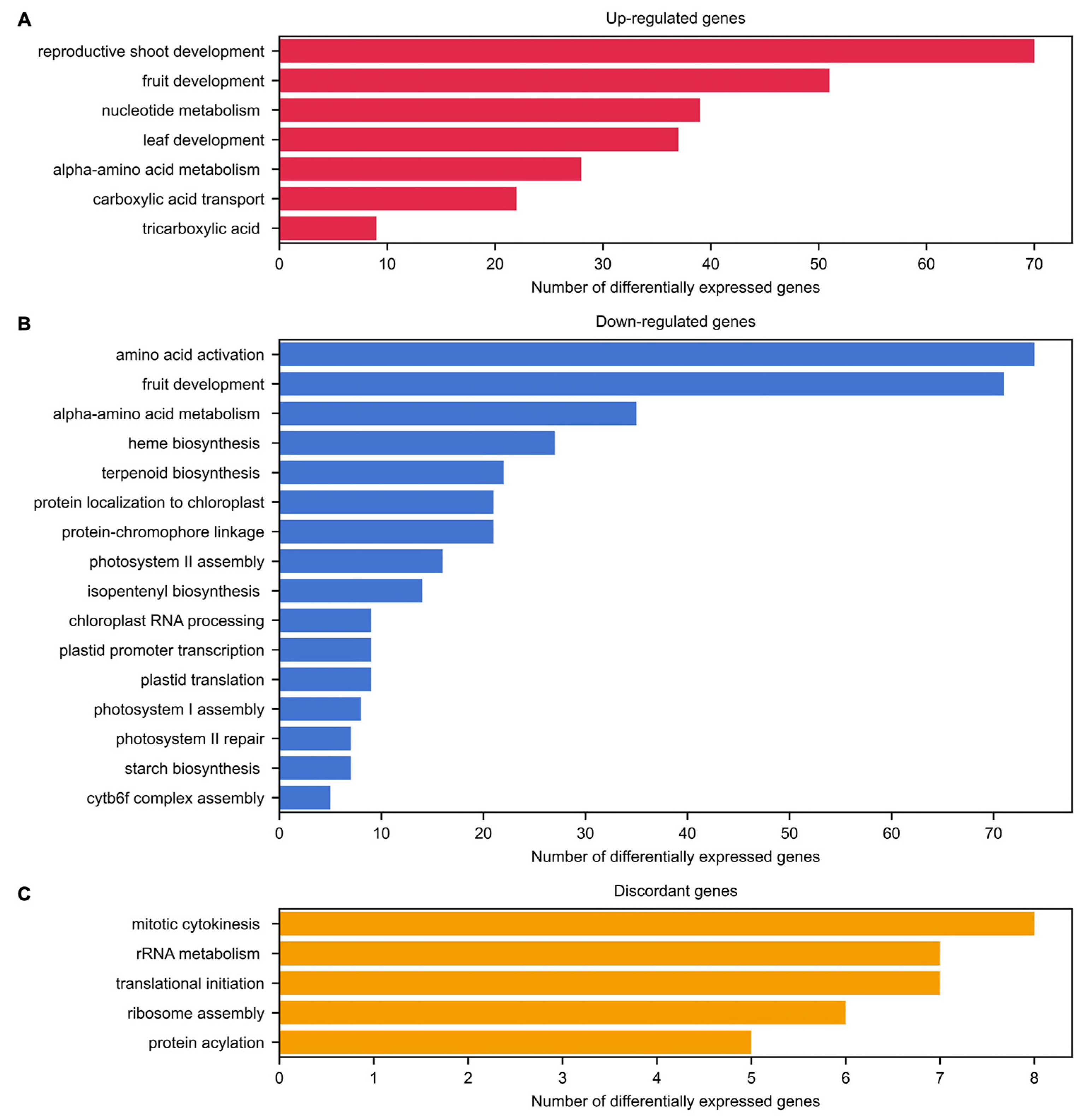
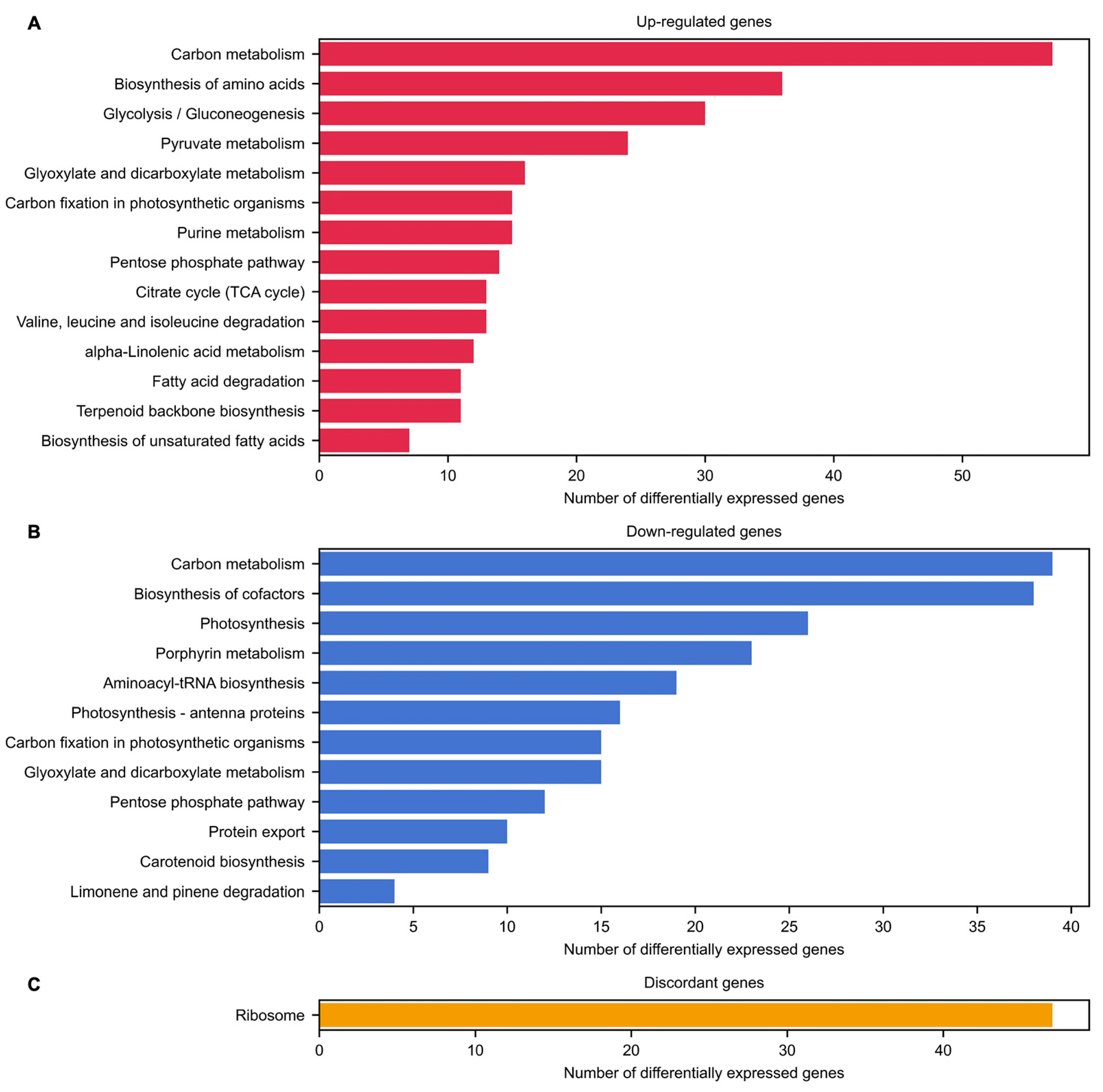
2.3. Biological Processes and Pathways Affected by Inflorescence Meristem Development
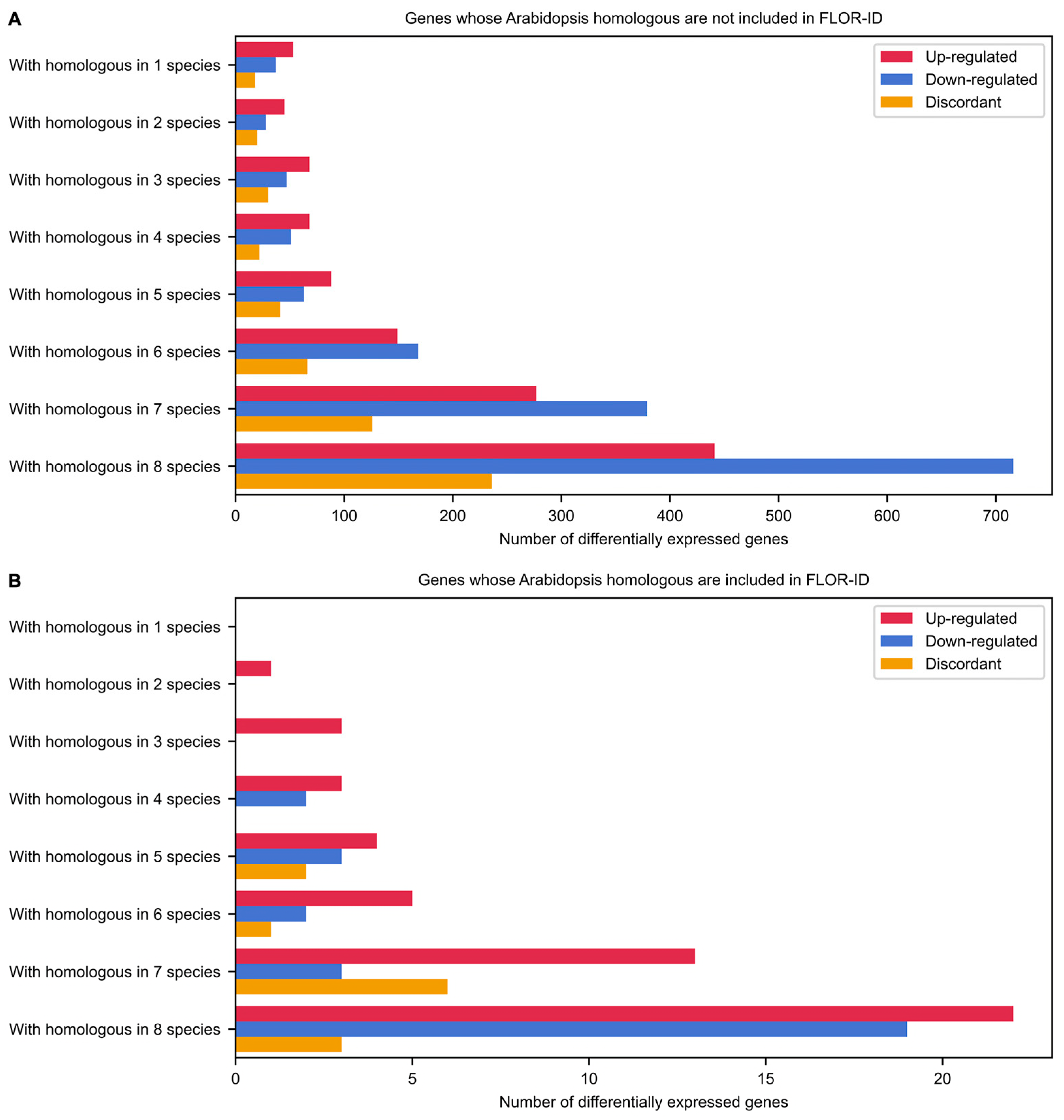
2.4. Evolutionarily Conserved Genes Involved in Flowering of the Common Bean
2.5. Candidate Marker Genes for Flowering Induction in the Common Bean
3. Discussion
4. Materials and Methods
4.1. Plant Growth and Tissue Collection
4.2. RNA-Seq Analysis
4.3. Identification of Differentially Expressed Genes
4.4. Biological Processes and Pathways Analyses
4.5. Evolutionary Conservation Analysis
4.6. Quantitative Real-Time PCR (qPCR) for Validation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muasya, R.M.; Lommen, W.J.M.; Struik, P.C. Differences in development of common bean (Phaseolus vulgaris L.) crops and pod fractions within a crop I. Seed growth and maturity. Field Crops Res. 2002, 75, 63–78. [Google Scholar] [CrossRef]
- Li, P.; Tao, Z.; Dean, C. Phenotypic evolution through variation in splicing of the noncoding RNA COOLAIR. Genes Dev. 2015, 29, 696–701. [Google Scholar] [CrossRef] [Green Version]
- Eshed, Y.; Lippman, Z.B. Revolutions in agriculture chart a course for targeted breeding of old and new crops. Science 2019, 366, eaax0025. [Google Scholar] [CrossRef]
- Srikanth, A.; Schmid, M. Regulation of flowering time: All roads lead to Rome. Cell Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar] [CrossRef] [PubMed]
- Garner, W.W.; Allard, H.A. Agricultural United States Department of Agriculture and for the Association. J. Agric. Res. 1920, 18, 553–606. [Google Scholar]
- Corbesier, L.; Coupland, G. The quest for florigen: A review of recent progress. J. Exp. Bot. 2006, 57, 3395–3403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, Y.; Weigel, D. Move on up, it’s time for change—Mobile signals controlling photoperiod-dependent flowering. Genes Dev. 2007, 21, 2371–2384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clavijo Michelangeli, J.A.; Bhakta, M.; Gezan, S.A.; Boote, K.J.; Vallejos, C.E. From flower to seed: Identifying phenological markers and reliable growth functions to model reproductive development in the common bean (Phaseolus vulgaris L.). Plant Cell Environ. 2013, 36, 2046–2058. [Google Scholar] [CrossRef] [PubMed]
- Benlloch, R.; Berbel, A.; Ali, L.; Gohari, G.; Millán, T.; Madueño, F. Genetic control of inflorescence architecture in legumes. Front. Plant Sci. 2015, 6, 543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prusinkiewicz, P.; Erasmus, Y.; Lane, B.; Harder, L.D.; Coen, E. Evolution and Development of Inflorescence Architectures. Science 2007, 316, 1452–1456. [Google Scholar] [CrossRef] [Green Version]
- Bartlett, M.E.; Thompson, B. Meristem identity and phyllotaxis in inflorescence development. Front. Plant Sci. 2014, 5, 508. [Google Scholar] [CrossRef]
- Blázquez, M.A.; Weigel, D. Integration of floral inductive signals in Arabidopsis. Nature 2000, 404, 889–892. [Google Scholar] [CrossRef] [PubMed]
- Araki, T. Transition from vegetative to reproductive phase. Curr. Opin. Plant Biol. 2001, 4, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Benlloch, R.; Berbel, A.; Serrano-Mislata, A.; Madueño, F. Floral initiation and inflorescence architecture: A comparative view. Ann. Bot. 2007, 100, 659–676. [Google Scholar] [CrossRef] [PubMed]
- Wellmer, F.; Riechmann, J.L. Gene networks controlling the initiation of flower development. Trends Genet. 2010, 26, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Andrés, F.; Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012, 13, 627–639. [Google Scholar] [CrossRef]
- Posé, D.; Yant, L.; Schmid, M. The end of innocence: Flowering networks explode in complexity. Curr. Opin. Plant Biol. 2012, 15, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Bouché, F.; Lobet, G.; Tocquin, P.; Périlleux, C. FLOR-ID: An interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 2016, 44, D1167–D1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hecht, V.; Foucher, F.; Ferrándiz, C.; Macknight, R.; Navarro, C.; Morin, J.; Vardy, M.E.; Ellis, N.; Beltrán, J.P.; Rameau, C.; et al. Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 2005, 137, 1420–1434. [Google Scholar] [CrossRef] [Green Version]
- Nelson, M.N.; Berger, J.D.; Erskine, W. Flowering time control in annual legumes: Prospects in a changing global climate. CABI Rev. 2010, 5, 1–14. [Google Scholar] [CrossRef]
- Jung, C.H.; Wong, C.E.; Singh, M.B.; Bhalla, P.L. Comparative genomic analysis of soybean flowering genes. PLoS ONE 2012, 7, e38250. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.Y.; Kang, Y.J.; Lee, T.; Lee, S. Divergence of Flowering-Related Genes in Three Legume Species. Plant Genome 2013, 6, 841–856. [Google Scholar] [CrossRef] [Green Version]
- Young, N.D.; Bharti, A.K. Genome-enabled insights into legume biology. Annu. Rev. Plant Biol. 2012, 63, 283–305. [Google Scholar] [CrossRef] [PubMed]
- Liew, L.C.; Hecht, V.; Sussmilch, F.C.; Weller, J.L. The pea photoperiod response gene STERILE NODES is an ortholog of LUX ARRHYTHMO. Plant Physiol. 2014, 165, 648–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weller, J.L.; Ortega, R. Genetic control of flowering time in legumes. Front. Plant. Sci. 2015, 6, 207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weller, J.L.; Macknight, R.C. Functional Genomics and Flowering Time in Medicago truncatula: An Overview. Methods Mol. Biol. 2018, 1822, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Ruelens, P.; De Maagd, R.A.; Proost, S.; Theißen, G.; Geuten, K.; Kaufmann, K. FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nat. Commun. 2013, 4, 2280. [Google Scholar] [CrossRef] [Green Version]
- Hofer, J.; Turner, L.; Hellens, R.; Ambrose, M.; Matthews, P.; Michael, A.; Ellis, N. UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr. Biol. 1997, 7, 581–587. [Google Scholar] [CrossRef] [Green Version]
- Hecht, V.; Knowles, C.L.; Vander Schoor, J.K.; Liew, L.C.; Jones, S.E.; Lambert, M.J.M.; Weller, J.L. Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol. 2007, 144, 648–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liew, L.C.; Hecht, V.; Laurie, R.E.; Knowles, C.L.; Vander Schoor, J.K.; Macknight, R.C.; Weller, J.L. DIE NEUTRALIS and LATE BLOOMER 1 contribute to regulation of the pea circadian clock. Plant Cell 2009, 21, 3198–3211. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, S.; Xia, Z.; Hideshima, R.; Tsubokura, Y.; Sato, S.; Yamanaka, N.; Takahashi, R.; Anai, T.; Tabata, S.; Kitamura, K.; et al. A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 2011, 188, 395–407. [Google Scholar] [CrossRef]
- Ridge, S.; Deokar, A.; Lee, R.; Daba, K.; Macknight, R.C.; Weller, J.L.; Tar’an, B. The chickpea Early flowering 1 (Efl1) locus is an ortholog of Arabidopsis ELF3. Plant Physiol. 2017, 175, 802–815. [Google Scholar] [CrossRef] [Green Version]
- Rubenach, A.J.; Hecht, V.; Vander Schoor, J.K.; Liew, L.C.; Aubert, G.; Burstin, J.; Weller, J.L. EARLY FLOWERING3 Redundancy Fine-Tunes Photoperiod Sensitivity. Plant Physiol. 2017, 173, 2253–2264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weller, J.L.; Vander Schoor, J.K.; Perez-Wright, E.C.; Hecht, V.; González, A.M.; Capel, C.; Yuste-Lisbona, F.J.; Lozano, R.; Santalla, M. Parallel origins of photoperiod adaptation following dual domestications of common bean. J. Exp. Bot. 2019, 70, 1209–1219. [Google Scholar] [CrossRef]
- González, A.M.; Vander Schoor, J.K.; Fang, C.; Kong, F.; Wu, J.; Weller, J.L.; Santalla, M. Ancient relaxation of an obligate short-day requirement in common bean through loss of CONSTANS-like gene function. Curr. Biol. 2021, 31, 1643–1652. [Google Scholar] [CrossRef] [PubMed]
- Cober, E.R.; Tanner, J.W.; Voldeng, H.D. Soybean photoperiod-sensitivity loci respond differentially to light quality. Crop Sci. 1996, 36, 606–610. [Google Scholar] [CrossRef]
- Wong, A.C.S.; Hecht, V.F.G.; Picard, K.; Diwadkar, P.; Laurie, R.E.; Wen, J.; Mysore, K.; Macknight, R.C.; Weller, J.L. Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Front. Plant Sci. 2014, 5, 486. [Google Scholar] [CrossRef] [PubMed]
- González, A.M.; Yuste-Lisbona, F.J.; Weller, J.; Vander Schoor, J.K.; Lozano, R.; Santalla, M. Characterization of QTL and Environmental Interactions Controlling Flowering Time in Andean Common Bean (Phaseolus vulgaris L.). Front. Plant Sci. 2021, 11, 599462. [Google Scholar] [CrossRef] [PubMed]
- Fernández, F.; Paul, G.; Marceliano, L. Stages of Development of the Common Bean Plant, 1st ed.; Centro Internacional De Agricultura Tropical (CIAT): Cali, Colombia, 1986. [Google Scholar]
- Borghi, M.; Fernie, A.R. Floral metabolism of sugars and amino acids: Implications for pollinators’ preferences and seed and fruit set. Plant Physiol. 2017, 175, 1510–1524. [Google Scholar] [CrossRef] [Green Version]
- Ó’Maoiléidigh, D.S.; Wuest, S.E.; Rae, L.; Raganelli, A.; Ryan, P.T.; Kwaśniewska, K.; Das, P.; Lohan, A.J.; Loftus, B.; Graciet, E.; et al. Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell 2013, 25, 2482–2503. [Google Scholar] [CrossRef] [Green Version]
- Kahlau, S.; Bock, R. Plastid transcriptomics and translatomics of tomato fruit development and chloroplast-to-chromoplast differentiation: Chromoplast gene expression largely serves the production of a single protein. Plant Cell 2008, 20, 856–874. [Google Scholar] [CrossRef] [PubMed]
- Sawicki, M.; Courteaux, B.; Rabenoelina, F.; Baillieul, F.; Clement, C.; Ait Barka, E.; Jacquard, C.; Vaillant-Gaveau, N. Leaf vs. inflorescence: Differences in photosynthetic activity of grapevine. Photosynthetica 2017, 55, 58–68. [Google Scholar] [CrossRef]
- Schmitz, R.J.; Hong, L.; Fitzpatrick, K.E.; Amasino, R.M. DICER-LIKE 1 and DICER-LIKE 3 redundantly act to promote flowering via repression of FLOWERING LOCUS C in Arabidopsis thaliana. Genetics 2007, 176, 1359–1362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, O.; Borovsky, Y.; David-Schwartz, R.; Paran, I. Capsicum annuum S (CaS) promotes reproductive transition and is required for flower formation in pepper (Capsicum annuum). New Phytol. 2014, 202, 1014–1023. [Google Scholar] [CrossRef]
- Lemmon, Z.H.; Park, S.J.; Jiang, K.; Van Eck, J.; Schatz, M.C.; Lippman, Z.B. The evolution of inflorescence diversity in the nightshades and heterochrony during meristem maturation. Genome Res. 2016, 26, 1676–1686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seibert, T.; Abel, C.; Wahl, V. Flowering time and the identification of floral marker genes in Solanum tuberosum ssp. Andigena. J. Exp. Bot. 2020, 71, 986–996. [Google Scholar] [CrossRef]
- Goslin, K.; Zheng, B.; Serrano-Mislata, A.; Rae, L.; Ryan, P.T.; Kwaśniewska, K.; Thomson, B.; Ó’Maoiléidigh, D.S.; Madueño, F.; Wellmer, F.; et al. Transcription factor interplay between LEAFY and APETALA1/CAULIFLOWER during floral initiation. Plant Physiol. 2017, 174, 1097–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oda, A.; Fujiwara, S.; Kamada, H.; Coupland, G.; Mizoguchi, T. Antisense suppression of the Arabidopsis PIF3 gene does not affect circadian rhythms but causes early flowering and increases FT expression. FEBS Lett. 2004, 557, 259–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Suh, S.S.; Park, E.; Cho, E.; Ahn, J.H.; Kim, S.G.; Lee, J.S.; Kwon, Y.M.; Lee, I. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 2000, 14, 2366–2376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Zhang, Y.; Ma, Q.; Zhang, Z.; Xue, Y.; Bao, S.; Chong, K. SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. EMBO J. 2007, 26, 1934–1941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Y.; Kato, N.; Wang, W.; Li, J.; Chen, X. Two RNA binding proteins, HEN4 and HUA1 act in the processing of AGAMOUS pre-mRNA in Arabidopsis thaliana. Dev. Cell 2003, 4, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Blázquez, M.A.; Soowal, L.N.; Lee, I.; Weigel, D. LEAFY expression and flower initiation in Arabidopsis. Development 1997, 124, 3835–3844. [Google Scholar] [CrossRef] [PubMed]
- Winter, C.M.; Yamaguchi, N.; Wu, M.F.; Wagner, D. Transcriptional programs regulated by both LEAFY and APETALA1 at the time of flower formation. Physiol. Plant 2015, 155, 55–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parcy, F. Flowering: A time for integration. Int. J. Dev. Biol. 2005, 49, 585–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassidim, M.; Harir, Y.; Yakir, E.; Kron, I.; Green, R.M. Over-expression of CONSTANS-LIKE 5 can induce flowering in short-day grown Arabidopsis. Planta 2009, 230, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Amasino, R. Seasonal and developmental timing of flowering. Plant J. 2010, 61, 1001–1013. [Google Scholar] [CrossRef]
- Liu, K.; Feng, S.; Pan, Y.; Zhong, J.; Chen, Y.; Yuan, C.; Li, H. Transcriptome Analysis and Identification of Genes Associated with Floral Transition and Flower Development in Sugar Apple (Annona squamosa L.). Front. Plant Sci. 2016, 7, 1695. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Yu, C.; Luo, L.; Wan, H.; Zhen, N.; Xu, T.; Tan, J.; Pan, H.; Zhang, Q. Transcriptome of the floral transition in Rosa chinensis ‘Old Blush’. BMC Genom. 2017, 18, 199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jue, D.; Sang, X.; Liu, L.; Shu, B.; Wang, Y.; Liu, C.; Wang, Y.; Xie, J.; Shi, S. Comprehensive analysis of the longan transcriptome reveals distinct regulatory programs during the floral transition. BMC Genom. 2019, 20, 126. [Google Scholar] [CrossRef]
- Shim, J.S.; Kubota, A.; Imaizumi, T. Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol. 2017, 173, 5–15. [Google Scholar] [CrossRef]
- Sawa, M.; Nusinow, D.A.; Kay, S.A.; Imaizumi, T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 2007, 318, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, S.E.; Cagnola, J.I.; Crepy, M.; Yanovsky, M.J.; Casal, J.J. Balancing forces in the photoperiodic control of flowering. Photochem. Photobiol. Sci. 2011, 10, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.H.; Estrada, D.A.; Johnson, R.S.; Kim, S.K.; Lee, S.Y.; MacCoss, M.J.; Imaizumi, T. Distinct roles of FKF1, GIGANTEA, and ZEITLUPE proteins in the regulation of CONSTANS stability in Arabidopsis photoperiodic flowering. Proc. Natl. Acad. Sci. USA 2014, 111, 17672–17677. [Google Scholar] [CrossRef] [Green Version]
- Goretti, D.; Silvestre, M.; Collani, S.; Langenecker, T.; Méndez, C.; Madueño, F.; Schmid, M. TERMINAL FLOWER1 functions as a mobile transcriptional cofactor in the shoot apical meristem. Plant Physiol. 2020, 182, 2081–2095. [Google Scholar] [CrossRef] [Green Version]
- Takase, T.; Nishiyama, Y.; Tanihigashi, H.; Ogura, Y.; Miyazaki, Y.; Yamada, Y.; Kiyosue, T. LOV KELCH PROTEIN2 and ZEITLUPE repress Arabidopsis photoperiodic flowering under non-inductive conditions, dependent on FLAVIN-BINDING KELCH REPEAT F-BOX1. Plant J. 2011, 67, 608–621. [Google Scholar] [CrossRef]
- Nemoto, Y.; Nonoue, Y.; Yano, M.; Izawa, T. Hd1, a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J. 2016, 86, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Shaw, L.M.; Li, C.; Woods, D.P.; Alvarez, M.A.; Lin, H.; Lau, M.Y.; Chen, A.; Dubcovsky, J. Epistatic interactions between PHOTOPERIOD1, CONSTANS1 and CONSTANS2 modulate the photoperiodic response in wheat. PLoS Genet. 2020, 16, e1008812. [Google Scholar] [CrossRef]
- González-Schain, N.D.; Díaz-Mendoza, M.; Żurczak, M.; Suárez-López, P. Potato CONSTANS is involved in photoperiodic tuberization in a graft-transmissible manner. Plant J. 2012, 70, 678–690. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yu, X.; Li, K.; Klejnot, J.; Yang, H.; Lisiero, D.; Lin, C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 2008, 322, 1535–1539. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Carlsson, J.; Takeuchi, T.; Newton, L.; Farré, E.M. Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J. 2013, 76, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Adrian, J.; Pankin, A.; Hu, J.; Dong, X.; von Korff, M.; Turck, F. Induced and natural variation of promoter length modulates the photoperiodic response of FLOWERING LOCUS T. Nat. Commun. 2014, 5, 4558. [Google Scholar] [CrossRef] [Green Version]
- Zuo, Z.; Liu, H.; Liu, B.; Liu, X.; Lin, C. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr. Biol. 2011, 21, 841–847. [Google Scholar] [CrossRef] [Green Version]
- Ito, S.; Song, Y.H.; Josephson-Day, A.R.; Miller, R.J.; Breton, G.; Olmstead, R.G.; Imaizumi, T. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 3582–3587. [Google Scholar] [CrossRef] [Green Version]
- Golembeski, G.S.; Kinmonth-Schultz, H.A.; Song, Y.H.; Imaizumi, T. Photoperiodic flowering regulation in Arabidopsis thaliana. Adv. Bot. Res. 2014, 72, 1–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawamoto, N.; Sasabe, M.; Endo, M.; Machida, Y.; Araki, T. Calcium-dependent protein kinases responsible for the phosphorylation of a bZIP transcription factor FD crucial for the florigen complex formation. Sci. Rep. 2015, 5, 8341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Z.; Zhang, X.; Huang, P.; Huang, G.; Zhu, J.; Chen, F.; Miao, Y.; Liu, L.; Fu, Y.F.; Wang, X. Nup96 and HOS1 are mutually stabilized and gate CONSTANS protein level, conferring Long-Day photoperiodic flowering regulation in Arabidopsis. Plant Cell 2020, 32, 374–391. [Google Scholar] [CrossRef]
- Singh, S.P. A key for identification of different growth habits of Phaseolus vulgaris L. Annu. Rep. Bean Improv. Coop. 1982, 25, 92–95. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Schmutz, J.; McClean, P.E.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Jenkins, J.; Shu, S.; Song, Q.; Chavarro, C.; et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 2014, 46, 707–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. Feature Counts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Ruotti, V.; Stewart, R.M.; Thomson, J.A.; Dewey, C.N. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 2009, 26, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wald, B.Y.A. Sequential Tests of Statistical Hypotheses. Ann. Math. Stat. 1945, 16, 117–186. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium. Gene ontology consortium: Going forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [Green Version]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, 1178–1186. [Google Scholar] [CrossRef] [PubMed]
- Hommel, G. A Stagewise Rejective Multiple Test Procedure Based on a Modified Bonferroni Test. Biometrika 1988, 75, 383–386. [Google Scholar] [CrossRef]
- Lex, A.; Gehlenborg, N.; Strobelt, H.; Vuillemot, R.; Pfister, H. UpSet: Visualization of Intersecting Sets. IEEE Trans. Vis. Comput. Graph. 2014, 20, 1983–1992. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.J.; Soltis, P.S.; Bell, C.D.; Burleigh, J.G.; Soltis, D.E. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. USA 2010, 107, 4623–4628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winer, J.; Jung, C.K.S.; Shackel, I.; Williams, P.M. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal. Biochem. 1999, 270, 41–49. [Google Scholar] [CrossRef] [PubMed]


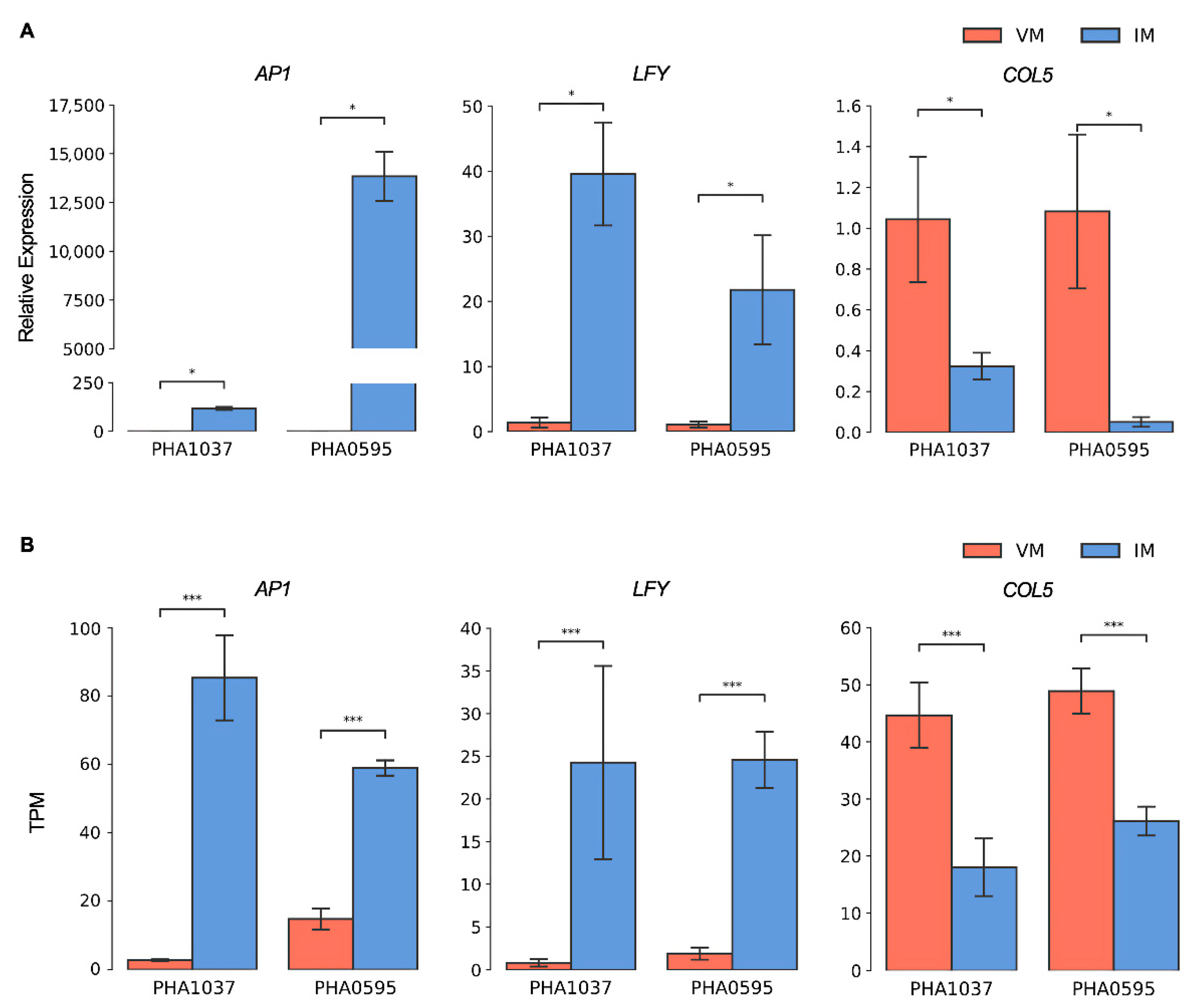
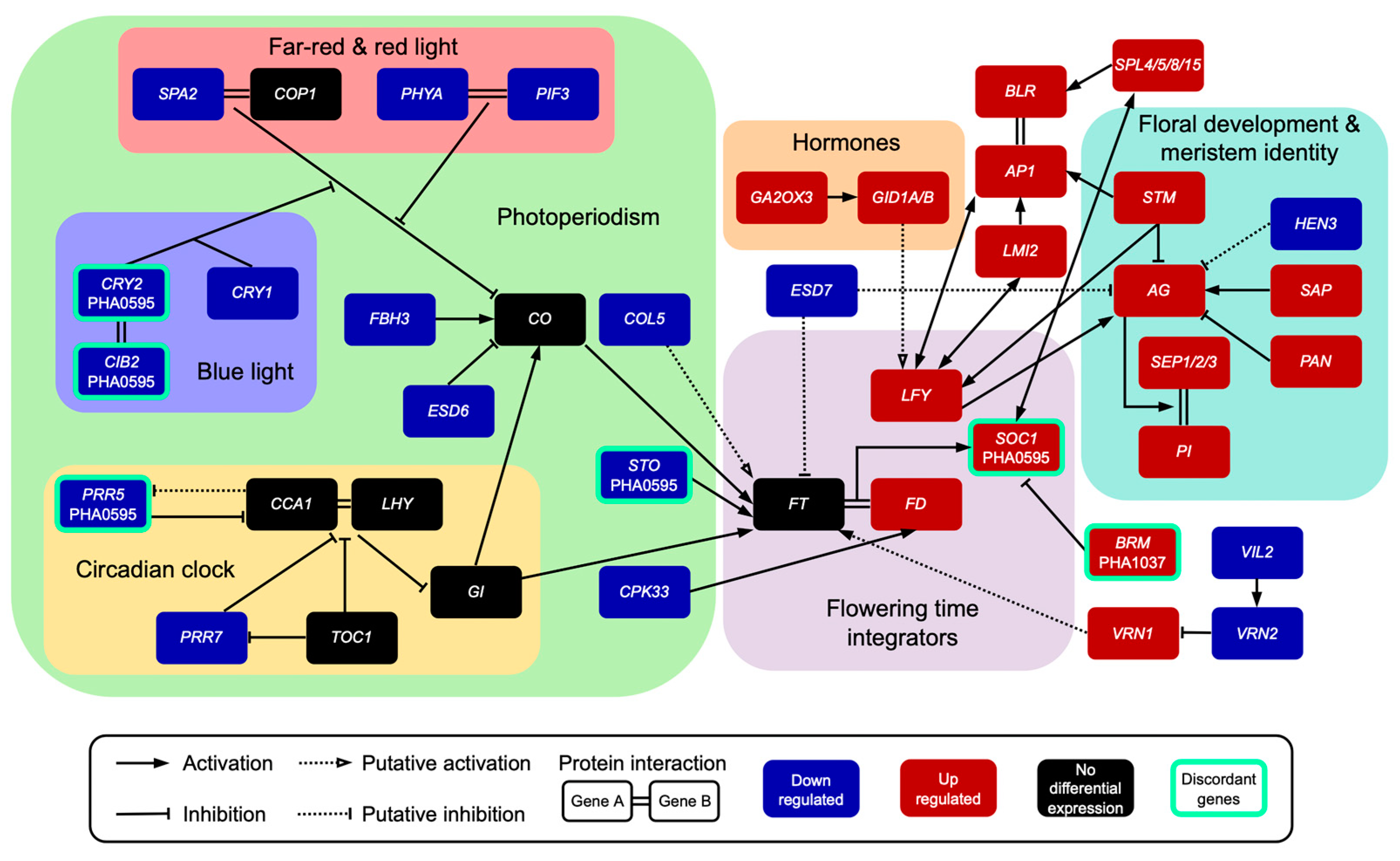
| Flowering and Flower Development Related Pathways | Up-Regulated | Down-Regulated | Discordant Genes |
|---|---|---|---|
| Photoperiod | FD (1) | COL5 (1), PHYA2 (1), CRY1 (1), PIF3 (1), PRR7 (1), EFS (1), FBH3 (1), SPA2 (1), CPK33 (1), ESD6 (1) | CRY2 (1, PHA1037), CIB2 (1, PHA1037), PRR5 (1, PHA1037), STO (1, PHA1037) |
| Vernalization | FRL2 (1), VRN1 (1), POB1 (1) | VIL2 (1), VRN2 (1) | |
| Autonomous | HTA11 (1) | HUB1 (1), ESD7 (1), GLK1 (1) | SKB1 (1, PHA0595), BRM (1, PHA1037), HULK1 (1, PHA1037), DCL1 (1, PHA1037) |
| Hormones | GA2OX3 (1), GID1 (2), GAS4 (1), ATH1 (2) | HIPP3 (1), GATA21 (2) | |
| Aging | SPLs (3) | TPS1 (1) | |
| Sugars | ADG1 (1), PGM1 (1) | ||
| Flowering Integrators | LFY (1), GIN2 (1) | SOC1 (1, PHA0595) | |
| Flower development and meristem identity | AP1 (1), LMI2 (1), BLR (2), SAP (1), SAP18 (1), STM (2), PAN (1), AG (2), PI (2), ARF3 (2), AGLs/SEP (3), STY1 (1) | HEN3 (1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González, A.M.; Lebrón, R.; Yuste-Lisbona, F.J.; Gómez-Martín, C.; Ortiz-Atienza, A.; Hackenberg, M.; Oliver, J.L.; Lozano, R.; Santalla, M. Decoding Gene Expression Signatures Underlying Vegetative to Inflorescence Meristem Transition in the Common Bean. Int. J. Mol. Sci. 2022, 23, 14783. https://doi.org/10.3390/ijms232314783
González AM, Lebrón R, Yuste-Lisbona FJ, Gómez-Martín C, Ortiz-Atienza A, Hackenberg M, Oliver JL, Lozano R, Santalla M. Decoding Gene Expression Signatures Underlying Vegetative to Inflorescence Meristem Transition in the Common Bean. International Journal of Molecular Sciences. 2022; 23(23):14783. https://doi.org/10.3390/ijms232314783
Chicago/Turabian StyleGonzález, Ana M., Ricardo Lebrón, Fernando J. Yuste-Lisbona, Cristina Gómez-Martín, Ana Ortiz-Atienza, Michael Hackenberg, José L. Oliver, Rafael Lozano, and Marta Santalla. 2022. "Decoding Gene Expression Signatures Underlying Vegetative to Inflorescence Meristem Transition in the Common Bean" International Journal of Molecular Sciences 23, no. 23: 14783. https://doi.org/10.3390/ijms232314783
APA StyleGonzález, A. M., Lebrón, R., Yuste-Lisbona, F. J., Gómez-Martín, C., Ortiz-Atienza, A., Hackenberg, M., Oliver, J. L., Lozano, R., & Santalla, M. (2022). Decoding Gene Expression Signatures Underlying Vegetative to Inflorescence Meristem Transition in the Common Bean. International Journal of Molecular Sciences, 23(23), 14783. https://doi.org/10.3390/ijms232314783






