New Insights into Dose-Dependent Effects of Curcumin on ARPE-19 Cells
Abstract
:1. Introduction
2. Results
2.1. Changes in Cell Viability and Cell Cycle Progression upon Curcumin Administration
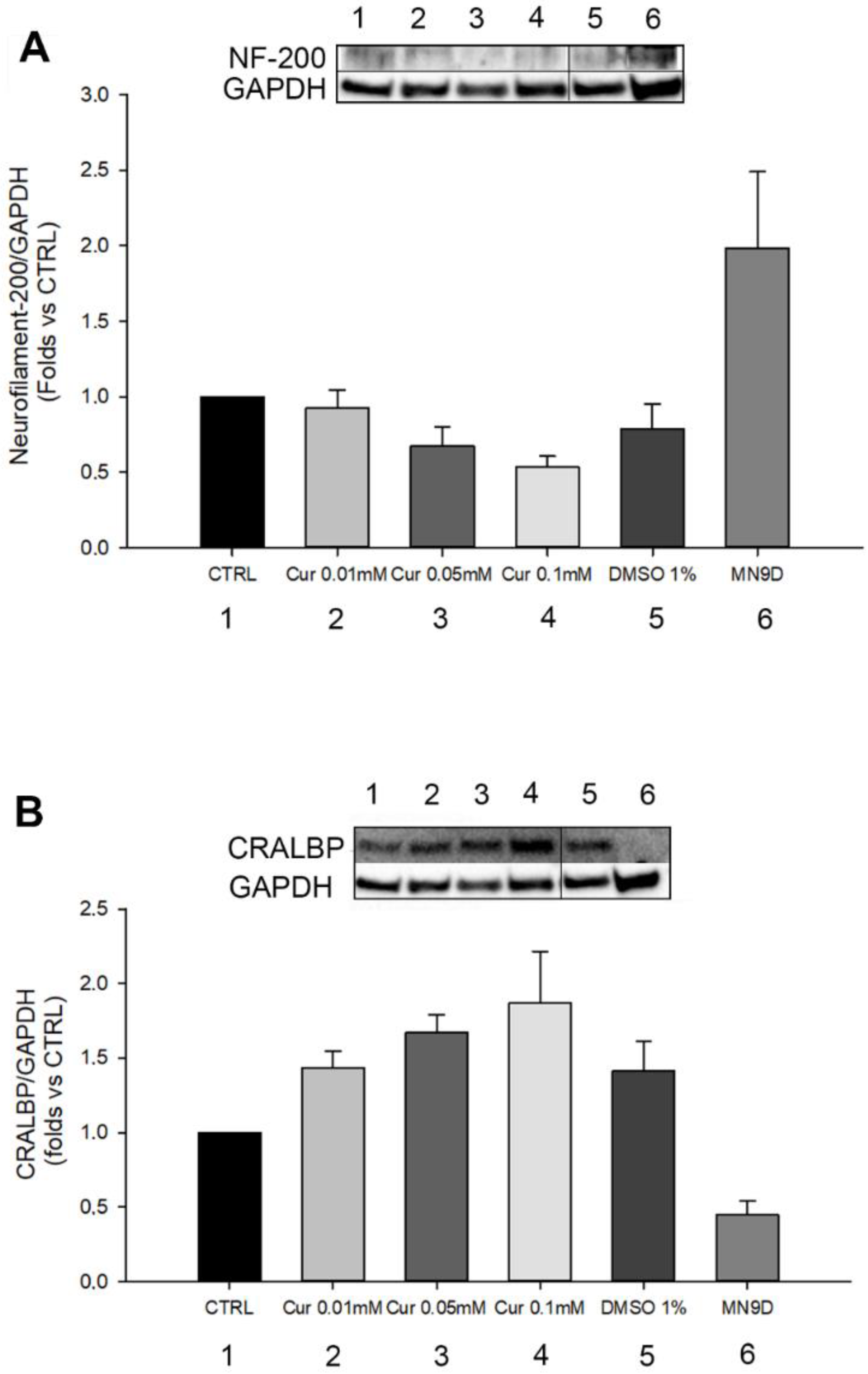
2.2. Morphological and Differentiation Analysis of Retinal Pigment Epithelial Cells after Curcumin Supplementation
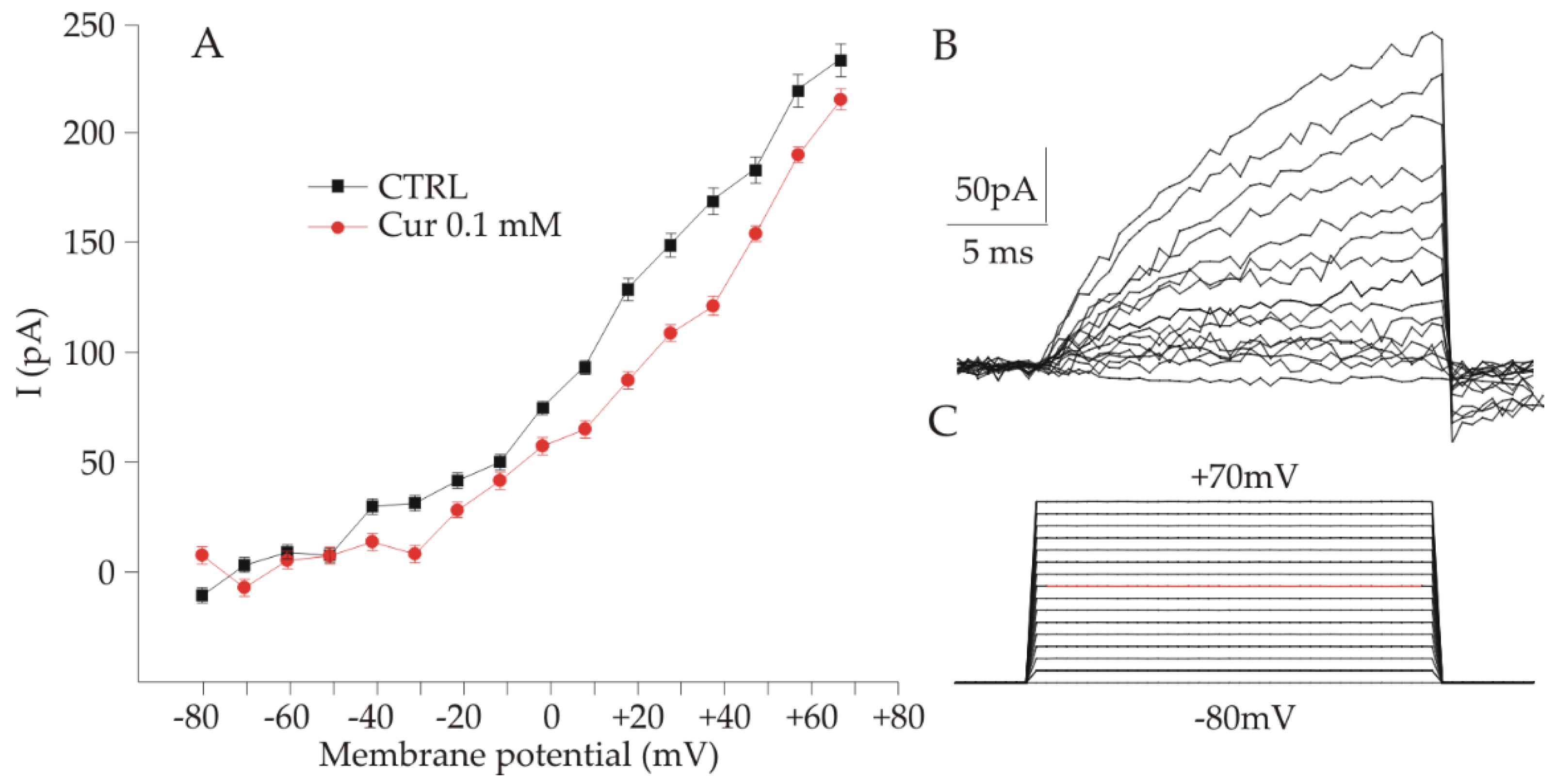
2.3. Electrophysiological Analysis of ARPE-19 Cells after Curcumin Supplementation
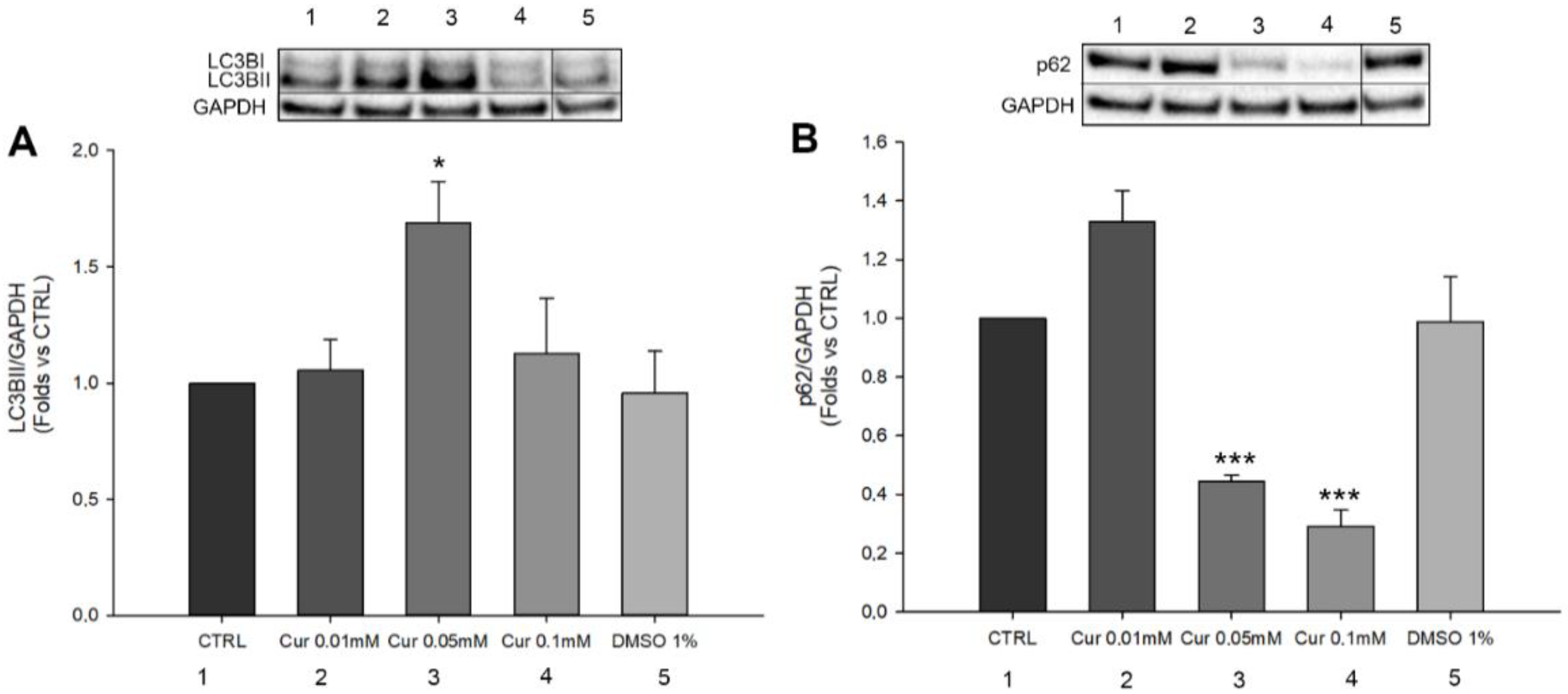
2.4. Curcumin Is Able to Induce Autophagic Flux Modulation in ARPE-19 Cells
3. Discussion
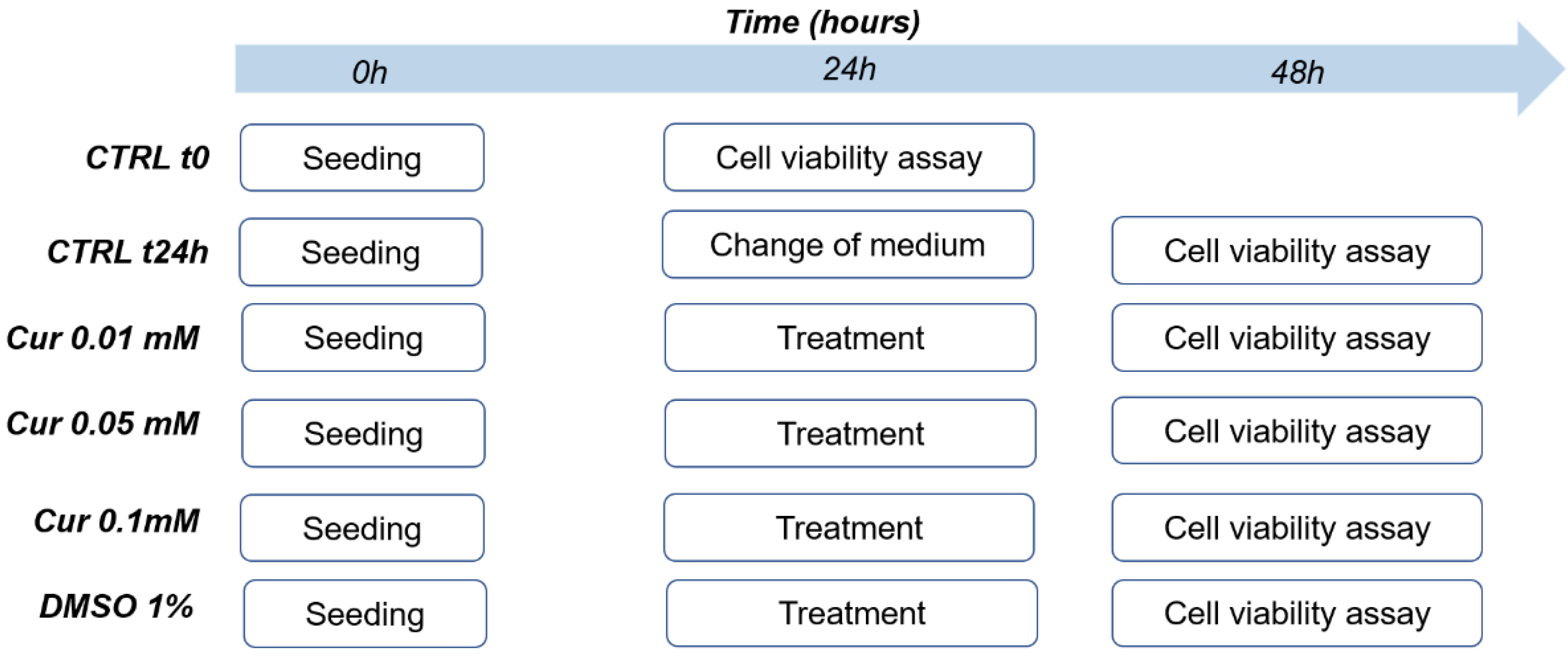
4. Materials and Methods
4.1. Cell Culture
4.2. Curcumin Treatment
4.3. Cell Viability Assay
4.4. Phalloidin Staining
4.5. Flow Cytometer Analysis
4.6. Western Blot
4.7. Patch Clamp Recordings
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tisi, A.; Feligioni, M.; Passacantando, M.; Ciancaglini, M.; Maccarone, R. The Impact of Oxidative Stress on Blood-Retinal Barrier Physiology in Age-Related Macular Degeneration. Cells 2021, 10, 64. [Google Scholar] [CrossRef] [PubMed]
- Strauss, O. The retinal pigment epithelium in visual function. Physiol. Rev. 2005, 85, 845–881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Idrees, S.; Sridhar, J.; Kuriyan, A.E. Proliferative Vitreoretinopathy: A Review. Int. Ophthalmol. Clin. 2019, 59, 221. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.; Liew, G.; Gopinath, B.; Wong, T.Y. Age-related macular degeneration. Lancet 2018, 392, 1147–1159. [Google Scholar] [CrossRef]
- Lestari, M.L.A.D.; Indrayanto, G. Curcumin. In Profiles of Drug Substances, Excipients and Related Methodology; Academic Press Inc.: New York, NY, USA, 2014; pp. 113–204. [Google Scholar]
- Perrone, L.; Squillaro, T.; Napolitano, F.; Terracciano, C.; Sampaolo, S.; Anna, M.; Melone, B. The Autophagy Signaling Pathway: A Potential Multifunctional Therapeutic Target of Curcumin in Neurological and Neuromuscular Diseases. Nutrients 2019, 11, 1881. [Google Scholar] [CrossRef] [Green Version]
- Hollborn, M.; Chen, R.; Wiedemann, P.; Reichenbach, A.; Bringmann, A. Cytotoxic Effects of Curcumin in Human Retinal Pigment Epithelial Cells. PLoS ONE 2013, 8, 59603. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; You, Z.P. Curcumin inhibits human retinal pigment epithelial cell proliferation. Int. J. Mol. Med. 2014, 34, 1013–1019. [Google Scholar] [CrossRef] [Green Version]
- Mandal, M.N.A.; Patlolla, J.M.R.; Zheng, L.; Agbaga, M.P.; Tran, J.T.A.; Wicker, L.; Kasus-Jacobi, A.; Elliott, M.H.; Rao, C.V.; Anderson, R.E. Curcumin protects retinal cells from light-and oxidant stress-induced cell death. Free Radic. Biol. Med. 2009, 46, 672–679. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.C.; Chang, W.C.; Hung, K.H.; Yang, D.M.; Cheng, Y.H.; Liao, Y.W.; Woung, L.C.; Tsai, C.Y.; Hsu, C.C.; Lin, T.C.; et al. The generation of induced pluripotent stem cells for macular degeneration as a drug screening platform: Identification of curcumin as a protective agent for retinal pigment epithelial cells against oxidative stress. Front. Aging Neurosci. 2014, 6, 191. [Google Scholar] [CrossRef] [Green Version]
- Pescosolido, N.; Giannotti, R.; Plateroti, A.M.; Pascarella, A.; Nebbioso, M. Curcumin: Therapeutical Potential in Ophtalmology. Planta Med. 2014, 80, 249–254. [Google Scholar] [CrossRef]
- Shakeri, A.; Cicero, A.F.G.; Panahi, Y.; Mohajeri, M.; Sahebkar, A. Curcumin: A naturally occurring autophagy modulator. J. Cell. Physiol. 2019, 234, 5643–5654. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wu, Y.; Zhou, D.; Xie, Y.; Zhou, Y.; Lu, Y.; Yang, R.; Liu, S. N-linoleyltyrosine protects PC12 cells against oxidative damage via autophagy: Possible involvement of CB1 receptor regulation. Int. J. Mol. Med. 2020, 46, 1827. [Google Scholar] [CrossRef] [PubMed]
- Intartaglia, D.; Giamundo, G.; Conte, I. Autophagy in the retinal pigment epithelium: A new vision and future challenges. FEBS J. 2021, 289, 7199–7212. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, T.A.; Green, D.R. Autophagy and phagocytosis converge for better vision. Autophagy 2013, 10, 165–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Pan, X.; Zhao, X.; Luo, J.; Xu, M.; Bai, D.; Hu, Y.; Liu, X.; Yu, Q.; Gao, D. Autophagy and Age-Related Eye Diseases. BioMed Res. Int. 2019, 2019, 5763658. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Zhao, X.; Guo, Q.; Feng, Y.; Ma, M.; Guo, W.; Dong, X.; Deng, C.; Li, C.; Song, X.; et al. Autophagy resists EMT process to maintain retinal pigment epithelium homeostasis. Int. J. Biol. Sci. 2019, 15, 507. [Google Scholar] [CrossRef] [Green Version]
- Miao, Q.; Xu, Y.; Yin, H.; Zhang, H.; Ye, J. KRT8 phosphorylation regulates the epithelial-mesenchymal transition in retinal pigment epithelial cells through autophagy modulation. J. Cell. Mol. Med. 2020, 24, 3217–3228. [Google Scholar] [CrossRef]
- Donato, L.; D’Angelo, R.; Alibrandi, S.; Rinaldi, C.; Sidoti, A.; Scimone, C. Effects of A2E-Induced Oxidative Stress on Retinal Epithelial Cells: New Insights on Differential Gene Response and Retinal Dystrophies. Antioxidants 2020, 9, 307. [Google Scholar] [CrossRef] [Green Version]
- Laher, I.; Guns, E.S.; Bucolo, C.; Platania, C.B.M.; Fidilio, A.; Lazzara, F.; Piazza, C.; Geraci, F.; Giurdanella, G.; Leggio, G.M.; et al. Retinal Protection and Distribution of Curcumin in Vitro and in Vivo. Front. Pharmacol. 2018, 9, 670. [Google Scholar]
- Park, S.I.; Lee, E.H.; Kim, S.R.; Jang, Y.P. Anti-apoptotic effects of Curcuma longa L. extract and its curcuminoids against blue light-induced cytotoxicity in A2E-laden human retinal pigment epithelial cells. J. Pharm. Pharmacol. 2017, 69, 334–340. [Google Scholar] [CrossRef]
- López-Malo, D.; Villarón-Casares, C.A.; Alarcón-Jiménez, J.; Miranda, M.; Díaz-Llopis, M.; Romero, F.J.; Villar, V.M. Curcumin as a Therapeutic Option in Retinal Diseases. Antioxidants 2020, 9, 48. [Google Scholar] [CrossRef] [PubMed]
- Menon, V.P.; Sudheer, A.R. Antioxidant and anti-inflammatory properties of curcumin. Adv. Exp. Med. Biol. 2007, 595, 105–125. [Google Scholar] [PubMed]
- Singh, A.K.; Sidhu, G.S.; Deepa, T.; Maheshwari, R.K. Curcumin inhibits the proliferation and cell cycle progression of human umbilical vein endothelial cell. Cancer Lett. 1996, 107, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Ning, Y.; Zeng, G.; Zhou, C.; Ding, X. Curcumin promotes cell cycle arrest and apoptosis of acute myeloid leukemia cells by inactivating AKT. Oncol. Rep. 2021, 45, 11. [Google Scholar] [CrossRef]
- Wang, J.; Wang, J.; Cai, Z.; Xu, C. The effect of curcumin on the differentiation, apoptosis and cell cycle of neural stem cells is mediated through inhibiting autophagy by the modulation of Atg7 and p62. Int. J. Mol. Med. 2018, 42, 2481–2488. [Google Scholar] [CrossRef] [Green Version]
- Guarnieri, S.; Pilla, R.; Morabito, C.; Sacchetti, S.; Mancinelli, R.; Fanò, G.; Mariggiò, M.A. Extracellular guanosine and GTP promote expression of differentiation markers and induce S-phase cell-cycle arrest in human SH-SY5Y neuroblastoma cells. Int. J. Dev. Neurosci. 2009, 27, 135–147. [Google Scholar] [CrossRef]
- Liu, X.; Yang, J.M.; Zhang, S.S.; Liu, X.Y.; Liu, D.X. Induction of cell cycle arrest at G1 and S phases and cAMP-dependent differentiation in C6 glioma by low concentration of cycloheximide. BMC Cancer 2010, 10, 684. [Google Scholar] [CrossRef] [Green Version]
- Shafei, E.V.; Kurinov, A.M.; Kuznetsova, A.V.; Aleksandrova, M.A. Reprogramming of Human Retinal Pigment Epithelial Cells under the effect of bFGF in vitro. Bull. Exp. Biol. Med. 2017, 163, 574–582. [Google Scholar] [CrossRef]
- Lueck, K.; Carr, A.J.F.; Stampoulis, D.; Gerke, V.; Rescher, U.; Greenwood, J.; Moss, S.E. Regulation of retinal pigment epithelial cell phenotype by Annexin A8. Sci. Rep. 2017, 7, 4638. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Fariss, R.N.; Kutty, R.K.; Nelson, R.; Wiggert, B. Fenretinide-induced neuronal differentiation of ARPE-19 human retinal pigment epithelial cells is associated with the differential expression of Hsp70, 14-3-3, Pax-6, Tubulin β-III, NSE, and Bag-1 proteins. Mol. Vis. 2006, 12, 1355–1363. [Google Scholar]
- Takahira, M.; Sakurai, M.; Sakurada, N.; Sugiyama, K. Swelling-activated potassium channel in porcine pigmented ciliary epithelial cells. Investig. Opthalmology Vis. Sci. 2011, 52, 5928–5932. [Google Scholar] [CrossRef] [PubMed]
- Lane, J.; Korolchuk, V.; Wilkinson, S.; de Zio, D.; Cecconi, F.; Mathiassen, S.G. Autophagy and the Cell Cycle: A Complex Landscape. Autophagy and the Cell Cycle: A Complex Landscape. Front Oncol. 2017, 7, 51. [Google Scholar]
- Anand, S.K.; Sharma, A.; Singh, N.; Kakkar, P. Entrenching role of cell cycle checkpoints and autophagy for maintenance of genomic integrity. DNA Repair 2020, 86, 102748. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, P.R.; Madanagopalan, V.G. Role of Curcumin in Retinal Diseases—A review. Graefe’s Arch. Clin. Exp. Ophthalmol. 2022, 260, 1457–1473. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.S.; Acharya, A.; Ray, R.S.; Agrawal, R.; Raghuwanshi, R.; Jain, P. Cellular and molecular mechanisms of curcumin in prevention and treatment of disease. Crit. Rev. Food Sci. Nutr. 2020, 60, 887–939. [Google Scholar] [CrossRef]
- Saleh, E.M.; El-awady, R.A.; Eissa, N.A.; Abdel-Rahman, W.M. Antagonism between curcumin and the topoisomerase II inhibitor etoposide. Cancer Biol. Ther. 2014, 13, 1058–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buyandelger, U.; Walker, D.G.; Taguchi, H.; Yanagisawa, D.; Tooyama, I. Novel fluorinated derivative of curcumin negatively regulates thioredoxin-interacting protein expression in retinal pigment epithelial and macrophage cells. Biochem. Biophys. Res. Commun. 2020, 532, 668–674. [Google Scholar] [CrossRef]
- Munia, I.; Gafray, L.; Bringer, M.A.; Goldschmidt, P.; Proukhnitzky, L.; Jacquemot, N.; Cercy, C.; Ramchani Ben Otman, K.; Errera, M.H.; Ranchon-Cole, I. Cytoprotective Effects of Natural Highly Bio-Available Vegetable Derivatives on Human-Derived Retinal Cells. Nutrients 2020, 12, 879. [Google Scholar] [CrossRef] [Green Version]
- Howell, J.C.; Chun, E.; Farrell, A.N.; Hur, E.Y.; Caroti, C.M.; Iuvone, P.M.; Haque, R. Global microRNA expression profiling: Curcumin (diferuloylmethane) alters oxidative stress-responsive microRNAs in human ARPE-19 cells. Mol. Vis. 2013, 19, 544–560. [Google Scholar]
- Demirci Kucuk, K.; Tokuc, E.O.; Aciksari, A.; Duruksu, G.; Yazir, Y.; Karabas, V.L. The effects of crocetin on oxidative stress induced ARPE-19 cells by H2O2. Exp. Eye Res. 2022, 226, 109305. [Google Scholar] [CrossRef]
- Xia, Z.H.; Zhang, S.Y.; Chen, Y.S.; Li, K.; Chen, W.B.; Liu, Y.Q. Curcumin anti-diabetic effect mainly correlates with its anti-apoptotic actions and PI3K/Akt signal pathway regulation in the liver. Food Chem. Toxicol. 2020, 146, 111803. [Google Scholar] [CrossRef] [PubMed]
- Muangnoi, C.; Sharif, U.; Ratnatilaka, P.; Bhuket, N.; Rojsitthisak, P.; Paraoan, L. Protective Effects of Curcumin Ester Prodrug, Curcumin Diethyl Disuccinate against H2O2-Induced Oxidative Stress in Human Retinal Pigment Epithelial Cells: Potential Therapeutic Avenues for Age-Related Macular Degeneration. Int. J. Mol. Sci. 2019, 20, 3367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engström, Y. Cell cycle regulators control stemness and differentiation. BioEssays 2021, 43, 2100123. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wen, S.; Chen, G.; Wang, S. Antiproliferative potential of piperine and curcumin in drug-resistant human leukemia cancer cells are mediated via autophagy and apoptosis induction, S-phase cell cycle arrest and inhibition of cell invasion and migration. J. BUON 2020, 25, 401–406. [Google Scholar] [PubMed]
- Keckeis, S.; Reichhart, N.; Roubeix, C.; Strauß, O. Anoctamin2 (TMEM16B) forms the Ca2+-activated Cl- channel in the retinal pigment epithelium. Exp. Eye Res. 2017, 154, 139–150. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Kamal Abdel-Aziz, A.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Princely Abudu, Y.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 2020, 17, 1–382. [Google Scholar]
- Liou, J.S.; Wu, Y.C.; Yen, W.Y.; Tang, Y.S.; Kakadiya, R.B.; Su, T.L.; Yih, L.H. Inhibition of autophagy enhances DNA damage-induced apoptosis by disrupting CHK1-dependent S phase arrest. Toxicol. Appl. Pharmacol. 2014, 278, 249–258. [Google Scholar] [CrossRef]
- Hewitt, G.; Carroll, B.; Sarallah, R.; Correia-Melo, C.; Ogrodnik, M.; Nelson, G.; Otten, E.G.; Manni, D.; Antrobus, R.; Morgan, B.A.; et al. SQSTM1/p62 mediates crosstalk between autophagy and the UPS in DNA repair. Autophagy 2016, 12, 1917–1930. [Google Scholar] [CrossRef]
- Williams, B.K.; di Nicola, M.; Acaba-Berrocal, L.A.; Milman, T.; Mashayekhi, A.; Lucio-Alvarez, J.A.; Eagle, R.C.; Shields, J.A.; Shields, C.L. Adenoma and Adenocarcinoma of the Retinal Pigment Epithelium: A Review of 51 Consecutive Patients. Ophthalmol. Retin. 2020, 4, 829–839. [Google Scholar] [CrossRef]

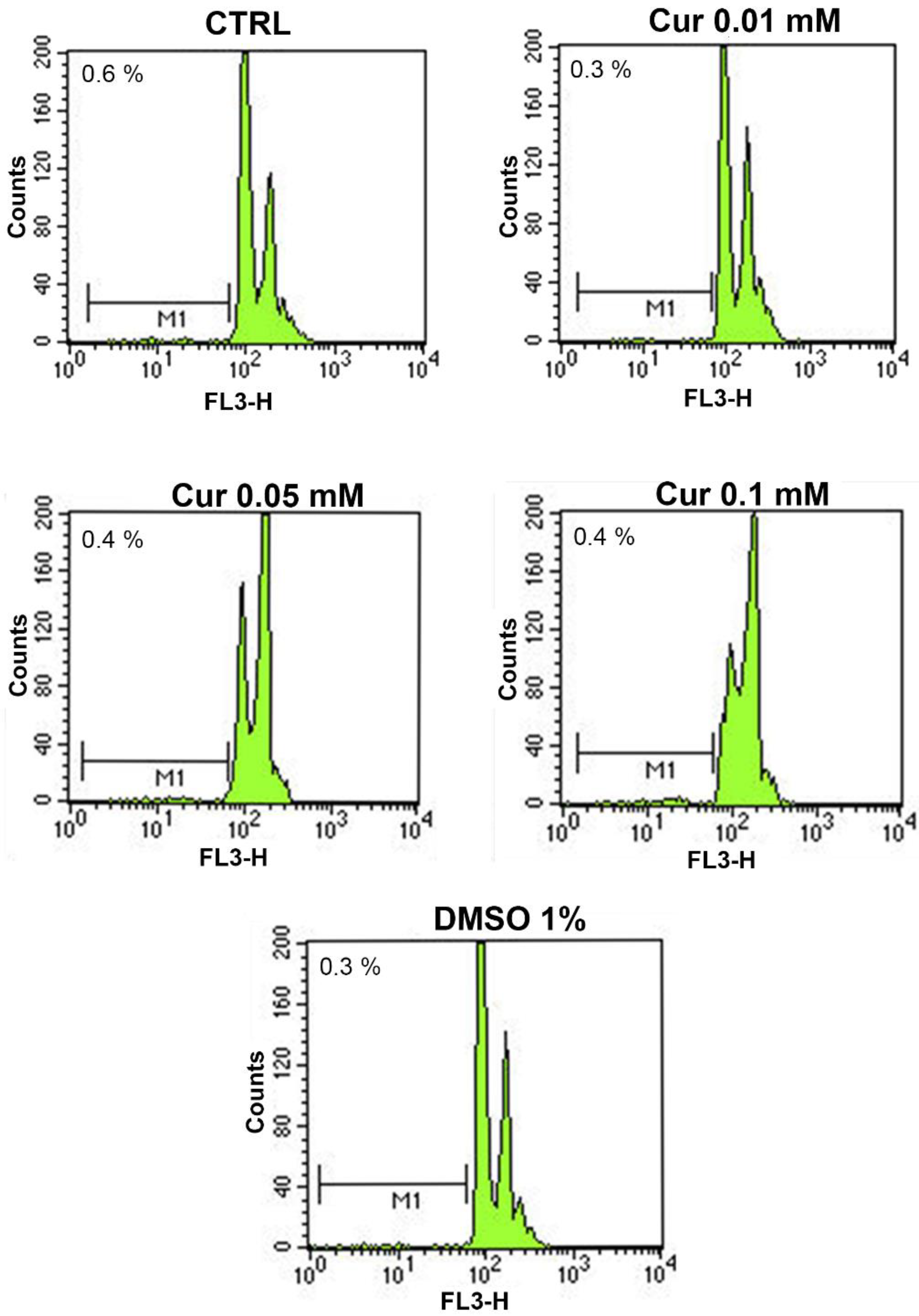
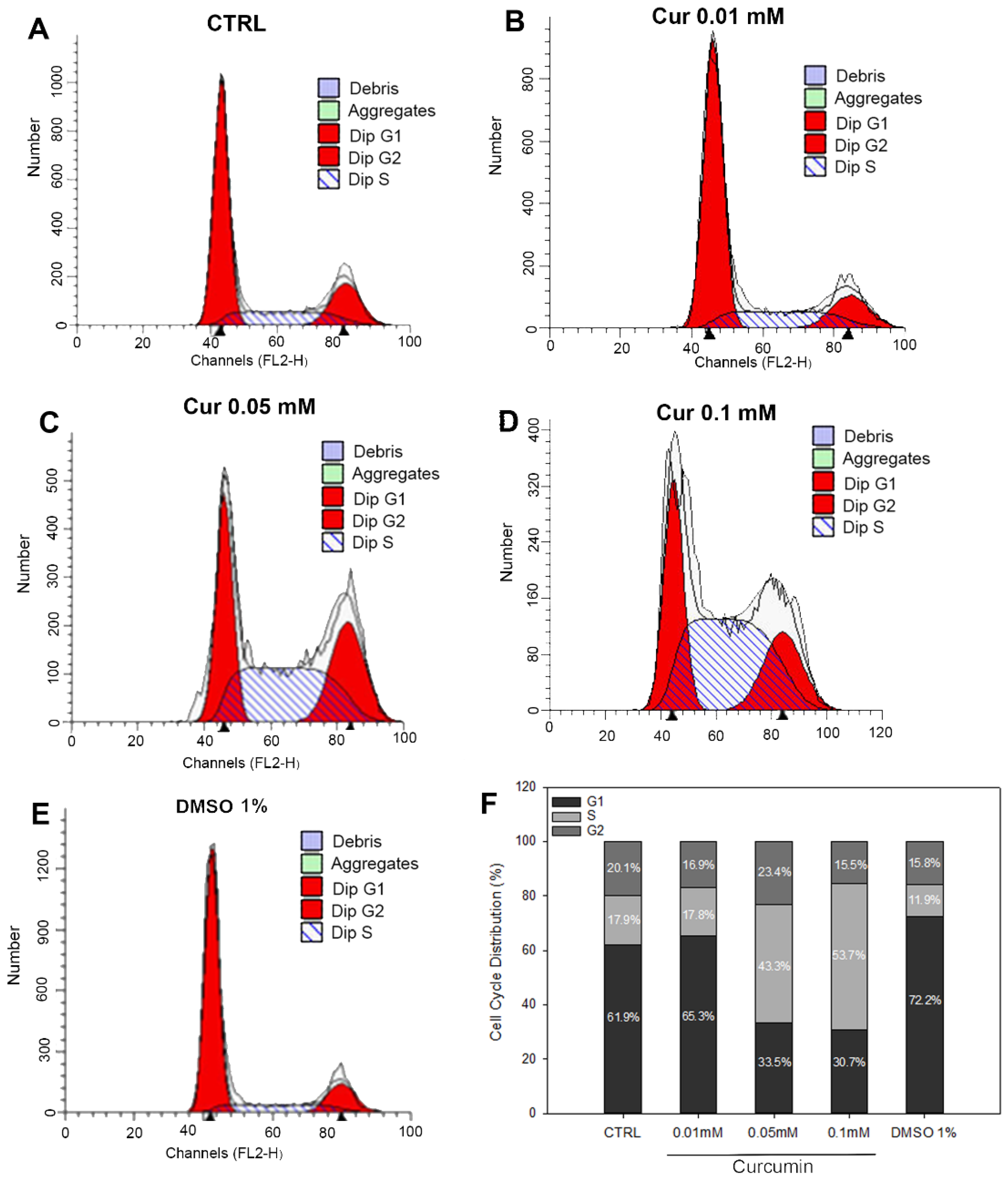

| Groups | Apoptosis Rate |
|---|---|
| CTRL | 0.6 ± 0.35 |
| Cur 0.01 mM | 0.3 ± 0.3 |
| Cur 0.05 mM | 0.4 ± 0.06 |
| Cur 0.1 mM | 0.4 ± 0.12 |
| DMSO 1% | 0.3 ± 0.03 |
| Groups | G0/G1 | S | G2/M |
|---|---|---|---|
| CTRL | 61.98 ± 2.89 | 17.92 ± 3.19 | 20.09 ± 0.30 |
| Cur 0.01 mM | 65.1 ± 1.02 | 17.82 ± 3.84 | 16.87 ± 2.81 |
| Cur 0.05 mM | 33.48 ± 2.83 * | 43.17 ± 0.75 * | 23.36 ± 3.58 |
| Cur 0.01 mM | 30.72 ± 1.45 * | 53.75 ± 3.23 * | 15.53 ± 4.64 |
| DMSO 1% | 72.23 ± 1.47 | 11.94 ± 2.45 | 15.84 ± 0.99 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carozza, G.; Tisi, A.; Capozzo, A.; Cinque, B.; Giovannelli, A.; Feligioni, M.; Flati, V.; Maccarone, R. New Insights into Dose-Dependent Effects of Curcumin on ARPE-19 Cells. Int. J. Mol. Sci. 2022, 23, 14771. https://doi.org/10.3390/ijms232314771
Carozza G, Tisi A, Capozzo A, Cinque B, Giovannelli A, Feligioni M, Flati V, Maccarone R. New Insights into Dose-Dependent Effects of Curcumin on ARPE-19 Cells. International Journal of Molecular Sciences. 2022; 23(23):14771. https://doi.org/10.3390/ijms232314771
Chicago/Turabian StyleCarozza, Giulia, Annamaria Tisi, Annamaria Capozzo, Benedetta Cinque, Aldo Giovannelli, Marco Feligioni, Vincenzo Flati, and Rita Maccarone. 2022. "New Insights into Dose-Dependent Effects of Curcumin on ARPE-19 Cells" International Journal of Molecular Sciences 23, no. 23: 14771. https://doi.org/10.3390/ijms232314771
APA StyleCarozza, G., Tisi, A., Capozzo, A., Cinque, B., Giovannelli, A., Feligioni, M., Flati, V., & Maccarone, R. (2022). New Insights into Dose-Dependent Effects of Curcumin on ARPE-19 Cells. International Journal of Molecular Sciences, 23(23), 14771. https://doi.org/10.3390/ijms232314771










