Potential and Metabolic Pathways of Eugenol in the Management of Xanthomonas perforans, a Pathogen of Bacterial Spot of Tomato
Abstract
1. Introduction
2. Results
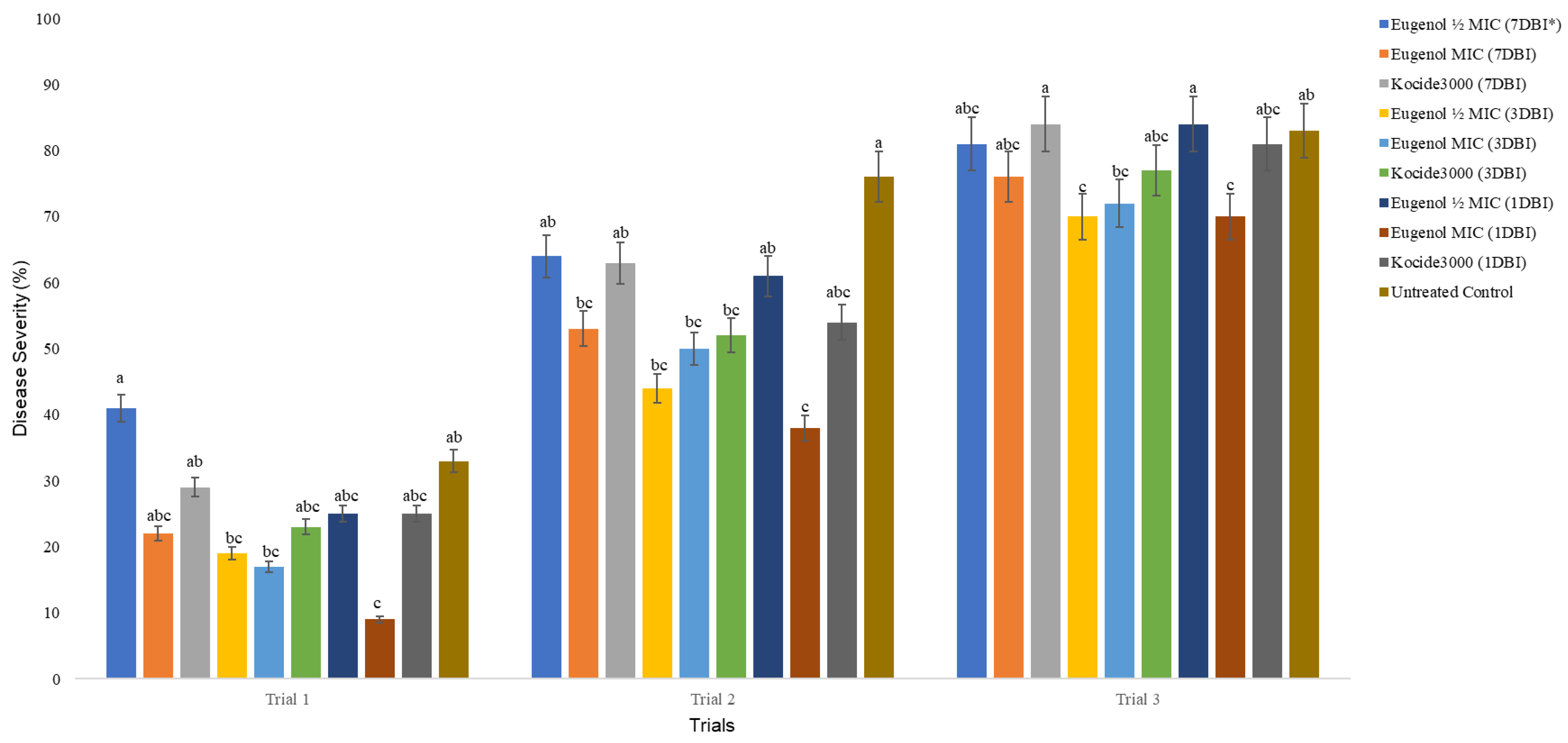
2.1. MIC, MBC, and Effects of Eugenol on Bacterial Spot in the Greenhouse
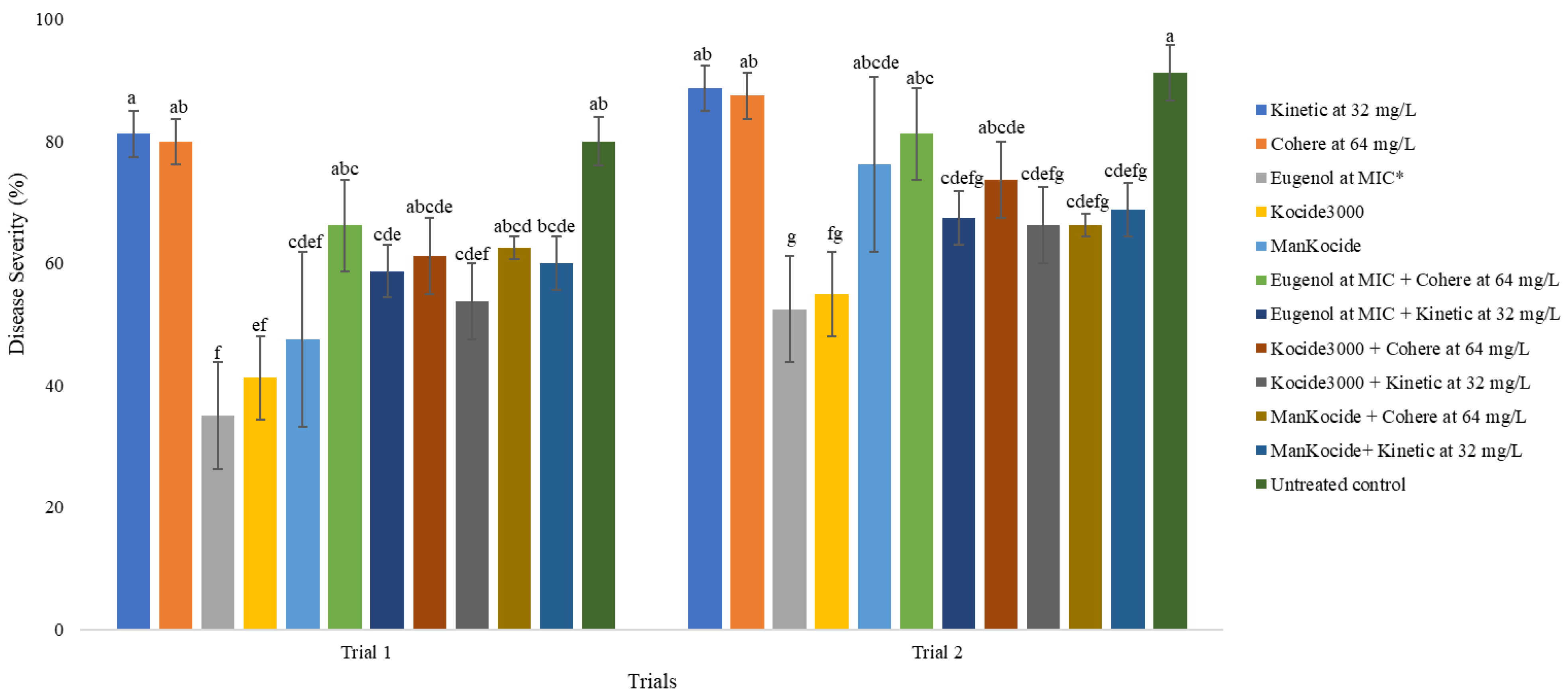
2.2. Effects of Eugenol, Surfactants, and Combination of Eugenol and Surfactants Applied as a Foliar Spray against X. perforans in the Greenhouse
2.3. Eugenol against Bacterial Spot of Tomato under Field Conditions
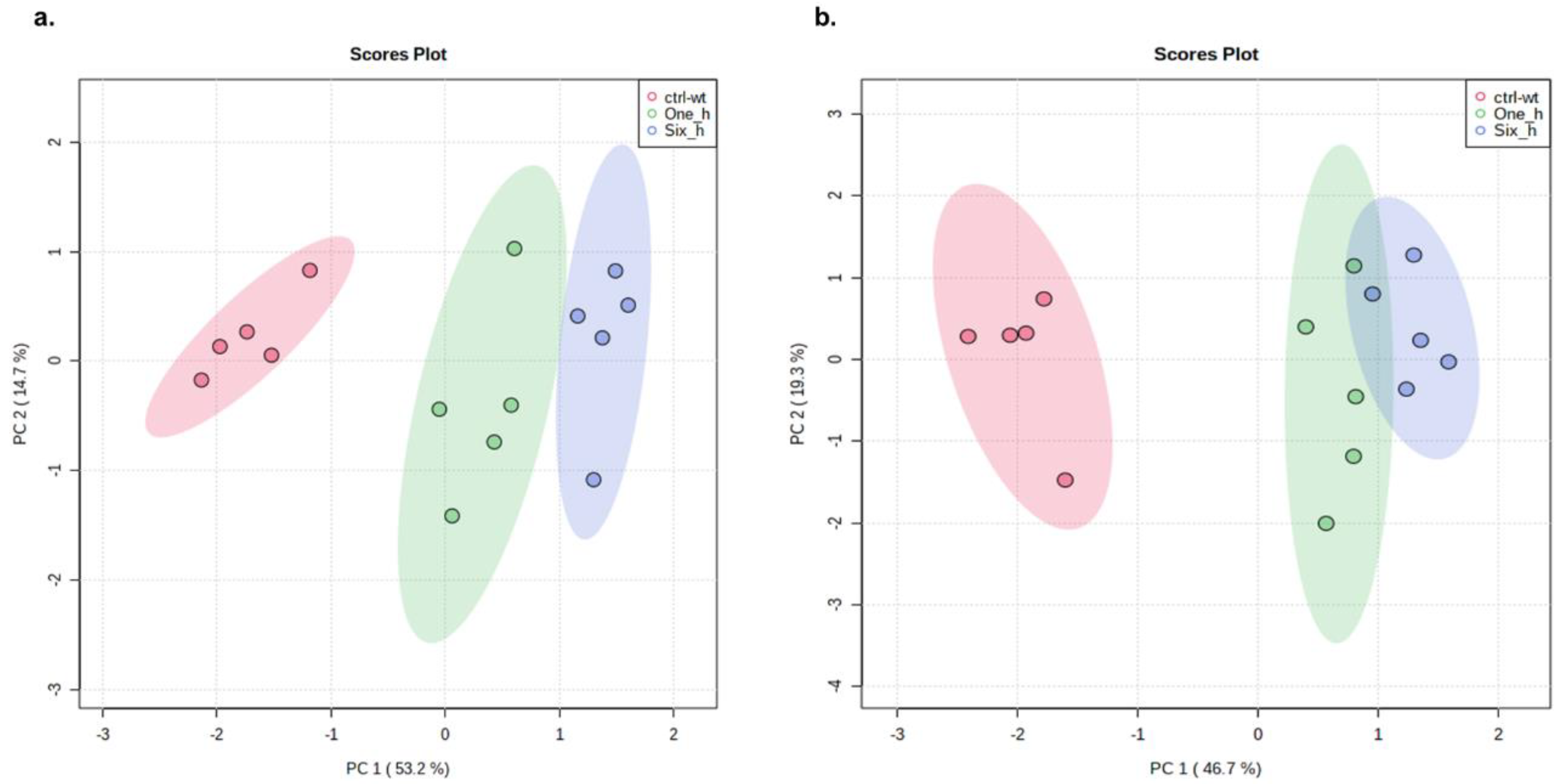
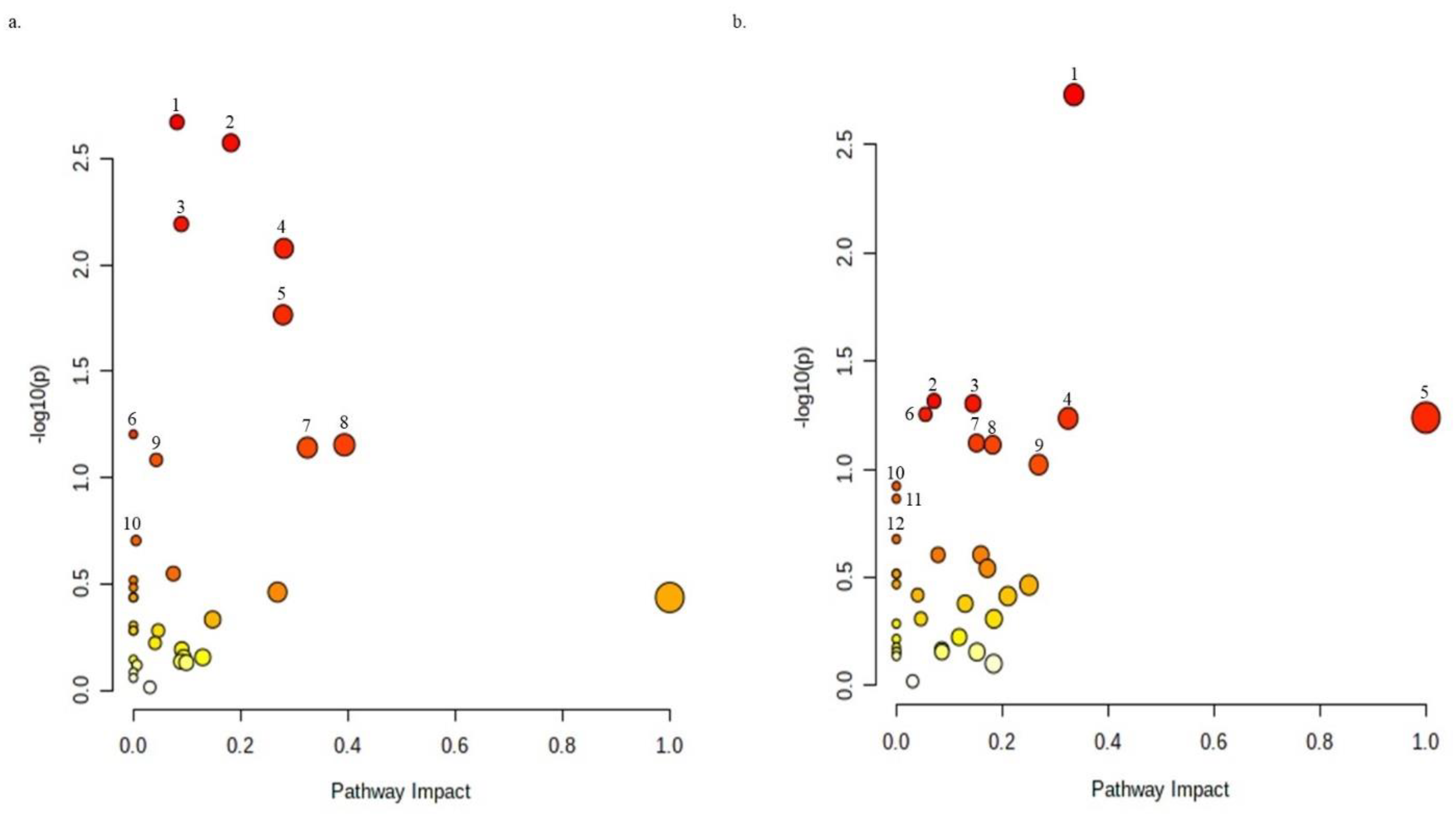
2.4. Metabolic Effect of Eugenol against X. perforans as Revealed by LC–MS Metabolomics
3. Discussion
3.1. Eugenol Improves X. perforans Management in Greenhouse and Field Conditions
3.2. Eugenol Regulates Metabolites in Pathways Critical for Bacterial Survival
4. Materials and Methods
4.1. Preparation of Stock Solutions of Eugenol
4.2. Determination of MIC and MBC of Small Molecules
4.3. Effects of Eugenol on X. perforans in the Greenhouse
4.4. Effects of Eugenol and Combination with Surfactants against X. perforans
4.5. Field Evaluation of Eugenol for Management of Bacterial Spot on Tomato
4.6. LC–MS Based Metabolomics Analyses of Effect of Eugenol on X. perforans
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Macielag, M.J. Chemical properties of antimicrobials and their uniqueness. In Antibiotic Discovery and Development; Dougherty, T.J., Pucci, M.J., Eds.; Springer: Boston, MA, USA, 2012; pp. 793–820. [Google Scholar]
- Gurevich, E.V.; Gurevich, V.V. Therapeutic potential of small molecules and engineered proteins. Handb. Exp. Pharmacol. 2014, 219, 1–12. [Google Scholar] [CrossRef]
- Filipowicz, W.; Jaskiewicz, L.; Kolb, F.A.; Pillai, R.S. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr. Opin. Struct. Biol. 2005, 15, 331–341. [Google Scholar] [CrossRef]
- Schneider-Poetsch, T.; Yoshida, M. Along the central dogma—Controlling gene expression with small molecules. Annu. Rev. Biochem. 2018, 87, 391–420. [Google Scholar] [CrossRef]
- Koonin, E.V. Does the central dogma still stand? Biol. Direct 2012, 7, 27. [Google Scholar] [CrossRef]
- Baker, D.D.; Alvi, K.A. Small-molecule natural products: New structures, new activities. Curr. Opin. Biotechnol. 2004, 15, 576–583. [Google Scholar] [CrossRef]
- Liu, Q.; Qiao, K.; Zhang, S. Potential of a small molecule carvacrol in management of vegetable diseases. Molecules 2019, 24, 1932. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhang, S.; Huang, Y.; Jones, J.B. Evaluation of a small molecule compound 3-indolylacetonitrile for control of bacterial spot on tomato. Crop Prot. 2019, 120, 7–12. [Google Scholar] [CrossRef]
- O’Neill, E.M.; Mucyn, T.S.; Patteson, J.B.; Finkel, O.M.; Chung, E.H.; Baccile, J.A.; Massolo, E.; Schroeder, F.C.; Dangl, J.L.; Li, B. Phevamine A, a small molecule that suppresses plant immune responses. Proc. Natl. Acad. Sci. USA 2018, 115, E9514–E9522. [Google Scholar] [CrossRef]
- Qiao, K.; Liu, Q.; Huang, Y.; Xia, Y.; Zhang, S. Management of bacterial spot of tomato caused by copper-resistant Xanthomonas perforans using a small molecule compound carvacrol. Crop Prot. 2020, 132, 105114. [Google Scholar] [CrossRef]
- Qiao, K.; Liu, Q.; Xia, Y.; Zhang, S. Evaluation of a small-molecule compound, N-acetylcysteine, for the management of bacterial spot of tomato caused by copper-resistant Xanthomonas perforans. Plant Dis. 2020, 105, 108–113. [Google Scholar] [CrossRef]
- Srivastava, V.; Deblais, L.; Kathayat, D.; Rotondo, F.; Helmy, Y.A.; Miller, S.A.; Rajashekara, G. Novel Small Molecule Growth Inhibitors of Xanthomonas spp. Causing Bacterial Spot of Tomato. Phytopathology 2021, 111, 940–953. [Google Scholar] [CrossRef] [PubMed]
- Tao, H.; Fan, S.S.; Jiang, S.; Xiang, X.; Yan, X.; Zhang, L.H.; Cui, Z.N. Small Molecule Inhibitors Specifically Targeting the Type III Secretion System of Xanthomonas oryzae on Rice. Int. J. Mol. Sci. 2019, 20, 971. [Google Scholar] [CrossRef]
- Shuab, R.; Lone, R.; Koul, K.K. Cinnamate and cinnamate derivatives in plants. Acta Physiol. Plant. 2016, 38, 64. [Google Scholar] [CrossRef]
- Muhlemann, J.K.; Woodworth, B.D.; Morgan, J.A.; Dudareva, N. The monolignol pathway contributes to the biosynthesis of volatile phenylpropenes in flowers. New Phytol. 2014, 204, 661–670. [Google Scholar] [CrossRef]
- Atkinson, R.G. Phenylpropenes: Occurrence, distribution, and biosynthesis in fruit. J. Agric. Food Chem. 2018, 66, 2259–2272. [Google Scholar] [CrossRef] [PubMed]
- Sellamuthu, R. Eugenol. In Encyclopedia of Toxicology, 3rd ed.; Wexler, P., Ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 539–541. [Google Scholar]
- Zhang, L.-L.; Zhang, L.-F.; Xu, J.-G.; Hu, Q.-P. Comparison study on antioxidant, DNA damage protective and antibacterial activities of eugenol and isoeugenol against several foodborne pathogens. Food Nutr. Res. 2017, 61, 1. [Google Scholar] [CrossRef]
- Koeduka, T.; Fridman, E.; Gang, D.R.; Vassão, D.G.; Jackson, B.L.; Kish, C.M.; Orlova, I.; Spassova, S.M.; Lewis, N.G.; Noel, J.P.; et al. Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a coniferyl alcohol ester. Proc. Natl. Acad. Sci. USA 2006, 103, 10128–10133. [Google Scholar] [CrossRef]
- Koeduka, T.; Louie, G.V.; Orlova, I.; Kish, C.M.; Ibdah, M.; Wilkerson, C.G.; Bowman, M.E.; Baiga, T.J.; Noel, J.P.; Dudareva, N.; et al. The multiple phenylpropene synthases in both Clarkia breweri and Petunia hybrida represent two distinct protein lineages. Plant J. 2008, 54, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Vassão, D.G.; Kim, S.-J.; Milhollan, J.K.; Eichinger, D.; Davin, L.B.; Lewis, N.G. A pinoresinol–lariciresinol reductase homologue from the creosote bush (Larrea tridentata) catalyzes the efficient in vitro conversion of p-coumaryl/coniferyl alcohol esters into the allylphenols chavicol/eugenol, but not the propenylphenols p-anol/isoeugenol. Arch. Biochem. Biophys. 2007, 465, 209–218. [Google Scholar] [CrossRef]
- Khalil, A.A.; Rahman, U.; Khan, M.R.; Sahar, A.; Mehmood, T.; Khan, M. Essential oil eugenol: Sources, extraction techniques and nutraceutical perspectives. RSC Adv. 2017, 7, 32669–32681. [Google Scholar] [CrossRef]
- Wagle, B.R.; Upadhyay, A.; Shrestha, S.; Arsi, K.; Upadhyaya, I.; Donoghue, A.M.; Donoghue, D.J. Pectin or chitosan coating fortified with eugenol reduces Campylobacter jejuni on chicken wingettes and modulates expression of critical survival genes. Poult. Sci. 2019, 98, 1461–1471. [Google Scholar] [CrossRef]
- Tripathi, A.K.; Mishra, S. Chapter 16—Plant Monoterpenoids (Prospective Pesticides). In Ecofriendly Pest Management for Food Security; Omkar, Ed.; Academic Press: Cambridge, MA, USA, 2016; pp. 507–524. [Google Scholar]
- Al-Shabib, N.A.; Husain, F.M.; Ahmad, I.; Baig, M.H. Eugenol inhibits quorum sensing and biofilm of toxigenic MRSA strains isolated from food handlers employed in Saudi Arabia. Biotechnol. Biotechnol. Equip. 2017, 31, 387–396. [Google Scholar] [CrossRef]
- Shah, S.; Gaikwad, S.; Nagar, S.; Kulshrestha, S.; Vaidya, V.; Nawani, N.; Pawar, S. Biofilm inhibition and anti-quorum sensing activity of phytosynthesized silver nanoparticles against the nosocomial pathogen Pseudomonas aeruginosa. Biofouling 2019, 35, 34–49. [Google Scholar] [CrossRef]
- Kamatou, G.P.; Vermaak, I.; Viljoen, A.M. Eugenol--from the remote Maluku Islands to the international market place: A review of a remarkable and versatile molecule. Molecules 2012, 17, 6953–6981. [Google Scholar] [CrossRef] [PubMed]
- Olea, A.F.; Bravo, A.; Martínez, R.; Thomas, M.; Sedan, C.; Espinoza, L.; Zambrano, E.; Carvajal, D.; Silva-Moreno, E.; Carrasco, H. Antifungal activity of eugenol derivatives against Botrytis Cinerea. Molecules 2019, 24, 1239. [Google Scholar] [CrossRef]
- Wang, C.; Fan, Y. Eugenol enhances the resistance of tomato against tomato yellow leaf curl virus. J. Sci. Food Agric. 2014, 94, 677–682. [Google Scholar] [CrossRef]
- Ojimelukwe, P.; Adler, C. Toxicity and repellent effects of eugenol, thymol, linalool, menthol and other pure compounds on Dinoderus bifloveatus (Coleoptera: Bostrichidae). J. Sustain. Agric. 2000, 2, 47–54. [Google Scholar]
- Huang, Y.; Ho, S.-H.; Lee, H.-C.; Yap, Y.-L. Insecticidal properties of eugenol, isoeugenol and methyleugenol and their effects on nutrition of Sitophilus zeamais Motsch. (Coleoptera: Curculionidae) and Tribolium castaneum (Herbst) (Coleoptera: Tenebrionidae). J. Stored Prod. Res. 2002, 38, 403–412. [Google Scholar] [CrossRef]
- Ajayi, O.E.; Appel, A.G.; Fadamiro, H.Y. Fumigation toxicity of essential oil monoterpenes to Callosobruchus maculatus (Coleoptera: Chrysomelidae: Bruchinae). J. Insects 2014, 2014, 917212. [Google Scholar] [CrossRef]
- Mohanty, S.K.; Reddy, P.R.; Sridhar, R. Modification of susceptibility in rice to bacterial leaf blight by phenols and related substances. Curr. Sci. 1982, 51, 980–982. [Google Scholar]
- Talreja, S.S.; Nerurkar, A.S. Small molecules cause virulence attenuation of Xanthomonas oryzae pv. oryzae, the pathogen causing bacterial blight of rice. Eur. J. Plant Pathol. 2018, 151, 229–241. [Google Scholar] [CrossRef]
- da Cruz Silva, G.; de Oliveira Filho, J.G.; de Mori Morselli Ribeiro, M.; de Souza, C.W.O.; Ferreira, M.D. Antibacterial activity of nanoemulsions based on essential oils compounds against species of Xanthomonas that cause citrus canker. Biointerface Res. Appl. Chem. 2021, 12, 1835–1846. [Google Scholar] [CrossRef]
- Wang, S.; Chen, J.; Shi, J.; Wang, Z.; Hu, D.; Song, B. Novel cinnamic acid derivatives containing the 1,3,4-oxadiazole moiety: Design, synthesis, antibacterial activities, and mechanisms. J. Agric. Food Chem. 2021, 69, 40. [Google Scholar] [CrossRef]
- Potnis, N.; Timilsina, S.; Strayer, A.; Shantharaj, D.; Barak, J.D.; Paret, M.L.; Vallad, G.E.; Jones, J.B. Bacterial spot of tomato and pepper: Diverse Xanthomonas species with a wide variety of virulence factors posing a worldwide challenge. Mol. Plant Pathol. 2015, 16, 907–920. [Google Scholar] [CrossRef] [PubMed]
- Osdaghi, E.; Jones, J.B.; Sharma, A.; Goss, E.M.; Abrahamian, P.; Newberry, E.A.; Potnis, N.; Carvalho, R.; Choudhary, M.; Paret, M.L.; et al. A centenary for bacterial spot of tomato and pepper. Mol. Plant Pathol. 2021, 22, 1500–1519. [Google Scholar] [CrossRef]
- Jibrin, M.O.; Timilsina, S.; Minsavage, G.V.; Vallad, G.E.; Roberts, P.D.; Goss, E.M.; Jones, J.B. Bacterial Spot of Tomato and Pepper in Africa: Diversity, Emergence of T5 Race, and Management. Front. Microbiol. 2022, 13, 835647. [Google Scholar] [CrossRef]
- Stall, R.E.; Jones, J.B.; Minsavage, G.V. Durability of resistance in tomato and pepper to xanthomonads causing bacterial spot. Annu. Rev. Phytopathol. 2009, 47, 265–284. [Google Scholar] [CrossRef]
- Ritchie, D.F. Bacterial spot of pepper and tomato. Plant Health Instr. 2000. [Google Scholar] [CrossRef]
- Klein-Gordon, J.M.; Xing, Y.; Garrett, K.A.; Abrahamian, P.; Paret, M.L.; Minsavage, G.V.; Strayer-Scherer, A.L.; Fulton, J.C.; Timilsina, S.; Jones, J.B.; et al. Assessing changes and associations in the Xanthomonas perforans population across Florida commercial tomato fields via a statewide survey. Phytopathology 2020, 111, 1029–1041. [Google Scholar] [CrossRef]
- Jibrin, M.O.; Potnis, N.; Timilsina, S.; Minsavage, G.V.; Vallad, G.E.; Roberts, P.D.; Jones, J.B.; Goss, E.M. Genomic inference of recombination-mediated evolution in Xanthomonas euvesicatoria and X. perforans. Appl. Environ. Microbiol. 2018, 84, e00136-18. [Google Scholar] [CrossRef]
- Newberry, E.A.; Bhandari, R.; Minsavage, G.V.; Timilsina, S.; Jibrin, M.O.; Kemble, J.; Sikora, E.J.; Jones, J.B.; Potnis, N. Independent Evolution with the Gene Flux Originating from Multiple Xanthomonas Species Explains Genomic Heterogeneity in Xanthomonas perforans. Appl. Environ. Microbiol. 2019, 85, e00885-19. [Google Scholar] [CrossRef]
- Jibrin, M.O.; Liu, Q.; Guingab-Cagmat, J.; Jones, J.B.; Garrett, T.J.; Zhang, S. Metabolomics Insights into Chemical Convergence in Xanthomonas perforans and Metabolic Changes Following Treatment with the Small Molecule Carvacrol. Metabolites 2021, 11, 879. [Google Scholar] [CrossRef]
- Picchi, S.C.; de Souza ESilva, M.; Saldanha, L.L.; Ferreira, H.; Takita, M.A.; Caldana, C.; de Souza, A.A. GC-TOF/MS-based metabolomics analysis to investigate the changes driven by N-Acetylcysteine in the plant-pathogen Xanthomonas citri subsp. citri. Sci. Rep. 2021, 11, 15558. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC–HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 2022, 17, 1735–1761. [Google Scholar] [CrossRef]
- Barboza, J.N.; da Silva Maia Bezerra Filho, C.; Silva, R.O.; Medeiros, J.V.R.; de Sousa, D.P. An overview on the anti-inflammatory potential and antioxidant profile of eugenol. Oxidative Med. Cell. Longev. 2018, 2018, 3957262. [Google Scholar] [CrossRef] [PubMed]
- Guzman, J.D. Natural cinnamic acids, synthetic derivatives and hybrids with antimicrobial activity. Molecules 2014, 19, 9292. [Google Scholar] [CrossRef] [PubMed]
- Kowalska-Krochmal, B.; Dudek-Wicher, R. The minimum inhibitory concentration of antibiotics: Methods, interpretation, clinical relevance. Pathogens 2021, 10, 165. [Google Scholar] [CrossRef]
- Kampf, G.; Hollingsworth, A. Comprehensive bactericidal activity of an ethanol-based hand gel in 15 seconds. Ann. Clin. Microbiol. Antimicrob. 2008, 7, 2. [Google Scholar] [CrossRef]
- Bajpai, V.K.; Cho, M.J.; Kang, S.C. Control of Plant Pathogenic Bacteria of Xanthomonas spp. by the Essential Oil and Extracts of Metasequoia glyptostroboides Miki ex Hu In vitro and In vivo. J. Phytopathol. 2010, 158, 479–486. [Google Scholar] [CrossRef]
- Dadasoglu, F.; Aydin, T.; Kotan, R.; Cakir, A.; Ozer, H.; Kordali, S.; Cakmacki, R.; Dikbas, N.; Mate, E. Antibacterial activities of extracts and essential oils of three origanum species against plant pathogenic bacteria and their potential use as seed disinfectants. J. Plant Pathol. 2011, 93, 271–282. [Google Scholar]
- Lucas, G.C.; Alves, E.; Pereira, R.B.; Perina, F.J.; de Souza, R.A. Antibacterial activity of essential oils on Xanthomonas vesicatoria and control of bacterial spot in tomato. Pesq. Agropec. Bras. 2012, 47, 351–359. [Google Scholar] [CrossRef]
- Khalil Bagy, H.; Abo-Elyousr, K. Antibacterial activity of some essential oils on bacterial spot disease of tomato plant caused by Xanthomonas axonopodis pv. Vesicatoria. Int. J. Phytopathol. 2019, 8, 53–61. [Google Scholar] [CrossRef]
- Lo Cantore, P.; Shanmugaiah, V.; Iacobellis, N.S. Antibacterial activity of essential oil components and their potential use in seed disinfection. J. Agric. Food Chem. 2009, 57, 9454–9461. [Google Scholar] [CrossRef]
- Jibrin, M.O.; Liu, Q.; Jones, J.B.; Zhang, S. Surfactants in plant disease management: A brief review and case studies. Plant Pathol. 2021, 70, 495–510. [Google Scholar] [CrossRef]
- Hartl, J.; Kiefer, P.; Kaczmarczyk, A.; Mittelviefhaus, M.; Meyer, F.; Vonderach, T.; Hattendorf, B.; Jenal, U.; Vorholt, J.A. Untargeted metabolomics links glutathione to bacterial cell cycle progression. Nat. Metab. 2020, 2, 153–166. [Google Scholar] [CrossRef]
- Fenves, A.Z.; Kirkpatrick, H.M.; 3rd Patel, V.V.; Sweetman, L.; Emmett, M. Increased anion gap metabolic acidosis as a result of 5-oxoproline (pyroglutamic acid): A role for acetaminophen. Clin. J. Am. Soc. Nephrol. CJASN 2006, 1, 441–447. [Google Scholar] [CrossRef]
- Duewall, J.L.; Fenves, A.Z.; Richey, D.S.; Tran, L.D.; Emmett, M. 5-Oxoproline (pyroglutamic) acidosis associated with chronic acetaminophen use. Proceedings 2010, 23, 19–20. [Google Scholar] [CrossRef] [PubMed]
- Felux, A.K.; Denger, K.; Weiss, M.; Cook, A.M.; Schleheck, D. Paracoccus denitrificans PD1222 utilizes hypotaurine via transamination followed by spontaneous desulfination to yield acetaldehyde and, finally, acetate for growth. J. Bacteriol. 2013, 195, 2921–2930. [Google Scholar] [CrossRef]
- Amorim Franco, T.M.; Blanchard, J.S. Bacterial Branched-Chain Amino Acid Biosynthesis: Structures, Mechanisms, and Drugability. Biochemistry 2017, 56, 5849–5865. [Google Scholar] [CrossRef]
- Luo, H.Z.; Guan, Y.; Yang, R.; Qian, G.L.; Yang, X.H.; Wang, J.S.; Jia, A.Q. Growth inhibition and metabolomic analysis of Xanthomonas oryzae pv. oryzae treated with resveratrol. BMC Microbiol. 2020, 20, 117. [Google Scholar] [CrossRef]
- Baker, B.P.; Grant, J.A. Eugenol Profile. 2018. Available online: https://ecommons.cornell.edu/bitstream/handle/1813/56125/eugenol-MRP-NYSIPM.pdf?sequence=1 (accessed on 30 August 2022).
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]


| Treatments | Trial 1 | Trial 2 | ||
|---|---|---|---|---|
| Severity | AUDPC | Severity | AUDPC | |
| Untreated Control | 19.0 a | 212.6 a | 49.4 a | 560.3 a |
| Kocide3000 | 15.6 b | 136.8 b | 46.3 ab | 475.0 b |
| ManKocide | 13.1 bc | 122.0 bc | 44.2 bc | 441.0 bc |
| Eugenol | 12.1 c | 108.0 c | 42.3 bcd | 424.1 cd |
| Eugenol + Cohere | 11.7 c | 104.6 c | 38.5 d | 380.2 d |
| LSD0.05 | 2.58 | 27.8 | 4.0 | 45.3 |
| S/N | Pathway Name | −log(p) | Metabolites Expressed in Regulated Pathways |
|---|---|---|---|
| Positive Ion phase | |||
| 1 | Glutathione metabolism | 2.6706 | Glycine (D), L-Ornithine (D), Putrescine (D), 5-Oxoproline (U), L-Cysteine (D) |
| 2 | Aminoacyl-tRNA biosynthesis | 2.5735 | L-Glutamine (U), L-Cysteine, Glycine (D), L-Serine (U), L-methionine (U), L-Lysine (D), L-Isoleucine (U), L-Proline (D), |
| 3 | Pyrimidine metabolism | 2.1917 | Cytosine, L-Glutamine (U), CMP (D), Cytidine (U), Uridine (U), Uracil (D), Deoxycytidine (U) |
| 4 | Arginine and proline metabolism | 2.0773 | L-Ornithine (D), L-Proline (D), Creatine (D), Creatinine (U), Putrescine D), N-Acetylputrescine (U) |
| 5 | Glycine, serine, and threonine metabolism | 1.7648 | L-Serine (U), Glycine (D), L-Cystathionine (U), L-Cysteine, Creatine (D), Ectoine (U), |
| 6 | Cyanoamino acid metabolism | 1.203 | L-Asparagine (U), Glycine (D), L-Serine (U) |
| 7 | Cysteine and methionine metabolism | 1.1538 | L-Cystine (D), L-Serine (U), L-Cystathionine (U), L-Cysteine (D), L-methionine (U). |
| 8 | Arginine biosynthesis | 1.1407 | L-Glutamine (U), L-Citrulline, L-Ornithine |
| 9 | Sulfur metabolism | 1.083 | L-Serine (U), Taurine (D), L-Cysteine (D) |
| 10 | Glyoxylate and dicarboxylate metabolism | 0.70377 | Glycine (D), L-Serine (U), L-Glutamine (U). |
| Negative Ion Phase | |||
| 1 | Sulfur metabolism | 2.7316 | L-Serine (U), Sulfate (D), Succinate (D), Sulfite (D), Taurine (D). |
| 2 | Glyoxylate and dicarboxylate metabolism | 1.3151 | (S)-Malate (D), L-Serine (U), L-Glutamine (U), Succinate (D) |
| 3 | Purine metabolism | 1.3032 | Hypoxanthine (D), Xanthine (D), L-Glutamine (U), D-Ribose-5-phosphate (D), Sulfate (D), GMP (D), AMP (D) |
| 4 | Pyrimidine metabolism | 1.2534 | L-Glutamine (U), CMP (D), Cytidine (U), Uridine (U), Uracil (D) |
| 5 | Taurine and hypotaurine metabolism | 1.2383 | Taurine (D), Sulfite (D) |
| 6 | Arginine biosynthesis | 1.2352 | L-Glutamine (U), L-Ornithine (U), L-Citrulline (U) |
| 7 | Pantothenate and CoA biosynthesis | 1.1208 | Uracil (D), Pantothenate (U), Pantetheine (D) |
| 8 | Aminoacyl-tRNA biosynthesis | 1.1126 | L-Histidine (U), L-Glutamine (U), L-Serine (U), L-Methionine (U), L-Proline (D) |
| 9 | Alanine, aspartate, and glutamate metabolism | 1.0208 | L-Asparagine (U), Succinate (D), L-Glutamine (U) |
| 10 | Monobactam biosynthesis | 0.92173 | L-Serine (U), Sulfate (D) |
| 11 | Beta-alanine metabolism | 0.863 | Uracil (D), Pantothenate (U) |
| 12 | Cyanoamino acid metabolism | 0.67626 | L-Asparagine (U), L-Serine (U) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jibrin, M.O.; Liu, Q.; Garrett, T.J.; Jones, J.B.; Zhang, S. Potential and Metabolic Pathways of Eugenol in the Management of Xanthomonas perforans, a Pathogen of Bacterial Spot of Tomato. Int. J. Mol. Sci. 2022, 23, 14648. https://doi.org/10.3390/ijms232314648
Jibrin MO, Liu Q, Garrett TJ, Jones JB, Zhang S. Potential and Metabolic Pathways of Eugenol in the Management of Xanthomonas perforans, a Pathogen of Bacterial Spot of Tomato. International Journal of Molecular Sciences. 2022; 23(23):14648. https://doi.org/10.3390/ijms232314648
Chicago/Turabian StyleJibrin, Mustafa Ojonuba, Qingchun Liu, Timothy J. Garrett, Jeffrey B. Jones, and Shouan Zhang. 2022. "Potential and Metabolic Pathways of Eugenol in the Management of Xanthomonas perforans, a Pathogen of Bacterial Spot of Tomato" International Journal of Molecular Sciences 23, no. 23: 14648. https://doi.org/10.3390/ijms232314648
APA StyleJibrin, M. O., Liu, Q., Garrett, T. J., Jones, J. B., & Zhang, S. (2022). Potential and Metabolic Pathways of Eugenol in the Management of Xanthomonas perforans, a Pathogen of Bacterial Spot of Tomato. International Journal of Molecular Sciences, 23(23), 14648. https://doi.org/10.3390/ijms232314648








