Osteoblast Demineralization Induced by Oxidized High-Density Lipoprotein via the Inflammatory Pathway Is Suppressed by Adiponectin
Abstract
:1. Introduction
2. Results
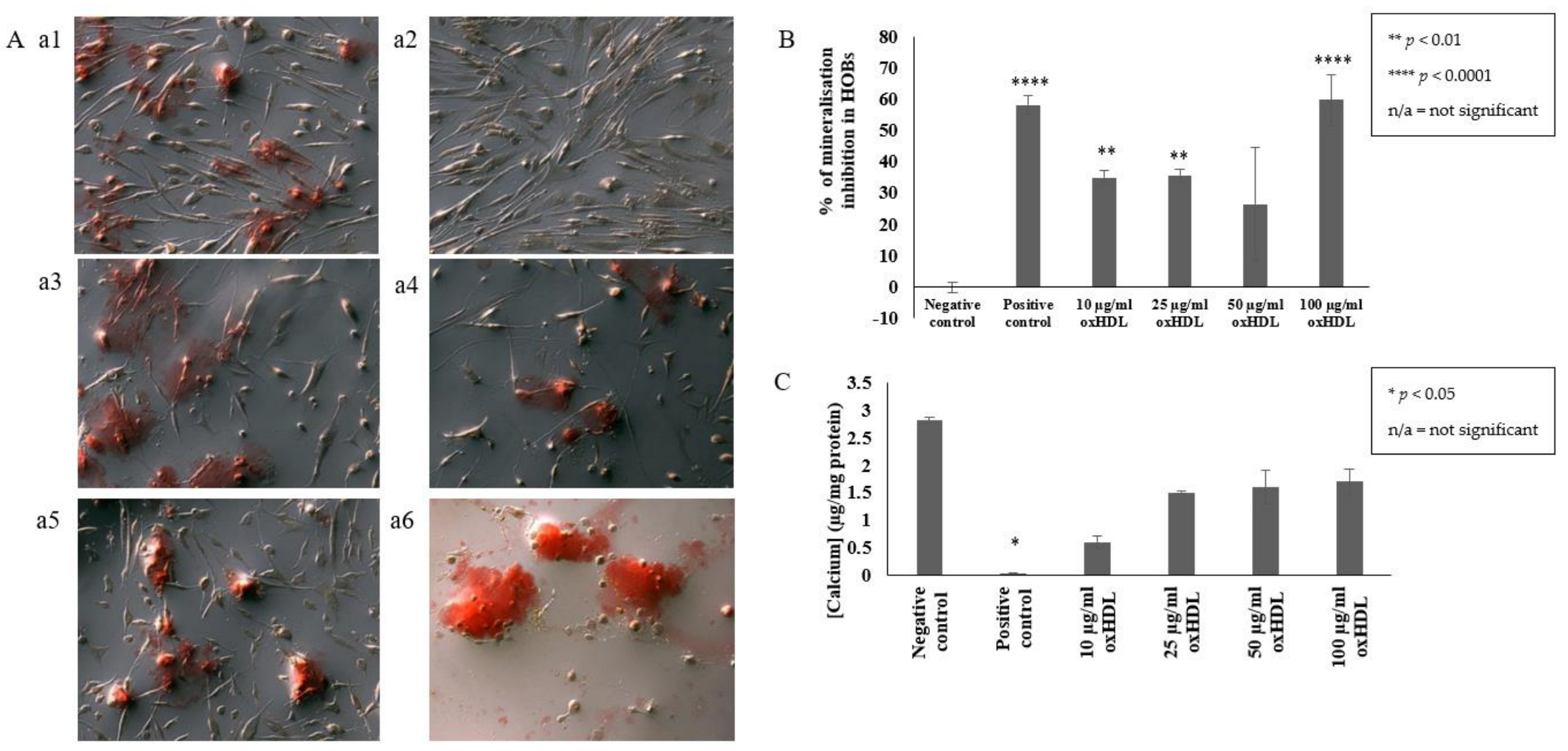
2.1. OxHDL Reduces the Formation of Mineral Nodules and Calcium Incorporation in HOBs
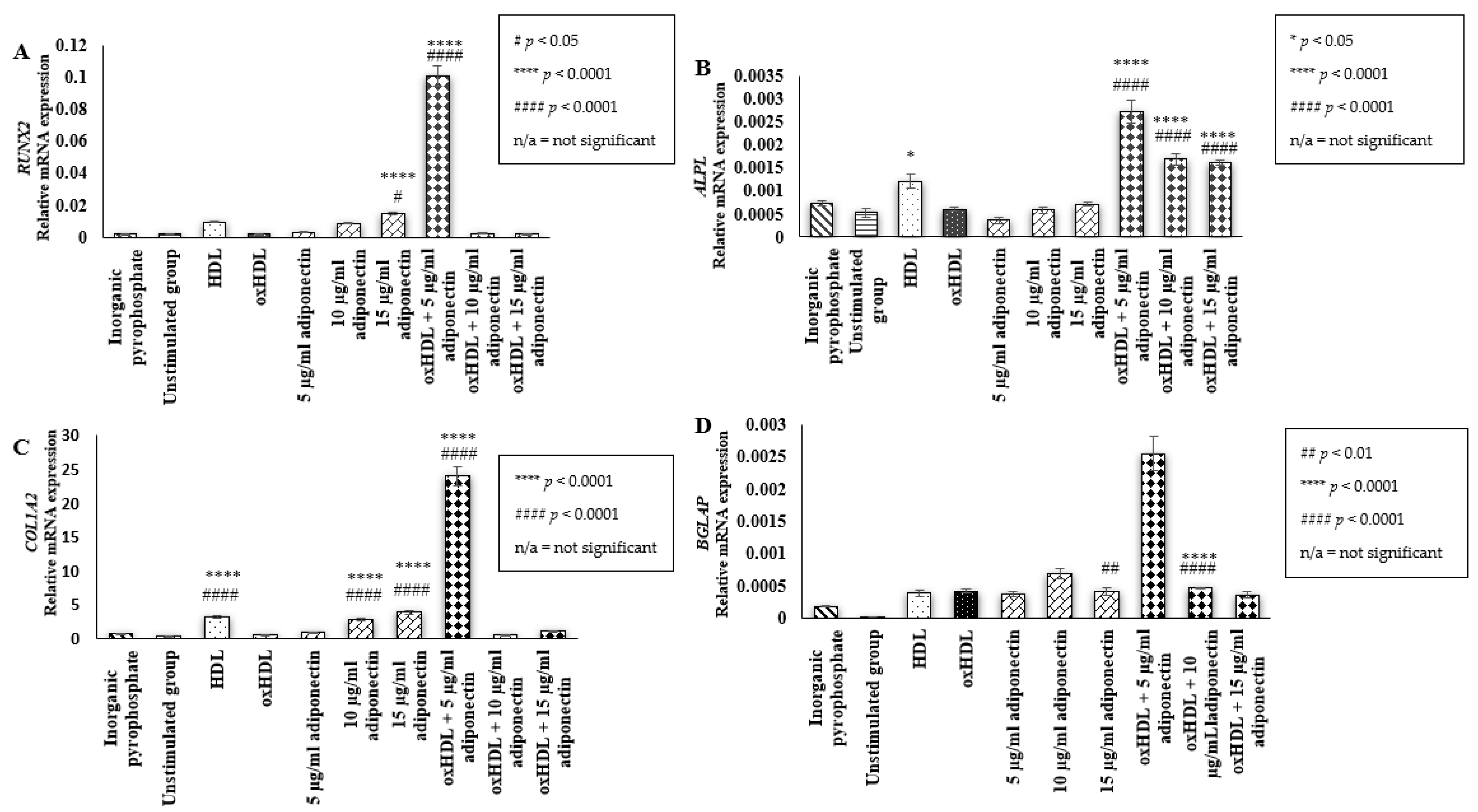
2.2. Effect of HDL, oxHDL, Adiponectin, and Co-Incubation of oxHDL with Adiponectin on the Expression of Bone Mineralization Markers (RUNX2, ALPL, COL1A2, BGLAP)
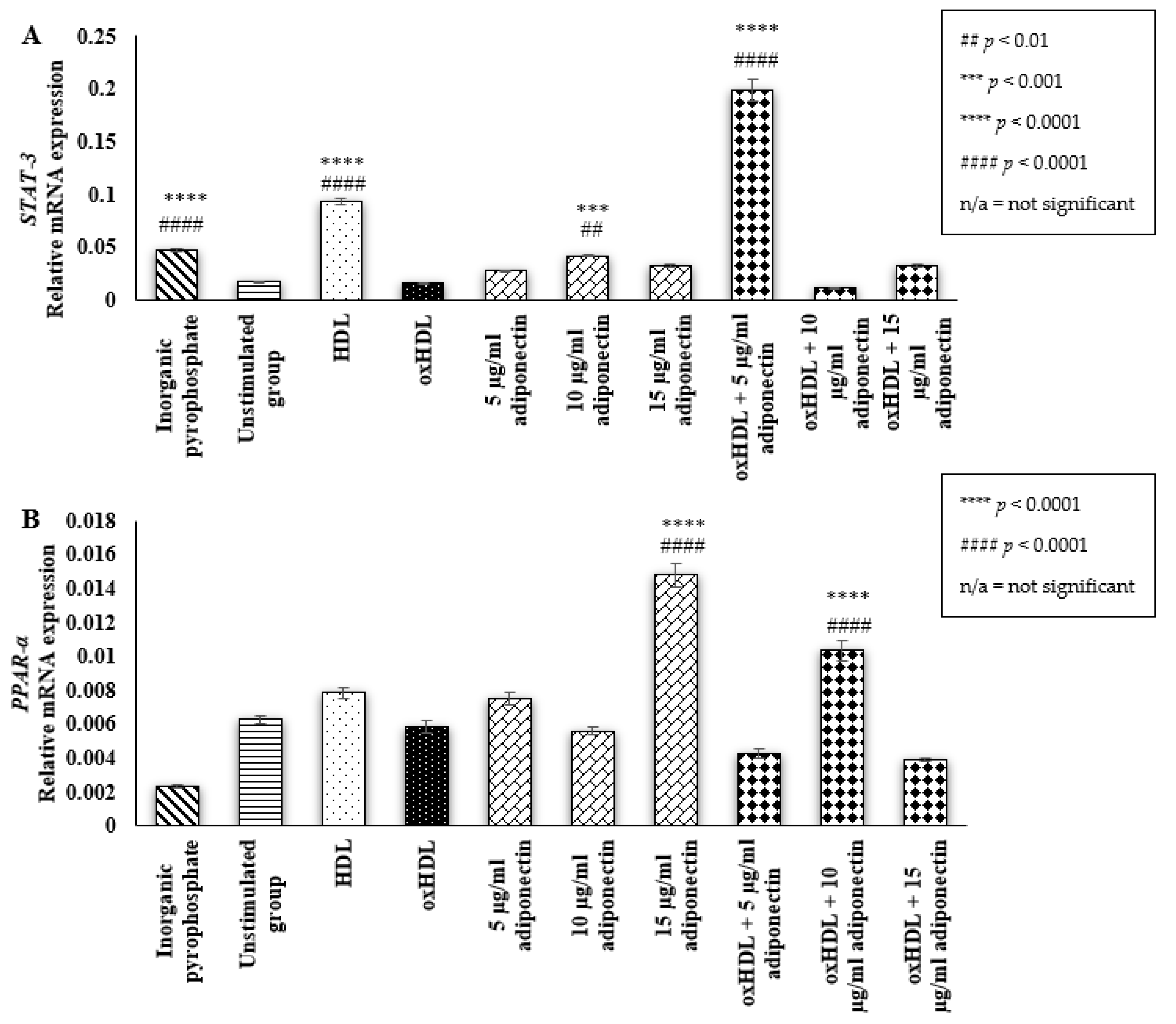
2.3. Effect of HDL, oxHDL, Adiponectin, and Co-Incubation of oxHDL with Adiponectin on the Expression of Osteoblastic Transcription Factor (STAT-3 and PPAR-α)
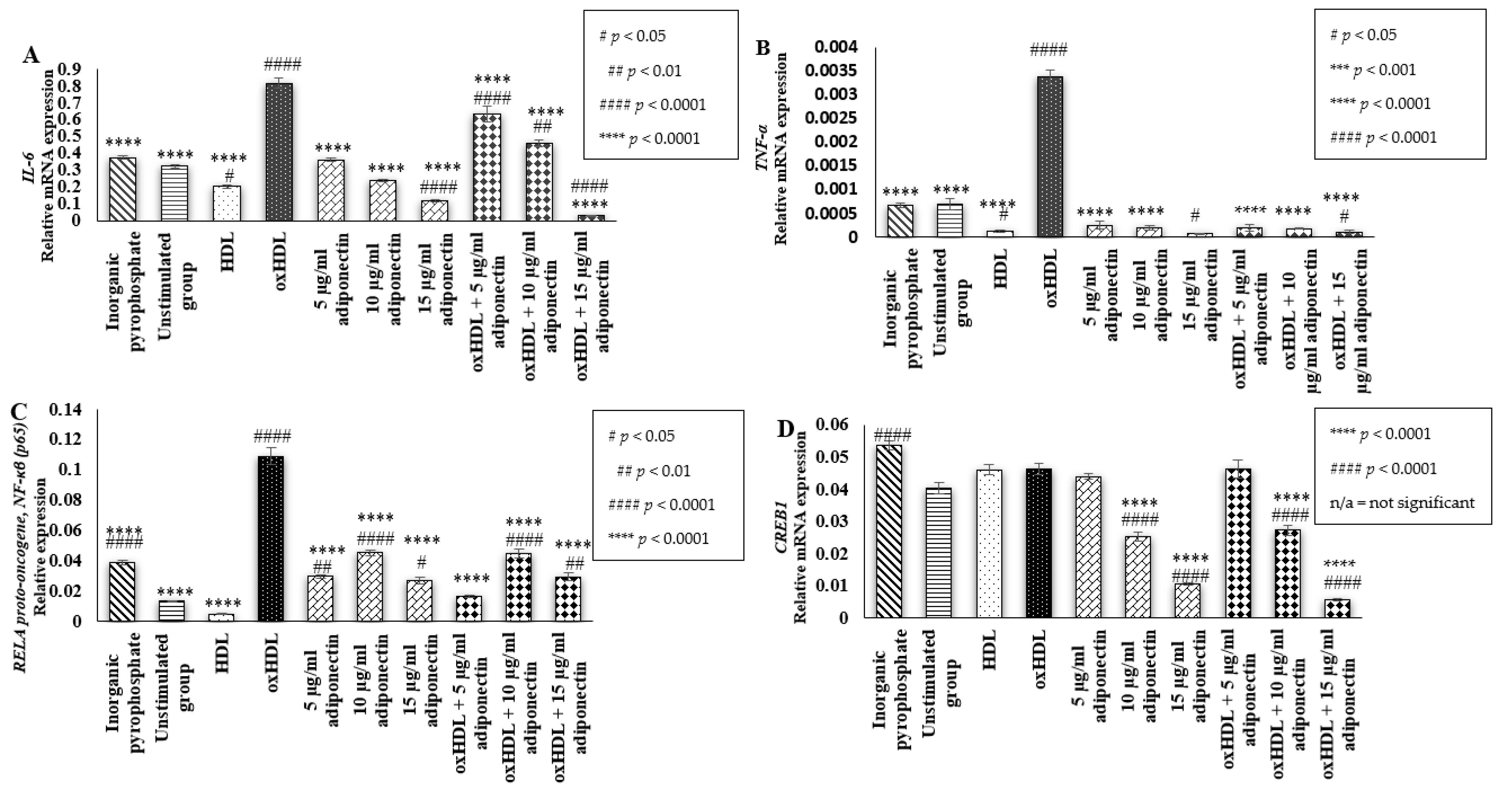
2.4. Effects of HDL, oxHDL, Adiponectin, and Co-Incubation of oxHDL with Adiponectin on the Expression of Inflammatory Markers (IL-6, TNF-α, RELA Proto-Oncogene NF-κβ Subunit (p65), and CREB1)
3. Discussion
4. Materials and Methods
4.1. Culturing and Maintenance of HOBs
4.2. Oxidation of HDL
4.3. Mineralization Detection via Alizarin Red Staining
4.4. Detection of Calcium Deposition Using the Calcium Colorimetric Assay
4.5. Quantification of Bone Mineralization (ALPL, COL1A2, BGLAP, and RUNX2), Osteoblast-Related Transcription Factors (STAT-3 and PPAR-α), and Inflammatory (IL-6, TNF-α, RELA Proto-Oncogene, NF-kB Subunit (P65), and CREB) Markers Using Quantitative Reverse Transcription-Polymerase Chain Reaction (RT-qPCR)
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blair, H.C.; Larrouture, Q.C.; Li, Y.; Lin, H.; Beer-Stoltz, D.; Liu, L.; Tuan, R.S.; Robinson, L.J.; Schlesinger, P.H.; Nelson, D.J. Osteoblast differentiation and bone matrix formation in vivo and in vitro. Tissue Eng. Part B Rev. 2017, 23, 268–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Florencio-Silva, R.; Sasso, G.R.D.S.; Sasso-Cerri, E.; Simões, M.J.; Cerri, P.S. Biology of bone tissue: Structure, function, and factors that influence bone cells. BioMed Res. Int. 2015, 2015, 421746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finkelstein, J.S.; Elaine, W.Y. Patient Education: Bone Density Testing (Beyond the Basics); UpToDate: Wolters Kluwer, The Netherlands, 2022. [Google Scholar]
- Salari, N.; Ghasemi, H.; Mohammadi, L.; Rabieenia, E.; Shohaimi, S.; Mohammadi, M. The global prevalence of osteoporosis in the world: A comprehensive systematic review and meta-analysis. J. Orthop. Surg. Res. 2021, 16, 609. [Google Scholar] [CrossRef]
- Tarantino, U.; Cariati, I.; Greggi, C.; Iundusi, R.; Gasbarra, E.; Iolascon, G.; Kurth, A.; Akesson, K.E.; Bouxsein, M.; Tranquilli Leali, P.; et al. Gaps and alternative surgical and non-surgical approaches in the bone fragility management: An updated review. Osteoporos. Int. 2022, 1–12. [Google Scholar] [CrossRef]
- Becker, D.J.; Kilgore, M.L.; Morrisey, M.A. The societal burden of osteoporosis. Curr. Rheumatol. Rep. 2010, 12, 186–191. [Google Scholar] [CrossRef]
- Byers, R.J.; Denton, J.; Hoyland, J.A.; Freemont, A.J. Differential patterns of osteoblast dysfunction in trabecular bone in patients with established osteoporosis. J. Clin. Pathol. 1997, 50, 760–764. [Google Scholar] [CrossRef] [Green Version]
- Demer, L.L. Vascular calcification and osteoporosis: Inflammatory responses to oxidized lipids. Int. J. Epidemiol. 2002, 31, 737–741. [Google Scholar] [CrossRef] [Green Version]
- Lacey, D.C.; Simmons, P.J.; Graves, S.E.; Hamilton, J.A. Pro-inflammatory cytokines inhibit osteogenic differentiation from stem cells: Implications for bone repair during inflammation. Osteoarthr. Cartil. 2009, 17, 735–742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, X.; McDonald, J.M. Disorders of Bone Remodeling. Annu. Rev. Pathol. 2011, 6, 121–145. [Google Scholar] [CrossRef] [Green Version]
- Parhami, F.; Morrow, A.D.; Balucan, J.; Leitinger, N.; Watson, A.D.; Tintut, Y.; Berliner, J.A.; Demer, L.L. Lipid oxidation products have opposite effects on calcifying vascular cell and bone cell differentiation. Arterioscler. Thromb. Vasc. Biol. 1997, 17, 680–687. [Google Scholar] [CrossRef]
- Tintut, Y.; Morony, S.; Demer, L.L. Hyperlipidemia promotes osteoclastic potential of bone marrow cells Ex Vivo. Arterioscler. Thromb. Vasc. Biol. 2004, 24, e6–e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazière, C.; Savitsky, V.; Galmiche, A.; Gomila, C.; Massy, Z.; Mazière, J.C. Oxidized low density lipoprotein inhibits phosphate signaling and phosphate-induced mineralization in Osteoblasts. Involvement of oxidative stress. Biochim. Biophys. Acta Mol. Basis Dis. 2010, 1802, 1013–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, F.; Ito, T. High-density lipoprotein (HDL) triglyceride and oxidized HDL: New lipid biomarkers of lipoprotein-related atherosclerotic cardiovascular disease. Antioxidants 2020, 9, 362. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.A.K. Dysfunctional HDL in diabetes mellitus and its role in the pathogenesis of cardiovascular disease. Mol. Cell. Biochem. 2017, 440, 167–187. [Google Scholar] [CrossRef] [PubMed]
- Rosenson, R.S., Jr.; Brewer, H.B.; Ansell, B.J.; Barter, P.; Chapman, M.J.; Heinecke, J.W.; Kontush, A.; Tall, A.R.; Webb, N.R. Dysfunctional HDL and atherosclerotic cardiovascular disease. Nat. Publ. Group 2015, 13, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Blair, H.C.; Kalyvioti, E.; Papachristou, N.I.; Tourkova, I.L.; Syggelos, S.A.; Deligianni, D.; Orkoula, M.G.; Kontoyannis, C.G.; Karavia, E.A.; Kypreos, K.E.; et al. Apolipoprotein A-1 regulates Osteoblast and lipoblast precursor cells in mice. Lab. Investig. 2016, 96, 763–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahim, S.; Abdullah, H.M.A.; Ali, Y.; Khan, U.I.; Ullah, W. Serum Apo A-1 and its role as a biomarker of coronary artery disease. Cureus 2016, 8, e941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martineau, C.; Martin-Falstrault, L.; Brissette, L.; Moreau, R. The atherogenic Scarb1 null mouse model shows a high bone mass phenotype. Am. J. Physiol. Endocrinol. Metab. 2014, 306, E48–E57. [Google Scholar] [CrossRef] [Green Version]
- Isales, C.M.; Zaidi, M.; Blair, H.C. ACTH is a novel regulator of bone mass. Ann. N. Y. Acad. Sci. 2010, 1192, 110–116. [Google Scholar] [CrossRef]
- Papachristou, N.I.; Blair, H.C.; Kypreos, K.E.; Papachristou, D.J. High-Density Lipoprotein (HDL) metabolism and bone mass. J. Endocrinol. 2017, 233, R95–R107. [Google Scholar] [CrossRef]
- Kirk, B.; Feehan, J.; Lombardi, G.; Duque, G. Muscle, bone, and fat crosstalk: The biological role of myokines, osteokines, and adipokines. Curr. Osteoporos. Rep. 2020, 18, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.V.; Scherer, P.E. Adiponectin, the past two decades. J. Mol. Cell Biol. 2016, 8, 93–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lubkowska, A.; Dobek, A.; Mieszkowski, J.; Garczynski, W.; Chlubek, D. Adiponectin as a biomarker of osteoporosis in postmenopausal women: Controversies. Dis. Markers 2014, 2014, 975178. [Google Scholar] [CrossRef] [PubMed]
- Pierce, J.L.; Begun, D.L.; Westendorf, J.J.; McGee-Lawrence, M.E. Defining osteoblast and adipocyte lineages in the bone marrow. Bone 2019, 118, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Hyun, W.L.; Sang, Y.K.; Kim, A.Y.; Eun, J.L.; Choi, J.Y.; Jae, B.K. Adiponectin stimulates Osteoblast differentiation through induction of COX2 in mesenchymal progenitor cells. Stem Cells 2009, 27, 2254–2262. [Google Scholar]
- Luo, X.H.; Guo, L.J.; Yuan, L.Q.; Xie, H.; Zhou, H.D.; Wu, X.P.; Liao, E.Y. Adiponectin stimulates human Osteoblasts proliferation and differentiation via the MAPK signaling pathway. Exp. Cell Res. 2005, 309, 99–109. [Google Scholar] [CrossRef]
- Komori, T. Regulation of osteoblast differentiation by Runx2. In Osteoimmunology; Springer: Boston, MA, USA, 2009; pp. 43–49. [Google Scholar]
- Li, J. JAK-STAT and bone metabolism. Jak-Stat 2013, 2, e23930. [Google Scholar] [CrossRef]
- Proshchalykin, M.Y.; Dathe, H.H. Peroxisome proliferator-activated receptor activators modulate the osteoblastic maturation of MC3T3-E1 pre-osteoblasts. Zootaxa 2012, 471, 119–124. [Google Scholar]
- Baum, R.; Gravallese, E.M. Impact of inflammation on the osteoblast in rheumatic diseases. Curr. Osteoporos. Rep. 2014, 12, 9–16. [Google Scholar] [CrossRef] [Green Version]
- Kao, R.; Lu, W.; Louie, A.; Nissenson, R. Cyclic AMP signaling in bone marrow stromal cells has reciprocal effects on the ability of mesenchymal stem cells to differentiate into mature osteoblasts versus mature adipocytes. Endocrine 2012, 42, 622–636. [Google Scholar] [CrossRef] [Green Version]
- Langdahl, B.; Ferrari, S.; Dempster, D.W. Bone modeling and remodeling: Potential as therapeutic targets for the treatment of osteoporosis. Ther. Adv. Musculoskelet. Dis. 2016, 8, 225–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orimo, H. The mechanism of mineralization and the role of alkaline phosphatase in health and disease. J. Nippon Med. Sch. 2010, 77, 4–12. [Google Scholar] [CrossRef]
- Chang, J.; Liu, F.; Lee, M.; Wu, B.; Ting, K.; Zara, J.N.; Soo, C.; Al Hezaimi, K.; Zou, W.; Chen, X.; et al. NF-κB inhibits osteogenic differentiation of mesenchymal stem cells by promoting β-catenin degradation. Proc. Natl. Acad. Sci. USA 2013, 110, 9469–9474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domazetovic, V.; Marcucci, G.; Iantomasi, T.; Brandi, M.L.; Vincenzini, M.T.; Vincenzini, M.T. Oxidative stress in bone remodeling: Role of antioxidants. Clin. Cases Miner. Bone Metab. 2017, 14, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Kaneshiro, S.; Ebina, K.; Shi, K.; Higuchi, C.; Hirao, M.; Okamoto, M.; Koizumi, K.; Morimoto, T.; Yoshikawa, H.; Hashimoto, J. IL-6 negatively regulates osteoblast differentiation through the SHP2/MEK2 and SHP2/Akt2 pathways in vitro. J. Bone Miner. Metab. 2014, 32, 378–392. [Google Scholar] [CrossRef]
- Ambrogini, E.; Que, X.; Wang, S.; Yamaguchi, F.; Weinstein, R.S.; Tsimikas, S.; Manolagas, S.C.; Witztum, J.L.; Jilka, R.L. Oxidation-specific epitopes restrain bone formation. Nat. Commun. 2018, 9, 2193. [Google Scholar] [CrossRef] [Green Version]
- Baker, R.G.; Hayden, M.S.; Ghosh, S. NF-κB, inflammation, and metabolic disease. Cell Metab. 2011, 13, 11–22. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.; Wang, Z.; Tang, E.; Fan, Z.; Mccauley, L.; Franceschi, R.; Guan, K.; Krebsbach, P.H.; Wang, C.Y. Inhibition of osteoblast functions by IKK/NF-κB in osteoporosis. Nature 2009, 15, 682–689. [Google Scholar]
- Boyce, B.F.; Yao, Z.; Xing, L. Functions of NF-κB in Bone. Ann. N. Y. Acad. Sci. 2010, 1192, 367. [Google Scholar] [CrossRef]
- Li, Y.; Li, A.; Strait, K.; Zhang, H.; Nanes, M.S.; Weitzmann, M.N. Endogenous TNFα lowers maximum peak bone mass and inhibits osteoblastic Smad activation through NF-κB. J. Bone Miner. Res. 2007, 22, 646–655. [Google Scholar] [CrossRef]
- Duttenhoefer, F.; Biswas, S.K.; Igwe, J.C.; Sauerbier, S.; Bierhaus, D.D.S.A. Sp1-dependent regulation of PPAR α in bone metabolism. Int. J. Oral Maxillofac. Implant. 2014, 29, e107–e116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.R.; Leung, W.N.; Li, G.; Kong, S.K.; Lu, X.; Wong, Y.M.; Chan, C.W. Osthole enhances osteogenesis in Osteoblasts by elevating transcription factor Osterix via cAMP/CREB signaling in vitro and in vivo. Nutrients 2017, 9, 588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishihara, S.; Ikeda, M.; Ozawa, H.; Akiyama, M.; Yamaguchi, S.; Nakahama, K.I. Role of cAMP in phenotypic changes of Osteoblasts. Biochem. Biophys. Res. Commun. 2018, 495, 941–946. [Google Scholar] [CrossRef]
- Itoh, S.; Udagawa, N.; Takahashi, N.; Yoshitake, F.; Narita, H.; Ebisu, S.; Ishihara, K. A critical role for interleukin-6 family-mediated Stat3 activation in Osteoblast differentiation and bone formation. Bone 2006, 39, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Gregory, C.A.; Gunn, W.G.; Peister, A.; Prockop, D.J. An Alizarin red-based assay of mineralization by adherent cells in culture: Comparison with cetylpyridinium chloride extraction. Anal. Biochem. 2004, 329, 77–84. [Google Scholar] [CrossRef]
| Gene Name | Gene Symbol | Gene Bank Accession Number | Primer Sequence |
|---|---|---|---|
| Alkaline phosphatase | ALPL | NM_000478.5 | F : CCAAGTACTGGCGAGACCAA |
| R : TGTGGAGACACCCATCCCAT | |||
| Bone gamma-carboxyglutamate protein | BGLAP | NM_199173.5 | F : TCACACTCCTCGCCCTATTG |
| R : CTCTTCACTACCTCGCTGCC | |||
| Runt-related transcription factor 2 | RUNX2 | NM_0010246303 | F : CCACCACTCACTACCACACC |
| R : AAGGGTCCACTCTGGCTTTG | |||
| Collagen type I alpha 2 chain | COL1A2 | NM_000089.3 | F : GGATGAGGAGACTGGCAACC |
| R : TGCCCTCAGCAACAAGTTCA | |||
| Interleukin 6 | IL-6 | NM_000600.4 | F : TGAGGAGACTTGCCTGGTGA |
| R : GCATTTGTGGTTGGGTCAGG | |||
| Tumor necrosis factor | TNF-α | NM_000594.3 | F: GGTCCTCTTCAAGGGCCAAG |
| R: TCCTCCTCACAGGGCAATGA | |||
| RELA proto-oncogene, NF-kB (p65) subunit | P65 | NM_021975.3 | F : AGGCTATCAGTCAGCGCATC |
| R : TCCCCACGCTGCTCTTCTAT | |||
| cAMP responsive element binding protein 1 | CREB1 | NM_004379.4 | F : CGAGAACCAGCAGAGTGGAG |
| R : CGGTGGGAGCAGATGATGTT | |||
| Signal transducer and activator of transcription 3 | STAT3 | NM_139276.2 | F : ACCATTGACCTGCCGATGTC |
| R : GTGAGGGACTCAAACTGCCC | |||
| Peroxisome proliferator activated receptor alpha | PPAR-α | NM_001001928.2 | F : AAGGCTGCAAGGGCTTCTTT |
| R : ACATCCCGACAGAAAGGCAC | |||
| Glyceraldehyde 3-phosphate dehydrogenase | GAPDH | NM_002046.6 | F : GGAGCGAGATCCCTCCAAAAT |
| R : GGCTGTTGTCATACTTCTCATGG | |||
| Hyperparathyroidism 1 | HRPT1 | NM_000194_2 | F : CCTGGCGTCGTGATTAGTGAT |
| R : AGACGTTCAGTCCTGTCCATAA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harun, N.H.; Froemming, G.R.A.; Mohd Ismail, A.; Nawawi, H.; Mokhtar, S.S.; Abd Muid, S. Osteoblast Demineralization Induced by Oxidized High-Density Lipoprotein via the Inflammatory Pathway Is Suppressed by Adiponectin. Int. J. Mol. Sci. 2022, 23, 14616. https://doi.org/10.3390/ijms232314616
Harun NH, Froemming GRA, Mohd Ismail A, Nawawi H, Mokhtar SS, Abd Muid S. Osteoblast Demineralization Induced by Oxidized High-Density Lipoprotein via the Inflammatory Pathway Is Suppressed by Adiponectin. International Journal of Molecular Sciences. 2022; 23(23):14616. https://doi.org/10.3390/ijms232314616
Chicago/Turabian StyleHarun, Noor Hanisa, Gabriele Ruth Anisah Froemming, Aletza Mohd Ismail, Hapizah Nawawi, Siti Shuhada Mokhtar, and Suhaila Abd Muid. 2022. "Osteoblast Demineralization Induced by Oxidized High-Density Lipoprotein via the Inflammatory Pathway Is Suppressed by Adiponectin" International Journal of Molecular Sciences 23, no. 23: 14616. https://doi.org/10.3390/ijms232314616







