Development of Alkaline Phosphatase-Fused Mouse Prion Protein and Its Application in Toxic Aβ Oligomer Detection
Abstract
1. Introduction
2. Results
2.1. Preparation and Enzymatic Activity Analysis of PrP–ALP
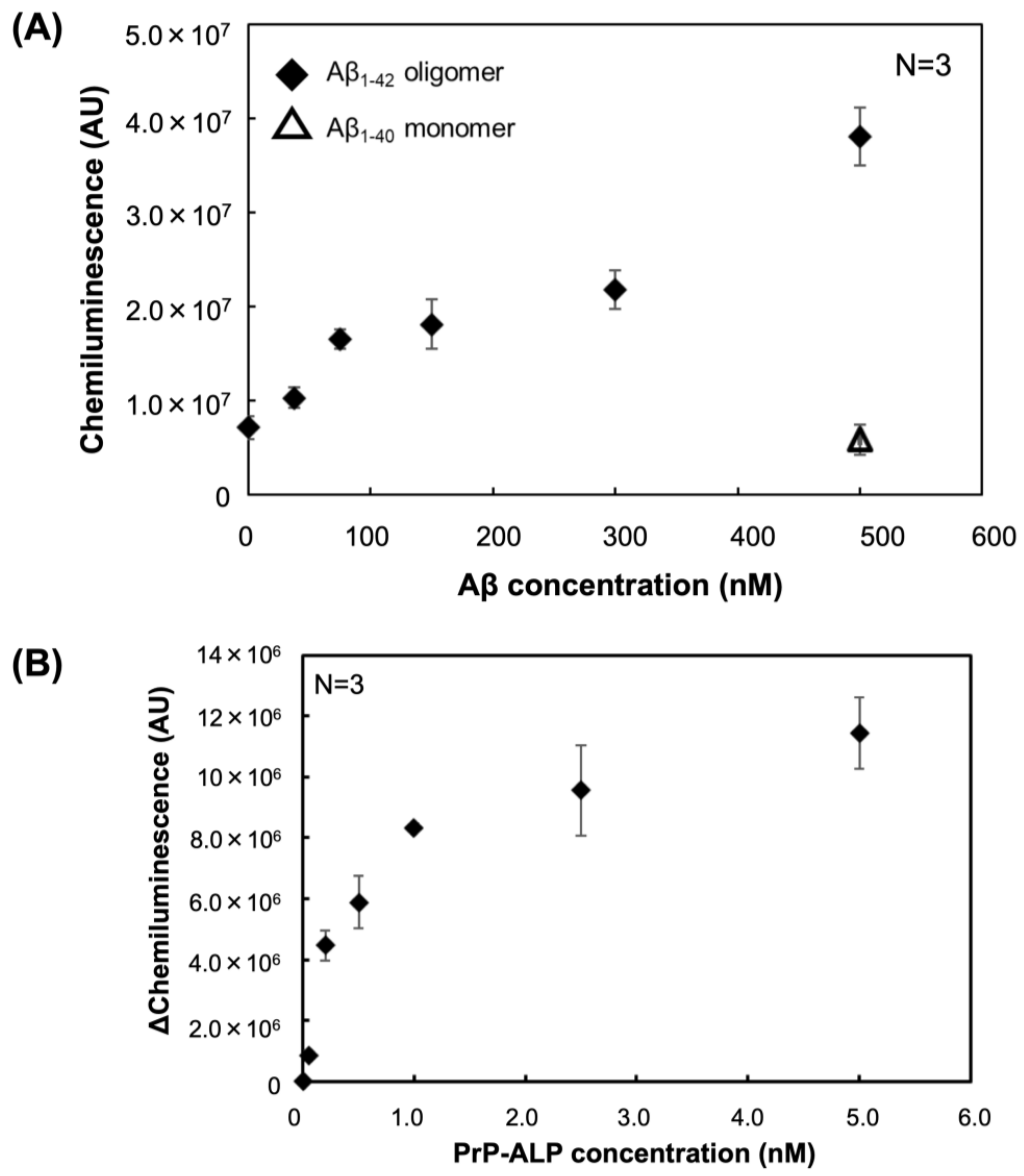
2.2. Evaluation of the Binding of PrP-ALP to Differently Aggregated Aβ
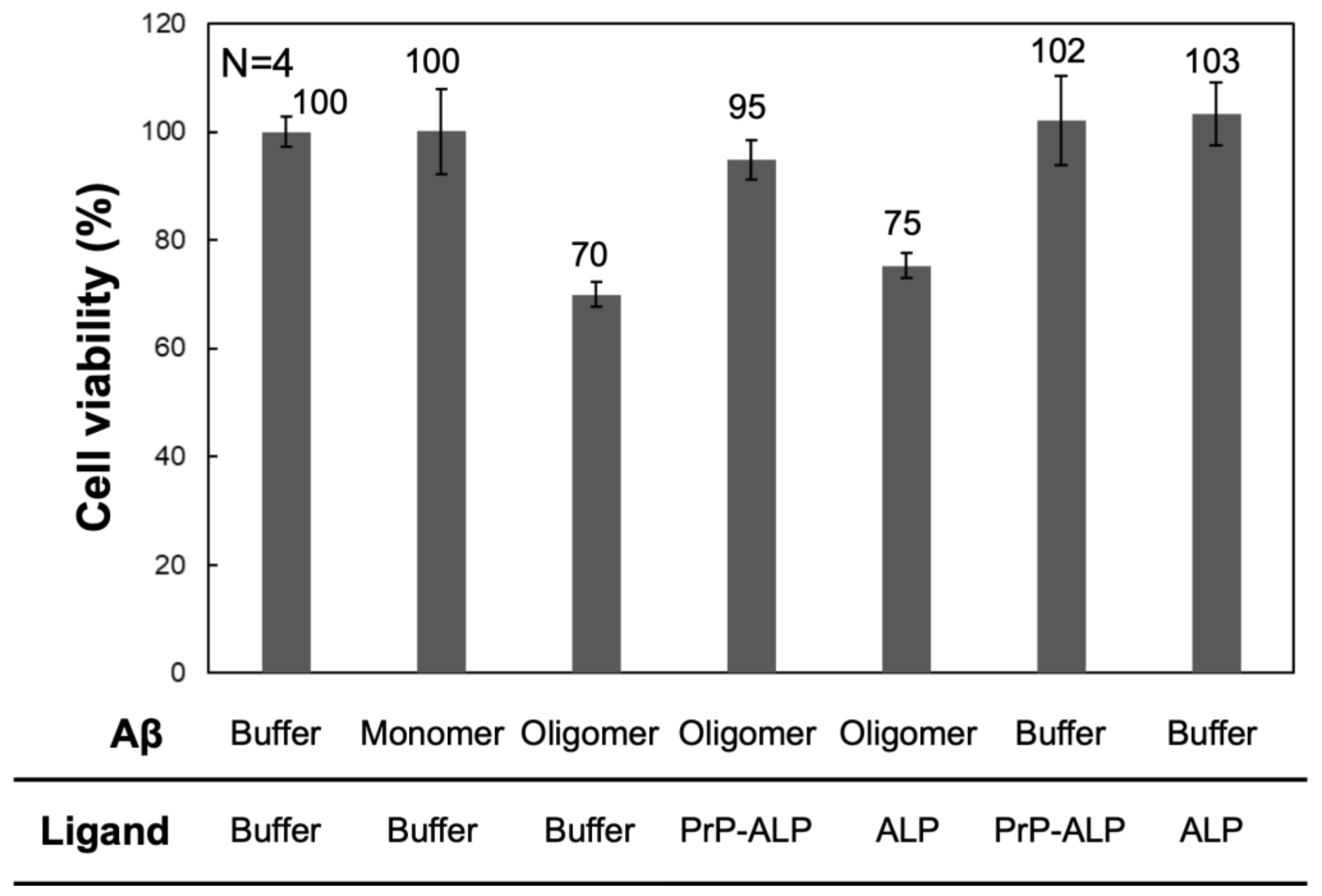
2.3. Aβ Oligomer Toxicity Neutralization with PrP-ALP
2.4. Aβ Oligomer Detection Based on Sandwich Plate Assay Using PrP–ALP
3. Discussion
4. Materials and Methods
4.1. Cloning, Expression, and Purification of PrP–ALP
4.2. Enzymatic Activity Assay
4.3. Aβ Preparation
4.4. Western Blotting Analysis for Prepared Aβ
4.5. Dot Blot Assay to Evaluate PrP–ALP Binding Aβ Species
4.6. Sandwich Plate Assay Using PrP–ALP to Detect Aβ Oligomers
4.7. Cell Culture and MTS Cell Viability Assay
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Hardy, J.; Selkoe, D.J. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science 2002, 297, 353–356. [Google Scholar] [CrossRef]
- Lambert, M.P.; Barlow, A.K.; Chromy, B.A.; Edwards, C.; Freed, R.; Liosatos, M.; Morgan, T.E.; Rozovsky, I.; Trommer, B.; Viola, K.L.; et al. Diffusible, nonfibrillar ligands derived from Abeta1–42 are potent central nervous system neurotoxins. Proc. Natl. Acad. Sci. USA 1998, 95, 6448–6453. [Google Scholar] [CrossRef]
- Walsh, D.M.; Tseng, B.P.; Rydel, R.E.; Podlisny, M.B.; Selkoe, D.J. The oligomerization of amyloid beta-protein begins intracellularly in cells derived from human brain. Biochemistry 2000, 39, 10831–10839. [Google Scholar] [CrossRef]
- Gong, Y.; Chang, L.; Viola, K.L.; Lacor, P.N.; Lambert, M.P.; Finch, C.E.; Krafft, G.A.; Klein, W.L. Alzheimer’s disease-affected brain: Presence of oligomeric A beta ligands (ADDLs) suggests a molecular basis for reversible memory loss. Proc. Natl. Acad. Sci. USA 2003, 100, 10417–10422. [Google Scholar] [CrossRef] [PubMed]
- Shankar, G.M.; Li, S.; Mehta, T.H.; Garcia-Munoz, A.; Shepardson, N.E.; Smith, I.; Brett, F.M.; Farrell, M.A.; Rowan, M.J.; Lemere, C.A.; et al. Amyloid-beta protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat. Med. 2008, 14, 837–842. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, A.; Matsumura, S.; Dezawa, M.; Tada, M.; Yanazawa, M.; Ito, A.; Akioka, M.; Kikuchi, S.; Sato, M.; Ideno, S.; et al. Isolation and characterization of patient-derived, toxic, high mass amyloid beta-protein (Abeta) assembly from Alzheimer disease brains. J. Biol. Chem. 2009, 284, 32895–32905. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.; Wang, Z.; Liu, W.; O’Malley, T.T.; Jin, M.; Willem, M.; Haass, C.; Frosch, M.P.; Walsh, D.M. Diffusible, highly bioactive oligomers represent a critical minority of soluble Abeta in Alzheimer’s disease brain. Acta Neuropathol. 2018, 136, 19–40. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; O’Malley, T.T.; Kanmert, D.; Jerecic, J.; Zieske, L.R.; Zetterberg, H.; Hyman, B.T.; Walsh, D.M.; Selkoe, D.J. A highly sensitive novel immunoassay specifically detects low levels of soluble Abeta oligomers in human cerebrospinal fluid. Alzheimers Res. Ther. 2015, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Sevigny, J.; Chiao, P.; Bussiere, T.; Weinreb, P.H.; Williams, L.; Maier, M.; Dunstan, R.; Salloway, S.; Chen, T.; Ling, Y.; et al. The antibody aducanumab reduces Abeta plaques in Alzheimer’s disease. Nature 2016, 537, 50–56. [Google Scholar] [CrossRef]
- Arndt, J.W.; Qian, F.; Smith, B.A.; Quan, C.; Kilambi, K.P.; Bush, M.W.; Walz, T.; Pepinsky, R.B.; Bussiere, T.; Hamann, S.; et al. Structural and kinetic basis for the selectivity of aducanumab for aggregated forms of amyloid-beta. Sci. Rep. 2018, 8, 6412. [Google Scholar] [CrossRef]
- Jin, M.; O’Nuallain, B.; Hong, W.; Boyd, J.; Lagomarsino, V.N.; O’Malley, T.T.; Liu, W.; Vanderburg, C.R.; Frosch, M.P.; Young-Pearse, T.; et al. An in vitro paradigm to assess potential anti-Abeta antibodies for Alzheimer’s disease. Nat. Commun. 2018, 9, 2676. [Google Scholar] [CrossRef] [PubMed]
- Kayed, R.; Head, E.; Thompson, J.L.; McIntire, T.M.; Milton, S.C.; Cotman, C.W.; Glabe, C.G. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 2003, 300, 486–489. [Google Scholar] [CrossRef] [PubMed]
- Lambert, M.P.; Velasco, P.T.; Chang, L.; Viola, K.L.; Fernandez, S.; Lacor, P.N.; Khuon, D.; Gong, Y.; Bigio, E.H.; Shaw, P.; et al. Monoclonal antibodies that target pathological assemblies of Abeta. J. Neurochem. 2007, 100, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Sebollela, A.; Cline, E.N.; Popova, I.; Luo, K.; Sun, X.; Ahn, J.; Barcelos, M.A.; Bezerra, V.N.; Lyra, E.S.N.M.; Patel, J.; et al. A human scFv antibody that targets and neutralizes high molecular weight pathogenic amyloid-beta oligomers. J. Neurochem. 2017, 142, 934–947. [Google Scholar] [CrossRef] [PubMed]
- Lauren, J.; Gimbel, D.A.; Nygaard, H.B.; Gilbert, J.W.; Strittmatter, S.M. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-beta oligomers. Nature 2009, 457, 1128–1132. [Google Scholar] [CrossRef]
- Chen, S.; Yadav, S.P.; Surewicz, W.K. Interaction between human prion protein and amyloid-beta (Abeta) oligomers: Role OF N-terminal residues. J. Biol. Chem. 2010, 285, 26377–26383. [Google Scholar] [CrossRef]
- Um, J.W.; Nygaard, H.B.; Heiss, J.K.; Kostylev, M.A.; Stagi, M.; Vortmeyer, A.; Wisniewski, T.; Gunther, E.C.; Strittmatter, S.M. Alzheimer amyloid-beta oligomer bound to postsynaptic prion protein activates Fyn to impair neurons. Nat. Neurosci. 2012, 15, 1227–1235. [Google Scholar] [CrossRef]
- Um, J.W.; Kaufman, A.C.; Kostylev, M.; Heiss, J.K.; Stagi, M.; Takahashi, H.; Kerrisk, M.E.; Vortmeyer, A.; Wisniewski, T.; Koleske, A.J.; et al. Metabotropic glutamate receptor 5 is a coreceptor for Alzheimer abeta oligomer bound to cellular prion protein. Neuron 2013, 79, 887–902. [Google Scholar] [CrossRef]
- Smith, L.M.; Kostylev, M.A.; Lee, S.; Strittmatter, S.M. Systematic and standardized comparison of reported amyloid-beta receptors for sufficiency, affinity, and Alzheimer’s disease relevance. J. Biol. Chem. 2019, 294, 6042–6053. [Google Scholar] [CrossRef]
- Kostylev, M.A.; Kaufman, A.C.; Nygaard, H.B.; Patel, P.; Haas, L.T.; Gunther, E.C.; Vortmeyer, A.; Strittmatter, S.M. Prion-Protein-interacting Amyloid-beta Oligomers of High Molecular Weight Are Tightly Correlated with Memory Impairment in Multiple Alzheimer Mouse Models. J. Biol. Chem. 2015, 290, 17415–17438. [Google Scholar] [CrossRef]
- Freir, D.B.; Nicoll, A.J.; Klyubin, I.; Panico, S.; Mc Donald, J.M.; Risse, E.; Asante, E.A.; Farrow, M.A.; Sessions, R.B.; Saibil, H.R.; et al. Interaction between prion protein and toxic amyloid beta assemblies can be therapeutically targeted at multiple sites. Nat. Commun. 2011, 2, 336. [Google Scholar] [CrossRef]
- Nicoll, A.J.; Panico, S.; Freir, D.B.; Wright, D.; Terry, C.; Risse, E.; Herron, C.E.; O’Malley, T.; Wadsworth, J.D.; Farrow, M.A.; et al. Amyloid-beta nanotubes are associated with prion protein-dependent synaptotoxicity. Nat. Commun. 2013, 4, 2416. [Google Scholar] [CrossRef]
- Fluharty, B.R.; Biasini, E.; Stravalaci, M.; Sclip, A.; Diomede, L.; Balducci, C.; La Vitola, P.; Messa, M.; Colombo, L.; Forloni, G.; et al. An N-terminal fragment of the prion protein binds to amyloid-beta oligomers and inhibits their neurotoxicity in vivo. J. Biol. Chem. 2013, 288, 7857–7866. [Google Scholar] [CrossRef]
- Zhang, X.-E.; Zhou, Y.-H.; Zhang, Z.-P.; Xu, H.-F.; Shao, W.-H.; Cass, A.E.G. Engineering, E. coli Alkaline Phosphatase Yields Changes of Catalytic Activity, Thermal Stability and Phosphate Inhibition. Biocatal. Biotransform. 2002, 20, 381–389. [Google Scholar] [CrossRef]
- Ogasawara, D.; Hasegawa, H.; Kaneko, K.; Sode, K.; Ikebukuro, K. Screening of DNA aptamer against mouse prion protein by competitive selection. Prion 2007, 1, 248–254. [Google Scholar] [CrossRef]
- Smith, L.M.; Strittmatter, S.M. Binding Sites for Amyloid-beta Oligomers and Synaptic Toxicity. Cold Spring Harb. Perspect. Med. 2017, 7, a024075. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, T.; Yanazawa, M.; Sasahara, T.; Kitamura, Y.; Hiroaki, H.; Fukazawa, Y.; Kii, I.; Nishiyama, T.; Kakita, A.; Takeda, H.; et al. Na, K-ATPase alpha3 is a death target of Alzheimer patient amyloid-beta assembly. Proc. Natl. Acad. Sci. USA 2015, 112, E4465–E4474. [Google Scholar] [CrossRef]
- Tsukakoshi, K.; Ikuta, Y.; Abe, K.; Yoshida, W.; Iida, K.; Ma, Y.; Nagasawa, K.; Sode, K.; Ikebukuro, K. Structural regulation by a G-quadruplex ligand increases binding abilities of G-quadruplex-forming aptamers. Chem. Commun. 2016, 52, 12646–12649. [Google Scholar] [CrossRef] [PubMed]
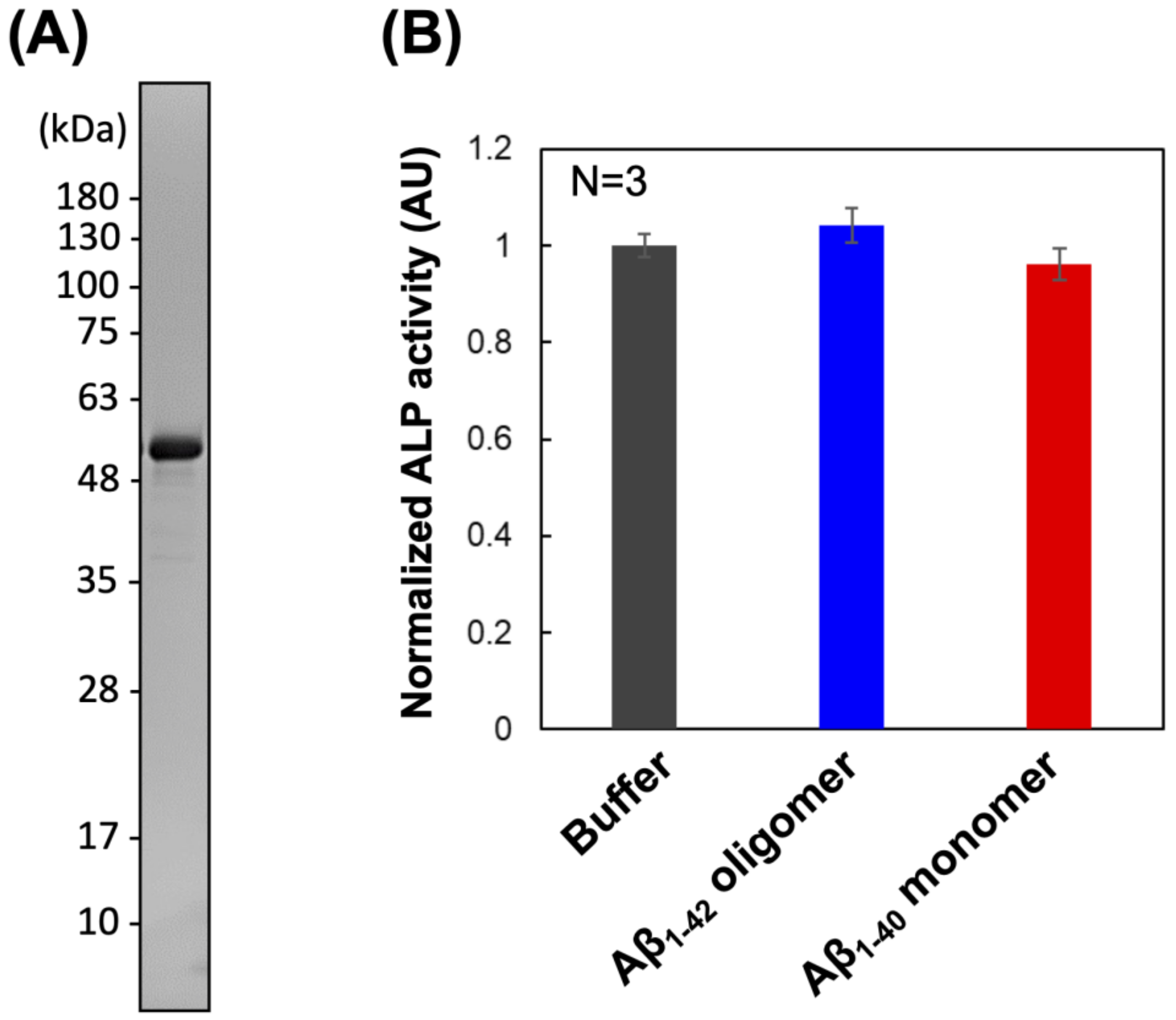


| Km (µM) | Vmax (U/nmol) | |
|---|---|---|
| PrP–ALP | 75 | 3.9 |
| ALP (D101S) | 76 | 6.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsukakoshi, K.; Kubo, R.; Ikebukuro, K. Development of Alkaline Phosphatase-Fused Mouse Prion Protein and Its Application in Toxic Aβ Oligomer Detection. Int. J. Mol. Sci. 2022, 23, 14588. https://doi.org/10.3390/ijms232314588
Tsukakoshi K, Kubo R, Ikebukuro K. Development of Alkaline Phosphatase-Fused Mouse Prion Protein and Its Application in Toxic Aβ Oligomer Detection. International Journal of Molecular Sciences. 2022; 23(23):14588. https://doi.org/10.3390/ijms232314588
Chicago/Turabian StyleTsukakoshi, Kaori, Rikako Kubo, and Kazunori Ikebukuro. 2022. "Development of Alkaline Phosphatase-Fused Mouse Prion Protein and Its Application in Toxic Aβ Oligomer Detection" International Journal of Molecular Sciences 23, no. 23: 14588. https://doi.org/10.3390/ijms232314588
APA StyleTsukakoshi, K., Kubo, R., & Ikebukuro, K. (2022). Development of Alkaline Phosphatase-Fused Mouse Prion Protein and Its Application in Toxic Aβ Oligomer Detection. International Journal of Molecular Sciences, 23(23), 14588. https://doi.org/10.3390/ijms232314588








