CRISPR/Cas9-Based Knock-Out of the PMR4 Gene Reduces Susceptibility to Late Blight in Two Tomato Cultivars
Abstract
1. Introduction
2. Results
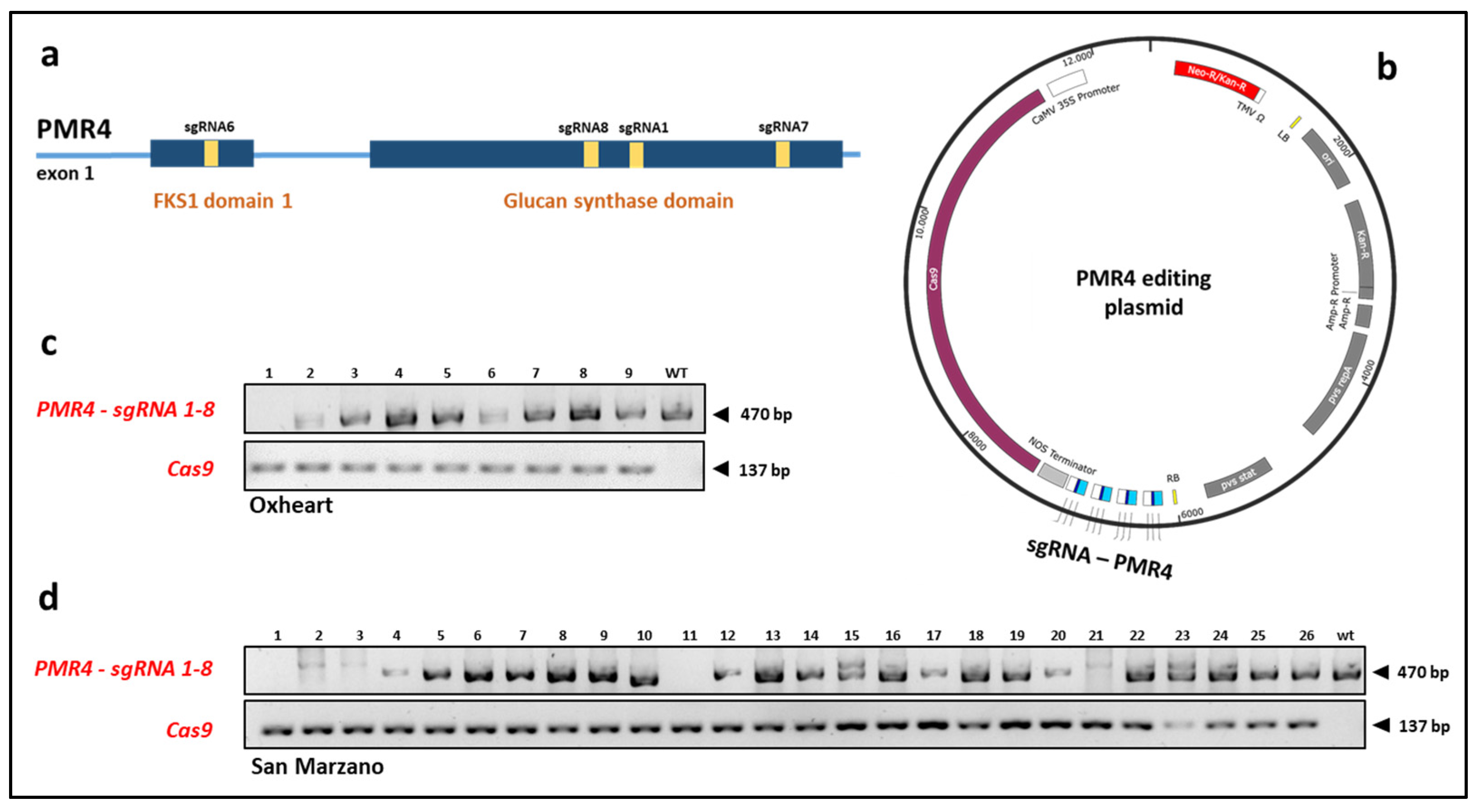
2.1. CRISPR/Cas9-Based Mutagenesis of SlPMR4
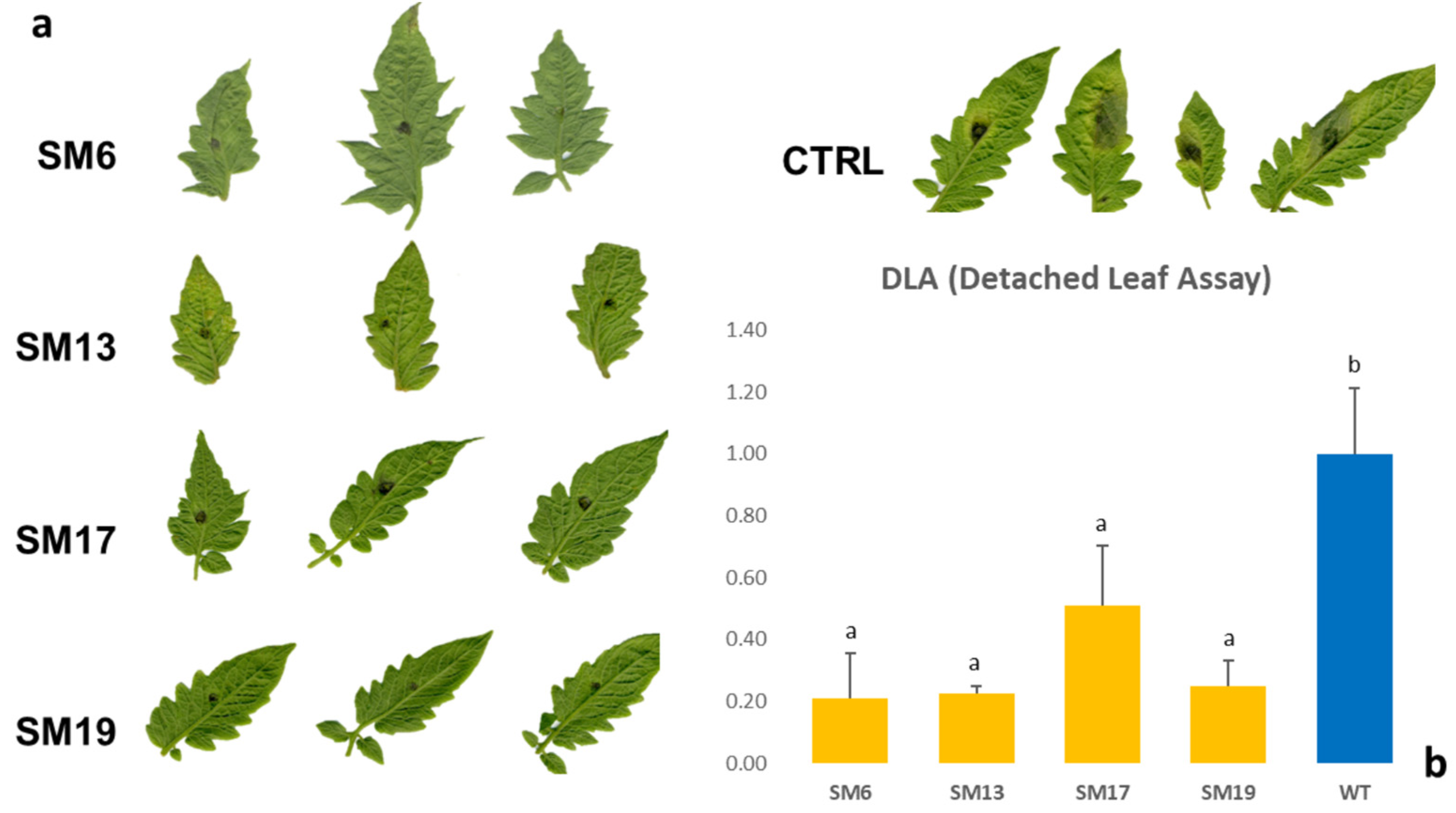
2.2. Reduced Susceptibility to Late Blight (LB)
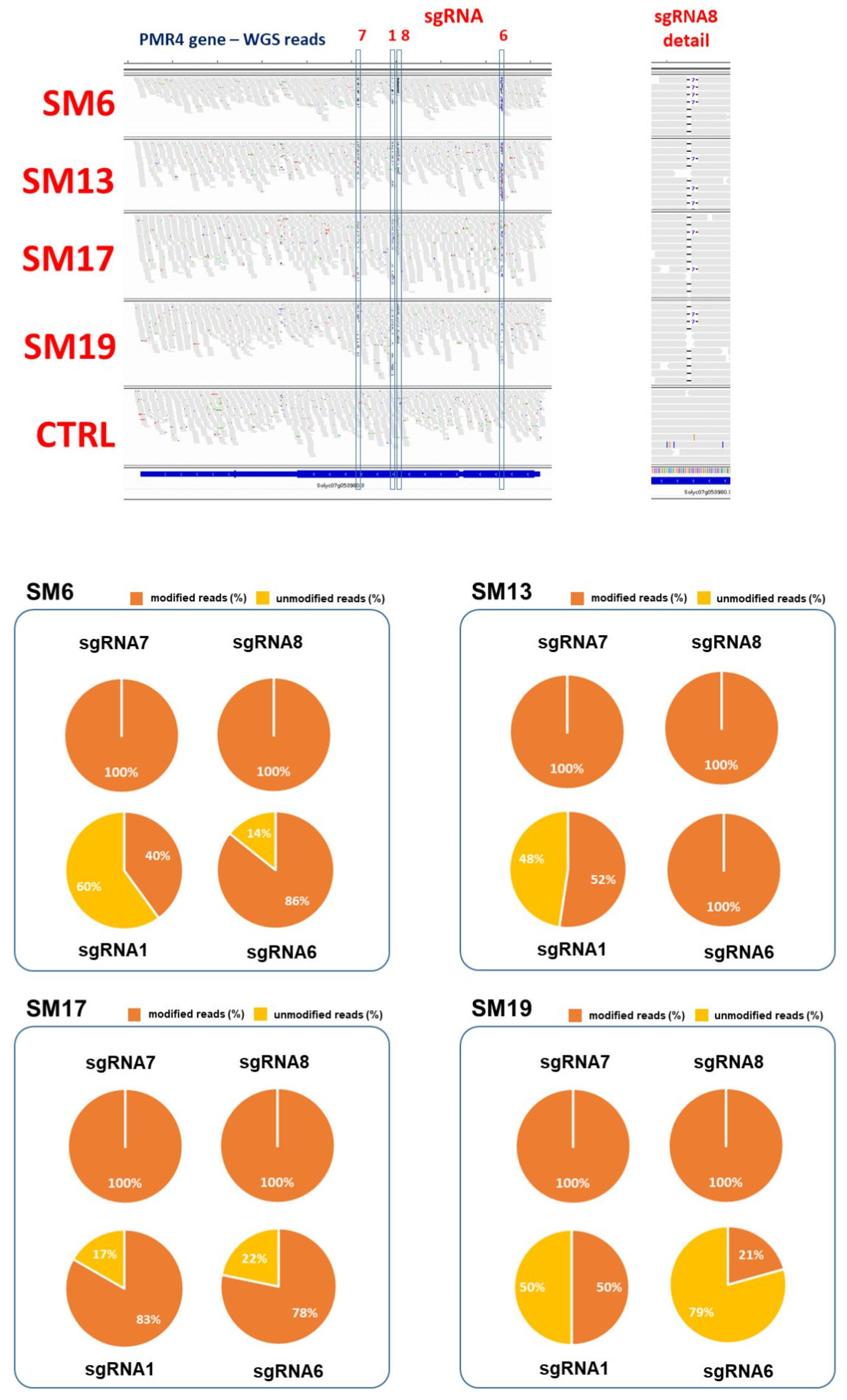
2.3. Genomics of Selected Pmr4 Mutants
2.4. Off-Target Events and SNP Analysis
3. Discussion
3.1. Reduced Susceptibility to LB in Tomato Cultivars Knocked-Out in the SlPMR4 Gene
3.2. Genome Editing Outcomes in Tomato Cultivars
3.3. Off-Target Absence and Emerging SNPs in Pmr4 Mutants
4. Materials and Methods
4.1. Plant Material and Transformation Vector
4.2. Plant Transformation, Regeneration, and Acclimation to Soil
4.3. Identification of Transformed Plants and Detection of the SlPMR4 Editing
4.4. Detached Leaf Assay with Phytophthora Infestans
4.5. Whole-Genome Sequencing
4.6. On- and Off-Target Analyses and SNP Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chaudhary, J.; Alisha, A.; Bhatt, V.; Chandanshive, S.; Kumar, N.; Mir, Z.; Kumar, A.; Yadav, S.K.; Shivaraj, S.M.; Sonah, H.; et al. Mutation Breeding in Tomato: Advances, Applicability and Challenges. Plants 2019, 8, 128. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Cheng, X.; Li, R.; Yao, J.; Li, Z.; Cheng, Y. Advances in application of genome editing in tomato and recent development of genome editing technology. Theor. Appl. Genet. 2021, 134, 2727–2747. [Google Scholar] [CrossRef] [PubMed]
- Ronga, D.; Francia, E.; Rizza, F.; Badeck, F.-W.; Caradonia, F.; Montevecchi, G.; Pecchioni, N. Changes in yield components, morphological, physiological and fruit quality traits in processing tomato cultivated in Italy since the 1930’s. Sci. Hortic. 2019, 257, 108726. [Google Scholar] [CrossRef]
- Schouten, H.J.; Tikunov, Y.; Verkerke, W.; Finkers, R.; Bovy, A.; Bai, Y.; Visser, R.G.F. Breeding Has Increased the Diversity of Cultivated Tomato in The Netherlands. Front. Plant Sci. 2019, 10, 1606. [Google Scholar] [CrossRef] [PubMed]
- Bergougnoux, V. The history of tomato: From domestication to biopharming. Biotechnol. Adv. 2014, 32, 170–189. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Ehsani, R.; Shi, Y.; de Castro, A.I.; Wang, S. Detection of multi-tomato leaf diseases (late blight, target and bacterial spots) in different stages by using a spectral-based sensor. Sci. Rep. 2018, 8, 2793. [Google Scholar] [CrossRef]
- Fry, W. Phytophthora infestans: The plant (and R gene) destroyer. Mol. Plant Pathol. 2008, 9, 385–402. [Google Scholar] [CrossRef]
- Nowicki, M.; Foolad, M.R.; Nowakowska, M.; Kozik, E.U. Potato and Tomato Late Blight Caused by Phytophthora infestans: An Overview of Pathology and Resistance Breeding. Plant Dis. 2012, 96, 4–17. [Google Scholar] [CrossRef]
- Laurindo, B.S.; Laurindo, R.D.F.; Fontes, P.P.; Vital, C.E.; Delazari, F.T.; Baracat-Pereira, M.C.; da Silva, D.J.H. Comparative analysis of constitutive proteome between resistant and susceptible tomato genotypes regarding to late blight. Funct. Integr. Genom. 2018, 18, 11–21. [Google Scholar] [CrossRef]
- Brouwer, D.J.; St. Clair, D.A. Fine mapping of three quantitative trait loci for late blight resistance in tomato using near isogenic lines (NILs) and sub-NILs. Theor. Appl. Genet. 2004, 108, 628–638. [Google Scholar] [CrossRef]
- Michalska, A.M.; Sobkowiak, S.; Flis, B.; Zimnoch-Guzowska, E. Virulence and aggressiveness of Phytophthora infestans isolates collected in Poland from potato and tomato plants identified no strong specificity. Eur. J. Plant Pathol. 2016, 144, 325–336. [Google Scholar] [CrossRef][Green Version]
- Huibers, R.P.; Loonen, A.E.H.M.; Gao, D.; Van den Ackerveken, G.; Visser, R.G.F.; Bai, Y. Powdery Mildew Resistance in Tomato by Impairment of SlPMR4 and SlDMR1. PLoS ONE 2013, 8, e67467. [Google Scholar] [CrossRef] [PubMed]
- Wastie, R. Breeding for resistance. Adv. Plant Pathol. 1991, 7, 193–224. [Google Scholar]
- van Schie, C.C.N.; Takken, F.L.W. Susceptibility Genes 101: How to Be a Good Host. Annu. Rev. Phytopathol. 2014, 52, 551–581. [Google Scholar] [CrossRef]
- Zaidi, S.S.-e.-A.; Mukhtar, M.S.; Mansoor, S. Genome Editing: Targeting Susceptibility Genes for Plant Disease Resistance. Trends Biotechnol. 2018, 36, 898–906. [Google Scholar] [CrossRef]
- Eckardt, N.A. Plant Disease Susceptibility Genes? Plant Cell 2002, 14, 1983–1986. [Google Scholar] [CrossRef]
- Pavan, S.; Jacobsen, E.; Visser, R.G.F.; Bai, Y. Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol. Breed. 2009, 25, 1. [Google Scholar] [CrossRef]
- Büschges, R.; Hollricher, K.; Panstruga, R.; Simons, G.; Wolter, M.; Frijters, A.; van Daelen, R.; van der Lee, T.; Diergaarde, P.; Groenendijk, J.; et al. The Barley Mlo Gene: A Novel Control Element of Plant Pathogen Resistance. Cell 1997, 88, 695–705. [Google Scholar] [CrossRef]
- Consonni, C.; Humphry, M.E.; Hartmann, H.A.; Livaja, M.; Durner, J.; Westphal, L.; Vogel, J.; Lipka, V.; Kemmerling, B.; Schulze-Lefert, P.; et al. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 2006, 38, 716–720. [Google Scholar] [CrossRef]
- Acevedo-Garcia, J.; Gruner, K.; Reinstädler, A.; Kemen, A.; Kemen, E.; Cao, L.; Takken, F.L.W.; Reitz, M.U.; Schäfer, P.; O’Connell, R.J.; et al. The powdery mildew-resistant Arabidopsis mlo2 mlo6 mlo12 triple mutant displays altered infection phenotypes with diverse types of phytopathogens. Sci. Rep. 2017, 7, 9319. [Google Scholar] [CrossRef]
- Vogel, J.; Somerville, S. Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc. Natl. Acad. Sci. USA 2000, 97, 1897–1902. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Wolters, A.-M.A.; Vossen, J.H.; Rouwet, M.E.; Loonen, A.E.H.M.; Jacobsen, E.; Visser, R.G.F.; Bai, Y. Silencing of six susceptibility genes results in potato late blight resistance. Transgenic Res. 2016, 25, 731–742. [Google Scholar] [CrossRef] [PubMed]
- Brooks, C.; Nekrasov, V.; Lippman, Z.B.; Van Eck, J. Efficient Gene Editing in Tomato in the First Generation Using the Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-Associated9 System. Plant Physiol. 2014, 166, 1292–1297. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, H.; Zhu, H. CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Hortic. Res. 2019, 6, 77. [Google Scholar] [CrossRef]
- Santillán Martínez, M.I.; Bracuto, V.; Koseoglou, E.; Appiano, M.; Jacobsen, E.; Visser, R.G.F.; Wolters, A.-M.A.; Bai, Y. CRISPR/Cas9-targeted mutagenesis of the tomato susceptibility gene PMR4 for resistance against powdery mildew. BMC Plant Biol. 2020, 20, 284. [Google Scholar] [CrossRef]
- Thomazella, D.P.T.; Seong, K.; Mackelprang, R.; Dahlbeck, D.; Geng, Y.; Gill, U.S.; Qi, T.; Pham, J.; Giuseppe, P.; Lee, C.Y.; et al. Loss of function of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. USA 2021, 118, e2026152118. [Google Scholar] [CrossRef]
- Foolad, M.R.; Sullenberger, M.T.; Ashrafi, H. Detached-Leaflet Evaluation of Tomato Germplasm for Late Blight Resistance and Its Correspondence to Field and Greenhouse Screenings. Plant Dis. 2015, 99, 718–722. [Google Scholar] [CrossRef]
- Jacobs, A.K.; Lipka, V.; Burton, R.A.; Panstruga, R.; Strizhov, N.; Schulze-Lefert, P.; Fincher, G.B. An Arabidopsis Callose Synthase, GSL5, Is Required for Wound and Papillary Callose Formation. Plant Cell 2003, 15, 2503–2513. [Google Scholar] [CrossRef]
- Nishimura, M.T.; Stein, M.; Hou, B.-H.; Vogel, J.P.; Edwards, H.; Somerville, S.C. Loss of a Callose Synthase Results in Salicylic Acid-Dependent Disease Resistance. Science 2003, 301, 969–972. [Google Scholar] [CrossRef]
- Flors, V.; Ton, J.; Van Doorn, R.; Jakab, G.; García-Agustín, P.; Mauch-Mani, B. Interplay between JA, SA and ABA signalling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola. Plant J. 2008, 54, 81–92. [Google Scholar] [CrossRef]
- Hashimoto, R.; Ueta, R.; Abe, C.; Osakabe, Y.; Osakabe, K. Efficient Multiplex Genome Editing Induces Precise, and Self-Ligated Type Mutations in Tomato Plants. Front. Plant Sci. 2018, 9, 916. [Google Scholar] [CrossRef]
- Jaganathan, D.; Ramasamy, K.; Sellamuthu, G.; Jayabalan, S.; Venkataraman, G. CRISPR for Crop Improvement: An Update Review. Front. Plant Sci. 2018, 9, 985. [Google Scholar] [CrossRef]
- Pan, C.; Ye, L.; Qin, L.; Liu, X.; He, Y.; Wang, J.; Chen, L.; Lu, G. CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci. Rep. 2016, 6, 24765. [Google Scholar] [CrossRef]
- Liu, X.; Yang, J.; Song, Y.; Zhang, X.; Wang, X.; Wang, Z. Effects of sgRNA length and number on gene editing efficiency and predicted mutations generated in rice. Crop J. 2022, 10, 577–581. [Google Scholar] [CrossRef]
- van Overbeek, M.; Capurso, D.; Carter, M.M.; Thompson, M.S.; Frias, E.; Russ, C.; Reece-Hoyes, J.S.; Nye, C.; Gradia, S.; Vidal, B. DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol. Cell 2016, 63, 633–646. [Google Scholar] [CrossRef]
- Chakrabarti, A.M.; Henser-Brownhill, T.; Monserrat, J.; Poetsch, A.R.; Luscombe, N.M.; Scaffidi, P. Target-Specific Precision of CRISPR-Mediated Genome Editing. Mol. Cell 2019, 73, 699–713.e6. [Google Scholar] [CrossRef]
- Liu, X.; Homma, A.; Sayadi, J.; Yang, S.; Ohashi, J.; Takumi, T. Sequence features associated with the cleavage efficiency of CRISPR/Cas9 system. Sci. Rep. 2016, 6, 19675. [Google Scholar] [CrossRef]
- Soyars, C.L.; Peterson, B.A.; Burr, C.A.; Nimchuk, Z.L. Cutting Edge Genetics: CRISPR/Cas9 Editing of Plant Genomes. Plant Cell Physiol. 2018, 59, 1608–1620. [Google Scholar] [CrossRef]
- Moreno-Mateos, M.A.; Vejnar, C.E.; Beaudoin, J.-D.; Fernandez, J.P.; Mis, E.K.; Khokha, M.K.; Giraldez, A.J. CRISPRscan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 2015, 12, 982–988. [Google Scholar] [CrossRef]
- Wada, N.; Ueta, R.; Osakabe, Y.; Osakabe, K. Precision genome editing in plants: State-of-the-art in CRISPR/Cas9-based genome engineering. BMC Plant Biol. 2020, 20, 234. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu Patrick, D.; Wu, X.; Jiang, W.; Marraffini Luciano, A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Nishimasu, H.; Ran, F.A.; Hsu, P.D.; Konermann, S.; Shehata, S.I.; Dohmae, N.; Ishitani, R.; Zhang, F.; Nureki, O. Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA. Cell 2014, 156, 935–949. [Google Scholar] [CrossRef]
- Semenova, E.; Jore, M.M.; Datsenko, K.A.; Semenova, A.; Westra, E.R.; Wanner, B.; van der Oost, J.; Brouns, S.J.J.; Severinov, K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. USA 2011, 108, 10098–10103. [Google Scholar] [CrossRef]
- Hahn, F.; Nekrasov, V. CRISPR/Cas precision: Do we need to worry about off-targeting in plants? Plant Cell Rep. 2019, 38, 437–441. [Google Scholar] [CrossRef]
- Manghwar, H.; Li, B.; Ding, X.; Hussain, A.; Lindsey, K.; Zhang, X.; Jin, S. CRISPR/Cas Systems in Genome Editing: Methodologies and Tools for sgRNA Design, Off-Target Evaluation, and Strategies to Mitigate Off-Target Effects. Adv. Sci. 2020, 7, 1902312. [Google Scholar] [CrossRef]
- Peterson, C.B.; Bogomolov, M.; Benjamini, Y.; Sabatti, C. TreeQTL: Hierarchical error control for eQTL findings. Bioinformatics 2016, 32, 2556–2558. [Google Scholar] [CrossRef]
- Nekrasov, V.; Wang, C.; Win, J.; Lanz, C.; Weigel, D.; Kamoun, S. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 2017, 7, 482. [Google Scholar] [CrossRef]
- Maioli, A.; Gianoglio, S.; Moglia, A.; Acquadro, A.; Valentino, D.; Milani, A.M.; Prohens, J.; Orzaez, D.; Granell, A.; Lanteri, S. Simultaneous CRISPR/Cas9 Editing of Three PPO Genes Reduces Fruit Flesh Browning in Solanum melongena L. Front. Plant Sci. 2020, 11, 607161. [Google Scholar] [CrossRef]
- Sung, W.; Ackerman, M.S.; Dillon, M.M.; Platt, T.G.; Fuqua, C.; Cooper, V.S.; Lynch, M. Evolution of the Insertion-Deletion Mutation Rate Across the Tree of Life. G3 2016, 6, 2583–2591. [Google Scholar] [CrossRef]
- Ossowski, S.; Schneeberger, K.; Lucas-Lledó, J.I.; Warthmann, N.; Clark, R.M.; Shaw, R.G.; Weigel, D.; Lynch, M. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 2010, 327, 92–94. [Google Scholar] [CrossRef]
- Miyao, A.; Nakagome, M.; Ohnuma, T.; Yamagata, H.; Kanamori, H.; Katayose, Y.; Takahashi, A.; Matsumoto, T.; Hirochika, H. Molecular spectrum of somaclonal variation in regenerated rice revealed by whole-genome sequencing. Plant Cell Physiol. 2012, 53, 256–264. [Google Scholar] [CrossRef]
- McCormick, S.; Niedermeyer, J.; Fry, J.; Barnason, A.; Horsch, R.; Fraley, R. Leaf disc transformation of cultivated tomato (L. esculentum) using Agrobacterium tumefaciens. Plant Cell Rep. 1986, 5, 81–84. [Google Scholar] [CrossRef]
- Brinkman, E.K.; Kousholt, A.N.; Harmsen, T.; Leemans, C.; Chen, T.; Jonkers, J.; van Steensel, B. Easy quantification of template-directed CRISPR/Cas9 editing. Nucleic Acids Res. 2018, 46, e58. [Google Scholar] [CrossRef]
- Caten, C.; Jinks, J. Spontaneous variability of single isolates of Phytophthora infestans. I. Cultural variation. Can. J. Bot. 1968, 46, 329–348. [Google Scholar] [CrossRef]
- Karki, H.S.; Halterman, D.A. Phytophthora infestans (Late blight) Infection Assay in a Detached Leaf of Potato. Bio-Protocol 2021, 11, e3926. [Google Scholar] [CrossRef]
- Foolad, M.R.; Sullenberger, M.T.; Ohlson, E.W.; Gugino, B.K. Response of accessions within tomato wild species, Solanum pimpinellifolium to late blight. Plant Breed. 2014, 133, 401–411. [Google Scholar] [CrossRef]
| Mutant | Mutational Status (%)-TIDE | |||
|---|---|---|---|---|
| sgRNA6 | sgRNA8 | sgRNA1 | sgRNA7 | |
| SM1 | 1.2 | - | - | - |
| SM2 | 2.5 | - | - | - |
| SM3 | 8.7 | - | - | - |
| SM4 | 38.5 | 89.4 | - | 98 |
| SM5 | 24.2 | - | - | 97.6 |
| SM6 | 40.2 | 95.1 | 4.1 | 98.9 |
| SM7 | 13.1 | 94.5 | 58.1 | - |
| SM8 | 34.6 | 95.5 | 33.6 | - |
| SM9 | - | 95.4 | 35.3 | - |
| SM12 | - | 93.3 | 9.1 | - |
| SM13 | 93.4 | 90.3 | 48.1 | 99 |
| SM14 | - | 92.8 | 3.4 | - |
| SM16 | - | 97.8 | 7.6 | - |
| SM17 | 64.6 | 94.2 | 76.2 | 97.9 |
| SM18 | - | 92.9 | 80.1 | - |
| SM19 | 1.5 | 93.1 | 31.3 | 99.3 |
| SM20 | 1.8 | 1.1 | 3.7 | 2.2 |
| SM22 | 69.3 | 94.2 | 9.7 | 98.3 |
| SM24 | - | 93.6 | 34.2 | - |
| SM25 | - | 94.2 | 5.2 | - |
| SM26 | - | 93.9 | 4.4 | - |
| OX1 | 55.3 | - | - | - |
| OX2 | 17.7 | 98.6 | 6.6 | 99 |
| OX3 | 26.8 | 98.2 | 56.7 | 99.3 |
| OX4 | 14.2 | 99 | 9.9 | 99 |
| OX9 | 13.5 | 95.9 | 4.7 | 99.3 |
| OX11 | 48.6 | 94.4 | 32.5 | 99.3 |
| Mutant | Alleles in sgRNA7 Region (%) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R2 | −18 | −10 | −8 | −7 | −6 | −5 | −3 | −2 | −1 | 1 | ref | Allelic State | ||
| SM6 | WGS | - | - | - | - | 100.0 | - | - | - | - | - | - | - | homozygous |
| Tide | 0.99 | - | - | - | 98.9 | - | - | - | - | - | - | - | ||
| SM13 | WGS | - | - | - | - | 100.0 | - | - | - | - | - | - | - | homozygous |
| Tide | 0.98 | - | - | - | 98.5 | - | - | - | - | - | - | - | ||
| SM17 | WGS | - | - | - | - | 100.0 | - | - | - | - | - | - | - | homozygous |
| Tide | 0.98 | - | - | - | 97.8 | - | - | - | - | - | - | - | ||
| SM19 | WGS | - | - | - | - | 100.0 | - | - | - | - | - | - | - | homozygous |
| Tide | 0.99 | - | - | - | 97.2 | - | - | - | - | - | - | - | ||
| SM-CTRL | WGS | - | - | - | - | - | - | - | - | - | - | - | 100.0 | wild type |
| Mutant | Alleles in sgRNA1 Region (%) | |||||||||||||
| R2 | −18 | −10 | −8 | −7 | −6 | −5 | −3 | −2 | −1 | 1 | ref | allelic state | ||
| SM6 | WGS | - | - | 10.0 | - | - | - | - | - | - | 23.3 | 69.0 | chimeric | |
| Tide | 0.96 | - | - | - | - | - | - | - | - | - | - | 92.2 | ||
| SM13 | WGS | - | - | - | - | - | 52.4 | - | - | - | - | - | 47.6 | heterozygous |
| Tide | 0.92 | - | - | - | - | 45.7 | - | - | - | - | - | 43.9 | ||
| SM17 | WGS | - | - | - | - | - | - | - | - | 62.5 | 8.3 | 8.3 | 20.8 | chimeric |
| Tide | 0.94 | - | - | - | - | - | - | - | 54.7 | 15.0 | 3.2 | 18.3 | ||
| SM19 | WGS | - | - | - | 43.7 | - | - | - | - | - | - | - | 56.3 | heterozygous |
| Tide | 0.91 | - | - | 27.6 | - | - | - | - | - | - | - | 60.0 | ||
| SM-CTRL | WGS | - | - | - | - | - | - | - | - | - | - | - | 100.0 | wild type |
| Mutant | Alleles in sgRNA8 Region (%) | |||||||||||||
| R2 | −18 | −10 | −8 | −7 | −6 | −5 | −3 | −2 | −1 | 1 | ref | allelic state | ||
| SM6 | WGS | - | - | - | - | 33.3 | - | - | - | 66.7 | - | - | - | biallelic |
| Tide | 0.95 | - | - | - | 46.4 | - | - | - | 48.3 | - | - | - | ||
| SM13 | WGS | - | - | - | - | 66.7 | - | - | - | 33.3 | - | - | - | biallelic |
| Tide | 0.9 | - | - | - | 43.4 | - | - | - | 45.8 | - | - | - | ||
| SM17 | WGS | - | - | - | - | 20.0 | - | - | - | 80.0 | - | - | - | biallelic |
| Tide | 0.94 | - | - | - | 43.2 | - | - | - | 50.1 | - | - | - | ||
| SM19 | WGS | - | - | - | - | 50.0 | - | - | - | 50.0 | - | - | - | biallelic |
| Tide | 0.93 | - | - | - | 45.1 | - | - | - | 47.7 | - | - | - | ||
| SM-CTRL | WGS | - | - | - | - | - | - | - | - | - | - | - | 100.0 | wild type |
| Mutant | Alleles in sgRNA6 Region (%) | |||||||||||||
| R2 | −18 | −10 | −8 | −7 | −6 | −5 | −3 | −2 | −1 | 1 | ref | allelic state | ||
| SM6 | WGS | - | - | - | - | 5.7 | - | 14.3 | - | 17.1 | - | 45.7 | 17.1 | chimeric |
| Tide | 0.95 | - | - | - | 11.4 | - | 12.0 | - | 16.8 | - | - | 54.8 | ||
| SM13 | WGS | - | 25.0 | - | - | - | - | 3.1 | 21.9 | - | 3.1 | 47 | - | chimeric |
| Tide | 0.93 | 27.5 | - | - | - | - | 12.0 | 8.1 | - | - | 38.8 | - | ||
| SM17 | WGS | - | - | - | - | - | 56.5 | - | - | - | - | 21.7 | 21.7 | chimeric |
| Tide | 0.93 | - | - | - | - | 31.7 | - | - | - | - | 31.7 | 28.2 | ||
| SM19 | WGS | - | - | - | - | - | - | - | - | - | - | 17.2 | 82.8 | heterozygous |
| Tide | 0.98 | - | - | - | - | - | - | - | - | - | - | 97.0 | ||
| SM-CTRL | WGS | - | - | - | - | - | - | - | - | - | - | - | 100.0 | wild type |
| sgRNAs | Off-Target All | Coding | Non-Coding | Obs. SNP/Indels |
|---|---|---|---|---|
| 7 | 4 | 1 | 3 | 0 |
| 1 | 5 | 2 | 3 | 0 |
| 8 | 6 | 0 | 6 | 0 |
| 6 | 10 | 4 | 6 | 0 |
| Total | 25 | 7 | 18 | 0 |
| Genotype | Plant Type | SNP | ||||||
|---|---|---|---|---|---|---|---|---|
| Genot. Specific | Homoz. | Heteroz. | In Exons | (%) | Per Mbp | Per Mbp (avg) | ||
| SM6 | edited | 8141 | 3318 | 4823 | 240 | 0.0010% | 9.6 | 8.95 |
| SM13 | edited | 9841 | 3554 | 6287 | 348 | 0.0013% | 8.0 | - |
| SM17 | edited | 8589 | 3491 | 5098 | 286 | 0.0011% | 9.1 | - |
| SM19 | edited | 8587 | 2945 | 5642 | 249 | 0.0011% | 9.1 | - |
| CTRL | in vitro | 7960 | 2969 | 4991 | 236 | 0.0010% | 9.8 | 9.04 |
| WT-1 | from seed | 8784 | 2925 | 5859 | 269 | 0.0011% | 8.9 | - |
| WT-2 | from seed | 9322 | 3236 | 6086 | 279 | 0.0012% | 8.4 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, R.; Maioli, A.; Yan, Z.; Bai, Y.; Valentino, D.; Milani, A.M.; Pompili, V.; Comino, C.; Lanteri, S.; Moglia, A.; et al. CRISPR/Cas9-Based Knock-Out of the PMR4 Gene Reduces Susceptibility to Late Blight in Two Tomato Cultivars. Int. J. Mol. Sci. 2022, 23, 14542. https://doi.org/10.3390/ijms232314542
Li R, Maioli A, Yan Z, Bai Y, Valentino D, Milani AM, Pompili V, Comino C, Lanteri S, Moglia A, et al. CRISPR/Cas9-Based Knock-Out of the PMR4 Gene Reduces Susceptibility to Late Blight in Two Tomato Cultivars. International Journal of Molecular Sciences. 2022; 23(23):14542. https://doi.org/10.3390/ijms232314542
Chicago/Turabian StyleLi, Ruiling, Alex Maioli, Zhe Yan, Yuling Bai, Danila Valentino, Anna Maria Milani, Valerio Pompili, Cinzia Comino, Sergio Lanteri, Andrea Moglia, and et al. 2022. "CRISPR/Cas9-Based Knock-Out of the PMR4 Gene Reduces Susceptibility to Late Blight in Two Tomato Cultivars" International Journal of Molecular Sciences 23, no. 23: 14542. https://doi.org/10.3390/ijms232314542
APA StyleLi, R., Maioli, A., Yan, Z., Bai, Y., Valentino, D., Milani, A. M., Pompili, V., Comino, C., Lanteri, S., Moglia, A., & Acquadro, A. (2022). CRISPR/Cas9-Based Knock-Out of the PMR4 Gene Reduces Susceptibility to Late Blight in Two Tomato Cultivars. International Journal of Molecular Sciences, 23(23), 14542. https://doi.org/10.3390/ijms232314542







