The Role of Italy in the Use of Advanced Plant Genomic Techniques on Fruit Trees: State of the Art and Future Perspectives
Abstract
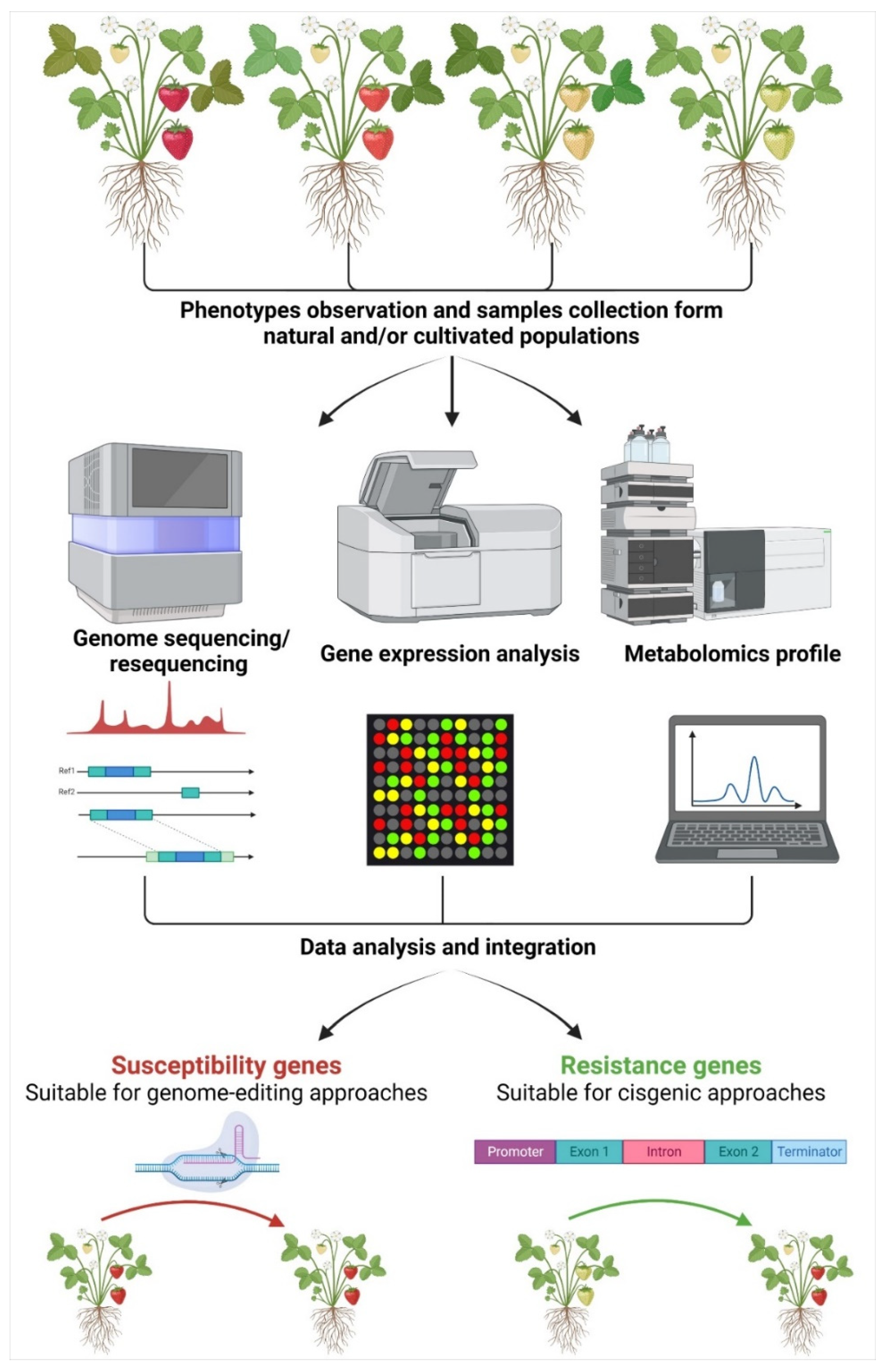
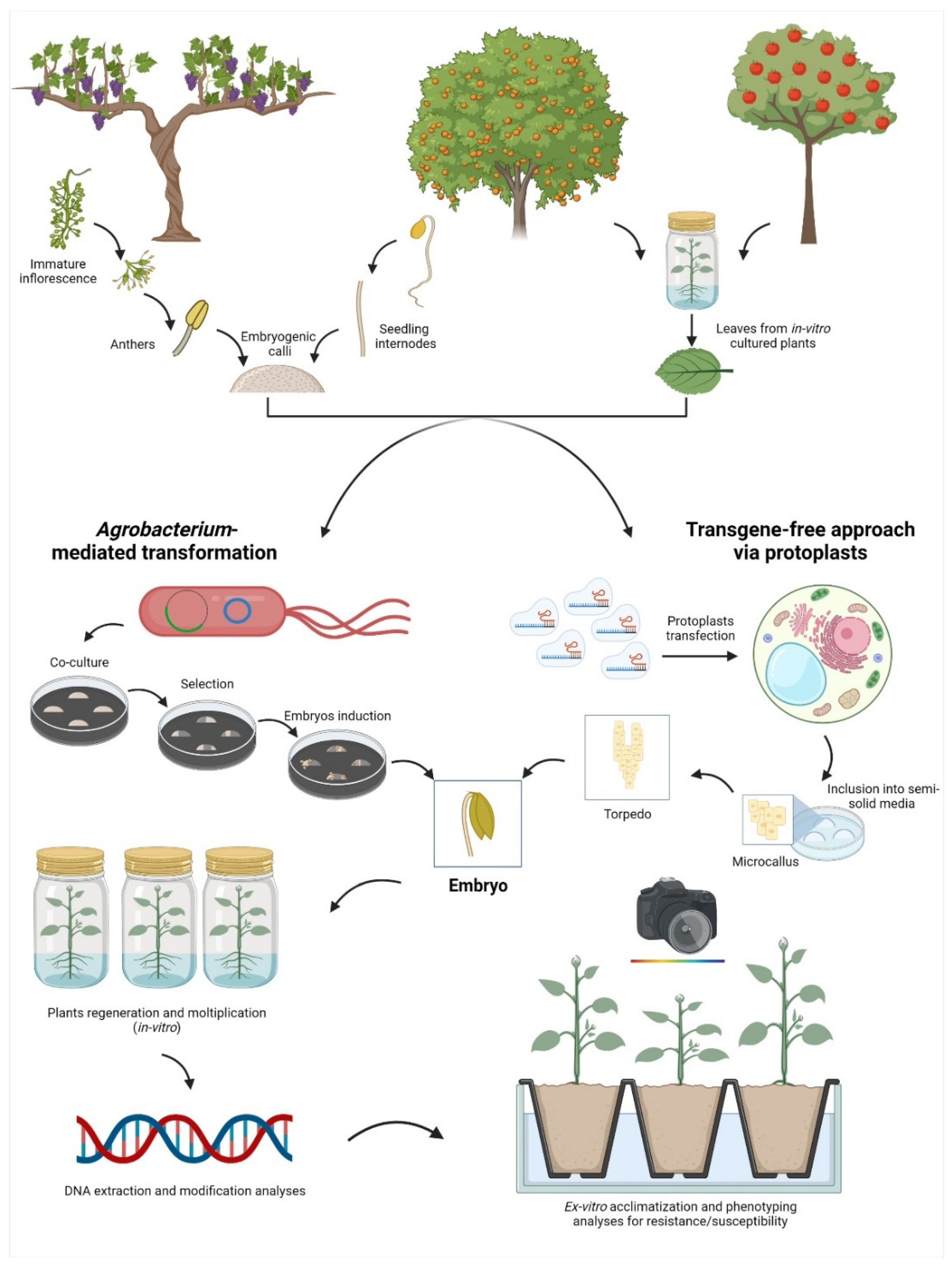
1. Application of New Genomic Techniques in Fruit Tree Species
2. Improving Citrus Fruit Quality by NGT Approaches—The CITRUS Contribution
2.1. Two Approaches to Produce Lycopene-Rich Citrus Varieties with Anthocyanins
2.2. The Production of Seedlessness Mandarin-like Fruits
3. VITECH: Improving Grapevine Genetics Features to Develop a Resilient and Sustainable Vineyard
3.1. Grapevine Tolerance to Powdery and Downy Mildews
3.2. Seedlessness Table Grape
3.3. Resistance to Drought
4. Chestnut: Genome Editing to Introduce the Tolerance to Biotic Stress
4.1. The Genome Editing of PDS Gene as Proof of Concept
4.2. A Successful Use of Chestnut Protoplasts to Produce Trangene-Free Edited Plants
5. BioSOSFru: A Large Project to Improve the Main Fruit Species Qualitative Features and Response to Biotic Stresses
5.1. Apple and Pear
5.1.1. A Cisgenic Approach to Introduce Scab and Fire Blight Resistances
5.1.2. The Genome Editing to Produce Apple and Pear Resistant to Fire Blight and Self-Compatible
5.2. Strawberry
5.2.1. An Example of Cisgenic/Intragenic Strawberry System
5.2.2. The Genome-Editing Approach Applied to Strawberry
5.3. Prunus
5.3.1. Resistance to Biotic Stress
5.3.2. Pillar Habitus
5.3.3. Early Flowering
5.4. Kiwifruit
An Attempt of Application of CRISPR/Cas9 to Produce Kiwifruit Resistant to Bacterial Canker
6. Regulatory Issues in Europe, with a Focus on Italy
6.1. Public Consultation Promoted by the European Commission
6.2. The Position of Italy
7. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arabska, E. From Farm to Fork: Human Health and Well-Being through Sustainable Agri-Food Systems. J. Life Econ. 2021, 8, 11–27. [Google Scholar]
- Eckert, E.; Kovalevska, O. Sustainability in the European Union: Analyzing the Discourse of the European Green Deal. J. Risk Financ. Manag. 2021, 14, 80. [Google Scholar] [CrossRef]
- Bazzano, L.A.; Serdula, M.K.; Liu, S. Dietary Intake of Fruits and Vegetables and Risk of Cardiovascular Disease. Curr. Atheroscler. Rep. 2003, 5, 492–499. [Google Scholar] [CrossRef] [PubMed]
- Rejman, K.; Górska-Warsewicz, H.; Kaczorowska, J.; Laskowski, W. Nutritional Significance of Fruit and Fruit Products in the Average Polish Diet. Nutrients 2021, 13, 2079. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.; Bayer, P.E.; Edwards, D. Climate Change and the Need for Agricultural Adaptation. Curr. Opin. Plant Biol. 2020, 56, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Kompas, T.; Pham, V.H.; Che, T.N. The Effects of Climate Change on GDP by Country and the Global Economic Gains from Complying with the Paris Climate Accord. Earths Future 2018, 6, 1153–1173. [Google Scholar] [CrossRef]
- Rahman, M.M.; Khan, I.; Field, D.L.; Techato, K.; Alameh, K. Powering Agriculture: Present Status, Future Potential, and Challenges of Renewable Energy Applications. Renew. Energy 2022, 188, 731–749. [Google Scholar] [CrossRef]
- Hackett, W. Juvenility and Maturity. In Cell and Tissue Culture in Forestry; Springer: Dordrecht, The Netherlands, 1987; Volume 24–26, pp. 216–231. [Google Scholar]
- Limera, C.; Sabbadini, S.; Sweet, J.B.; Mezzetti, B. New Biotechnological Tools for the Genetic Improvement of Major Woody Fruit Species. Front. Plant Sci. 2017, 8, 1418. [Google Scholar] [CrossRef]
- Giudice, G.; Moffa, L.; Varotto, S.; Cardone, M.F.; Bergamini, C.; De Lorenzis, G.; Velasco, R.; Nerva, L.; Chitarra, W. Novel and Emerging Biotechnological Crop Protection Approaches. Plant Biotechnol. J. 2021, 19, 1495–1510. [Google Scholar] [CrossRef]
- Lloyd, A.; Plaisier, C.L.; Carroll, D.; Drews, G.N. Targeted Mutagenesis Using Zinc-Finger Nucleases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 2232–2237. [Google Scholar] [CrossRef]
- Peer, R.; Rivlin, G.; Golobovitch, S.; Lapidot, M.; Gal-On, A.; Vainstein, A.; Tzfira, T.; Flaishman, M.A. Targeted Mutagenesis Using Zinc-Finger Nucleases in Perennial Fruit Trees. Planta 2015, 241, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Klocko, A.L.; Dow, M.; Ma, C.; Amarasinghe, V.; Strauss, S.H. Low Frequency of Zinc-Finger Nuclease-Induced Mutagenesis in Populus. Mol. Breed. 2016, 36, 121. [Google Scholar] [CrossRef]
- Min, T.; Hwarari, D.; Li, D.; Movahedi, A.; Yang, L. CRISPR-Based Genome Editing and Its Applications in Woody Plants. Int. J. Mol. Sci. 2022, 23, 10175. [Google Scholar] [CrossRef]
- Zaboikin, M.; Zaboikina, T.; Freter, C.; Srinivasakumar, N. Non-Homologous End Joining and Homology Directed DNA Repair Frequency of Double-Stranded Breaks Introduced by Genome Editing Reagents. PLoS ONE 2017, 12, e0169931. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liang, Z.; Zong, Y.; Wang, Y.; Liu, J.; Chen, K.; Qiu, J.-L.; Gao, C. Efficient and Transgene-Free Genome Editing in Wheat through Transient Expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016, 7, 12617. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.-L. Targeted Genome Modification of Crop Plants Using a CRISPR-Cas System. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef]
- Wang, M.; Lu, Y.; Botella, J.R.; Mao, Y.; Hua, K.; Zhu, J. Gene Targeting by Homology-Directed Repair in Rice Using a Geminivirus-Based CRISPR/Cas9 System. Mol. Plant 2017, 10, 1007–1010. [Google Scholar] [CrossRef]
- Yue, J.-J.; Yuan, J.-L.; Wu, F.-H.; Yuan, Y.-H.; Cheng, Q.-W.; Hsu, C.-T.; Lin, C.-S. Protoplasts: From Isolation to CRISPR/Cas Genome Editing Application. Front. Genome Ed. 2021, 3, 717017. [Google Scholar] [CrossRef]
- Mahmoud, L.M.; Kaur, P.; Stanton, D.; Grosser, J.W.; Dutt, M. A Cationic Lipid Mediated CRISPR/Cas9 Technique for the Production of Stable Genome Edited Citrus Plants. Plant Methods 2022, 18, 33. [Google Scholar] [CrossRef]
- Huang, X.; Wang, Y.; Wang, N. Highly Efficient Generation of Canker-Resistant Sweet Orange Enabled by an Improved CRISPR/Cas9 System. Front. Plant Sci. 2021, 12, 769907. [Google Scholar] [CrossRef]
- Parajuli, S.; Huo, H.; Gmitter, F.G.; Duan, Y.; Luo, F.; Deng, Z. Editing the CsDMR6 Gene in Citrus Results in Resistance to the Bacterial Disease Citrus Canker. Hortic. Res. 2022, 9, uhac082. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tu, M.; Wang, D.; Liu, J.; Li, Y.; Li, Z.; Wang, Y.; Wang, X. CRISPR/Cas9-mediated Efficient Targeted Mutagenesis in Grape in the First Generation. Plant Biotechnol. J. 2018, 16, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Li, M.-Y.; Jiao, Y.-T.; Wang, Y.-T.; Zhang, N.; Wang, B.-B.; Liu, R.-Q.; Yin, X.; Xu, Y.; Liu, G.-T. CRISPR/Cas9-Mediated VvPR4b Editing Decreases Downy Mildew Resistance in Grapevine (Vitis vinifera L.). Hortic. Res. 2020, 7, 149. [Google Scholar] [CrossRef] [PubMed]
- Wan, D.-Y.; Guo, Y.; Cheng, Y.; Hu, Y.; Xiao, S.; Wang, Y.; Wen, Y.-Q. CRISPR/Cas9-Mediated Mutagenesis of VvMLO3 Results in Enhanced Resistance to Powdery Mildew in Grapevine (Vitis vinifera). Hortic. Res. 2020, 7, 116. [Google Scholar] [CrossRef]
- Sunitha, S.; Rock, C.D. CRISPR/Cas9-Mediated Targeted Mutagenesis of TAS4 and MYBA7 Loci in Grapevine Rootstock 101-14. Transgenic Res. 2020, 29, 355–367. [Google Scholar] [CrossRef]
- Ren, C.; Guo, Y.; Kong, J.; Lecourieux, F.; Dai, Z.; Li, S.; Liang, Z. Knockout of VvCCD8 Gene in Grapevine Affects Shoot Branching. BMC Plant Biol. 2020, 20, 47. [Google Scholar] [CrossRef]
- Wang, Z.; Wong, D.C.J.; Wang, Y.; Xu, G.; Ren, C.; Liu, Y.; Kuang, Y.; Fan, P.; Li, S.; Xin, H. GRAS-Domain Transcription Factor PAT1 Regulates Jasmonic Acid Biosynthesis in Grape Cold Stress Response. Plant Physiol. 2021, 186, 1660–1678. [Google Scholar] [CrossRef]
- Corredoira, E.; San José, M.C.; Vieitez, A.; Allona, I.; Aragoncillo, C.; Ballester, A. Agrobacterium-Mediated Transformation of European Chestnut Somatic Embryos with a Castanea sativa (Mill.) Endochitinase Gene. New For. 2016, 47, 669–684. [Google Scholar] [CrossRef]
- Carlson, E.; Stewart, K.; Baier, K.; McGuigan, L.; Culpepper, T.; Powell, W. Pathogen-induced Expression of a Blight Tolerance Transgene in American Chestnut. Mol. Plant Pathol. 2022, 23, 370–382. [Google Scholar] [CrossRef]
- Zhou, H.; Bai, S.; Wang, N.; Sun, X.; Zhang, Y.; Zhu, J.; Dong, C. CRISPR/Cas9-Mediated Mutagenesis of MdCNGC2 in Apple Callus and VIGS-Mediated Silencing of MdCNGC2 in Fruits Improve Resistance to Botryosphaeria dothidea. Front. Plant Sci. 2020, 11, 575477. [Google Scholar] [CrossRef]
- Charrier, A.; Vergne, E.; Dousset, N.; Richer, A.; Petiteau, A.; Chevreau, E. Efficient Targeted Mutagenesis in Apple and First Time Edition of Pear Using the CRISPR-Cas9 System. Front. Plant Sci. 2019, 10, 40. [Google Scholar] [CrossRef] [PubMed]
- Malabarba, J.; Chevreau, E.; Dousset, N.; Veillet, F.; Moizan, J.; Vergne, E. New Strategies to Overcome Present CRISPR/Cas9 Limitations in Apple and Pear: Efficient Dechimerization and Base Editing. Int. J. Mol. Sci. 2020, 22, 319. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, P.; Bozorov, T.A.; Zhang, D. Application of CRISPR/Cas9 Technology in Wild Apple (Malus sieverii) for Paired Sites Gene Editing. Plant Methods 2021, 17, 79. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Dai, C.; Luo, H.; Han, Y.; Liu, Z.; Kang, C. Reporter Gene Expression Reveals Precise Auxin Synthesis Sites during Fruit and Root Development in Wild Strawberry. J. Exp. Bot. 2019, 70, 563–574. [Google Scholar] [CrossRef]
- Varkonyi-Gasic, E.; Wang, T.; Voogd, C.; Jeon, S.; Drummond, R.S.; Gleave, A.P.; Allan, A.C. Mutagenesis of Kiwifruit CENTRORADIALIS-like Genes Transforms a Climbing Woody Perennial with Long Juvenility and Axillary Flowering into a Compact Plant with Rapid Terminal Flowering. Plant Biotechnol. J. 2019, 17, 869–880. [Google Scholar] [CrossRef]
- Varkonyi-Gasic, E.; Wang, T.; Cooney, J.; Jeon, S.; Voogd, C.; Douglas, M.J.; Pilkington, S.M.; Akagi, T.; Allan, A.C. Shy Girl, a Kiwifruit Suppressor of Feminization, Restricts Gynoecium Development via Regulation of Cytokinin Metabolism and Signalling. New Phytol. 2021, 230, 1461–1475. [Google Scholar] [CrossRef]
- Herath, D.; Voogd, C.; Mayo-Smith, M.; Yang, B.; Allan, A.C.; Putterill, J.; Varkonyi-Gasic, E. CRISPR-Cas9-mediated Mutagenesis of Kiwifruit BFT Genes Results in an Evergrowing but Not Early Flowering Phenotype. Plant Biotechnol. J. 2022, 20, 2064–2076. [Google Scholar] [CrossRef]
- Schouten, H.J.; Krens, F.A.; Jacobsen, E. Do Cisgenic Plants Warrant Less Stringent Oversight? Nat. Biotechnol. 2006, 24, 753. [Google Scholar] [CrossRef]
- Schaart, J.G. Towards Consumer-Friendly Cisgenic Strawberries Which Are Less Susceptible to Botrytis cinerea; Wageningen University and Research: Wageningen, The Netherlands, 2004; ISBN 9798516028632. [Google Scholar]
- Holme, I.B.; Wendt, T.; Holm, P.B. Intragenesis and Cisgenesis as Alternatives to Transgenic Crop Development. Plant Biotechnol. J. 2013, 11, 395–407. [Google Scholar] [CrossRef]
- Hospital, F. Selection in Backcross Programmes. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1503–1511. [Google Scholar] [CrossRef]
- Wang, M.; Wang, S.; Liang, Z.; Shi, W.; Gao, C.; Xia, G. From Genetic Stock to Genome Editing: Gene Exploitation in Wheat. Trends Biotechnol. 2018, 36, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.-M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C. The Grapevine Genome Sequence Suggests Ancestral Hexaploidization in Major Angiosperm Phyla. Nature 2007, 449, 463. [Google Scholar] [PubMed]
- Velasco, R.; Zharkikh, A.; Troggio, M.; Cartwright, D.A.; Cestaro, A.; Pruss, D.; Pindo, M.; FitzGerald, L.M.; Vezzulli, S.; Reid, J. A High Quality Draft Consensus Sequence of the Genome of a Heterozygous Grapevine Variety. PLoS ONE 2007, 2, e1326. [Google Scholar] [CrossRef]
- Xu, Q.; Chen, L.-L.; Ruan, X.; Chen, D.; Zhu, A.; Chen, C.; Bertrand, D.; Jiao, W.-B.; Hao, B.-H.; Lyon, M.P. The Draft Genome of Sweet Orange (Citrus sinensis). Nat. Genet. 2013, 45, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Di Guardo, M.; Moretto, M.; Moser, M.; Catalano, C.; Troggio, M.; Deng, Z.; Cestaro, A.; Caruso, M.; Distefano, G.; La Malfa, S. The Haplotype-Resolved Reference Genome of Lemon (Citrus limon L. Burm f.). Tree Genet. Genomes 2021, 17, 46. [Google Scholar] [CrossRef]
- Verde, I.; Abbott, A.G.; Scalabrin, S.; Jung, S.; Shu, S.; Marroni, F.; Zhebentyayeva, T.; Dettori, M.T.; Grimwood, J.; Cattonaro, F. The High-Quality Draft Genome of Peach (Prunus persica) Identifies Unique Patterns of Genetic Diversity, Domestication and Genome Evolution. Nat. Genet. 2013, 45, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Verde, I.; Jenkins, J.; Dondini, L.; Micali, S.; Pagliarani, G.; Vendramin, E.; Paris, R.; Aramini, V.; Gazza, L.; Rossini, L. The Peach v2. 0 Release: High-Resolution Linkage Mapping and Deep Resequencing Improve Chromosome-Scale Assembly and Contiguity. BMC Genom. 2017, 18, 225. [Google Scholar] [CrossRef]
- Groppi, A.; Liu, S.; Cornille, A.; Decroocq, S.; Bui, Q.T.; Tricon, D.; Cruaud, C.; Arribat, S.; Belser, C.; Marande, W. Population Genomics of Apricots Unravels Domestication History and Adaptive Events. Nat. Commun. 2021, 12, 3956. [Google Scholar] [CrossRef]
- Jiang, F.; Zhang, J.; Wang, S.; Yang, L.; Luo, Y.; Gao, S.; Zhang, M.; Wu, S.; Hu, S.; Sun, H. The Apricot (Prunus armeniaca L.) Genome Elucidates Rosaceae Evolution and Beta-Carotenoid Synthesis. Hortic. Res. 2019, 6, 128. [Google Scholar] [CrossRef]
- Shirasawa, K.; Isuzugawa, K.; Ikenaga, M.; Saito, Y.; Yamamoto, T.; Hirakawa, H.; Isobe, S. The Genome Sequence of Sweet Cherry (Prunus avium) for Use in Genomics-Assisted Breeding. DNA Res. 2017, 24, 499–508. [Google Scholar] [CrossRef]
- Pinosio, S.; Marroni, F.; Zuccolo, A.; Vitulo, N.; Mariette, S.; Sonnante, G.; Aravanopoulos, F.A.; Ganopoulos, I.; Palasciano, M.; Vidotto, M. A Draft Genome of Sweet Cherry (Prunus avium L.) Reveals Genome-wide and Local Effects of Domestication. Plant J. 2020, 103, 1420–1432. [Google Scholar] [CrossRef] [PubMed]
- Kalyanaraman, A.; Fontana, P.; Bhatnagar, S.; Troggio, M.; Pruss, D.; Salvi, S.; Pindo, M.; Baldi, P.; Castelletti, S.; Cavaiuolo, M. The Genome of the Domesticated Apple (Malus x domestica Borkh.). Nat. Genet. 2010, 42, 833–839. [Google Scholar]
- Daccord, N.; Celton, J.-M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; Van de Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R. High-Quality de Novo Assembly of the Apple Genome and Methylome Dynamics of Early Fruit Development. Nat. Genet. 2017, 49, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Chagné, D.; Crowhurst, R.N.; Pindo, M.; Thrimawithana, A.; Deng, C.; Ireland, H.; Fiers, M.; Dzierzon, H.; Cestaro, A.; Fontana, P. The Draft Genome Sequence of European Pear (Pyrus communis L. ‘Bartlett’). PLoS ONE 2014, 9, e92644. [Google Scholar] [CrossRef]
- Linsmith, G.; Rombauts, S.; Montanari, S.; Deng, C.H.; Celton, J.-M.; Guérif, P.; Liu, C.; Lohaus, R.; Zurn, J.D.; Cestaro, A. Pseudo-Chromosome–Length Genome Assembly of a Double Haploid “Bartlett” Pear (Pyrus communis L.). Gigascience 2019, 8, giz138. [Google Scholar] [CrossRef]
- Wu, H.; Ma, T.; Kang, M.; Ai, F.; Zhang, J.; Dong, G.; Liu, J. A High-Quality Actinidia chinensis (Kiwifruit) Genome. Hortic. Res. 2019, 6, 117. [Google Scholar] [CrossRef]
- Shulaev, V.; Sargent, D.J.; Crowhurst, R.N.; Mockler, T.C.; Folkerts, O.; Delcher, A.L.; Jaiswal, P.; Mockaitis, K.; Liston, A.; Mane, S.P. The Genome of Woodland Strawberry (Fragaria vesca). Nat. Genet. 2011, 43, 109–116. [Google Scholar] [CrossRef]
- Edger, P.P.; Poorten, T.J.; VanBuren, R.; Hardigan, M.A.; Colle, M.; McKain, M.R.; Smith, R.D.; Teresi, S.J.; Nelson, A.D.; Wai, C.M. Origin and Evolution of the Octoploid Strawberry Genome. Nat. Genet. 2019, 51, 541–547. [Google Scholar] [CrossRef]
- Staton, M.; Addo-Quaye, C.; Cannon, N.; Sun, Y.; Zhebentyayeva, T.; Huff, M.; Fan, S.; Bellis, E.; Islam-Faridi, N.; Yu, J. The Chinese Chestnut Genome: A Reference for Species Restoration. BioRxiv 2019, 615047. [Google Scholar] [CrossRef]
- Shirasawa, K.; Nishio, S.; Terakami, S.; Botta, R.; Marinoni, D.T.; Isobe, S. Chromosome-Level Genome Assembly of Japanese Chestnut (Castanea crenata Sieb. et Zucc.) Reveals Conserved Chromosomal Segments in Woody Rosids. DNA Res. 2021, 28, dsab016. [Google Scholar] [CrossRef]
- Gambino, G.; Ruffa, P.; Vallania, R.; Gribaudo, I. Somatic Embryogenesis from Whole Flowers, Anthers and Ovaries of Grapevine (Vitis spp.). Plant Cell Tissue Organ. Cult. 2007, 90, 79–83. [Google Scholar] [CrossRef]
- Capriotti, L.; Limera, C.; Mezzetti, B.; Ricci, A.; Sabbadini, S. From Induction to Embryo Proliferation: Improved Somatic Embryogenesis Protocol in Grapevine for Italian Cultivars and Hybrid Vitis Rootstocks. Plant Cell Tissue Organ. Cult. PCTOC 2022, 151, 221–233. [Google Scholar] [CrossRef]
- Gribaudo, I.; Gambino, G.; Boccacci, P.; Perrone, I.; Cuozzo, D. A Multi-Year Study on the Regenerative Potential of Several Vitis Genotypes. Acta Hortic. 2017, 1155, 45–50. [Google Scholar] [CrossRef]
- Ricci, A.; Sabbadini, S.; Prieto, H.; Padilla, I.M.; Dardick, C.; Li, Z.; Scorza, R.; Limera, C.; Mezzetti, B.; Perez-Jimenez, M. Genetic Transformation in Peach (Prunus persica L.): Challenges and Ways Forward. Plants 2020, 9, 971. [Google Scholar] [CrossRef] [PubMed]
- Court of Justice of the European Union. Judgment of the Court (Grand Chamber) of 25 July 2018 (ECLI:EU:C:2018:583) © Curia, 15/12/2018, curia.europa.eu. Available online: https://curia.europa.eu/juris/document/document.jsf?docid=204387&doclang=EN (accessed on 20 December 2022).
- Nandy, S.; Srivastava, V. Site-specific Gene Integration in Rice Genome Mediated by the FLP–FRT Recombination System. Plant Biotechnol. J. 2011, 9, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.-J.; Cho, H.-S.; Lim, S.-H.; Shin, K.-S.; Lee, S.-M.; Lee, K.-J.; Kim, D.-H.; Cho, Y.-G. Auto-Excision of Selectable Marker Genes from Transgenic Tobacco via a Stress Inducible FLP/FRT Site-Specific Recombination System. Transgenic Res. 2009, 18, 455–465. [Google Scholar] [CrossRef]
- Sreekala, C.; Wu, L.; Gu, K.; Wang, D.; Tian, D.; Yin, Z. Excision of a Selectable Marker in Transgenic Rice (Oryza sativa L.) Using a Chemically Regulated Cre/LoxP System. Plant Cell Rep. 2005, 24, 86–94. [Google Scholar] [CrossRef]
- Cuellar, W.; Gaudin, A.; Solorzano, D.; Casas, A.; Nopo, L.; Chudalayandi, P.; Medrano, G.; Kreuze, J.; Ghislain, M. Self-Excision of the Antibiotic Resistance Gene NptII Using a Heat Inducible Cre-LoxP System from Transgenic Potato. Plant Mol. Biol. 2006, 62, 71–82. [Google Scholar] [CrossRef]
- Pompili, V.; Dalla Costa, L.; Piazza, S.; Pindo, M.; Malnoy, M. Reduced Fire Blight Susceptibility in Apple Cultivars Using a High-efficiency CRISPR/Cas9-FLP/FRT-based Gene Editing System. Plant Biotechnol. J. 2020, 18, 845–858. [Google Scholar] [CrossRef]
- Caruso, M.; Ferlito, F.; Licciardello, C.; Allegra, M.; Strano, M.C.; Di Silvestro, S.; Russo, M.P.; Paolo, D.P.; Caruso, P.; Las Casas, G. Pomological Diversity of the Italian Blood Orange Germplasm. Sci. Hortic. 2016, 213, 331–339. [Google Scholar] [CrossRef]
- Rapisarda, P.; Fabroni, S.; Peterek, S.; Russo, G.; Mock, H.-P. Juice of New Citrus Hybrids (Citrus clementina Hort. Ex Tan.× C. sinensis L. Osbeck) as a Source of Natural Antioxidants. Food Chem. 2009, 117, 212–218. [Google Scholar] [CrossRef]
- Recupero, G.R.; Russo, G.; Recupero, S.; Zurru, R.; Deidda, B.; Mulas, M. Horticultural Evaluation of New Citrus latipes Hybrids as Rootstocks for Citrus. HortScience 2009, 44, 595–598. [Google Scholar] [CrossRef]
- Recupero, G.R.; Russo, G.; Recupero, S. New Promising Citrus Triploid Hybrids Selected from Crosses between Monoembryonic Diploid Female and Tetraploid Male Parents. HortScience 2005, 40, 516–520. [Google Scholar] [CrossRef]
- Talon, M.; Caruso, M.; Gmitter, F.G., Jr. The Genus Citrus; Woodhead Publishing: Sawston, UK, 2020; ISBN 0-12-812217-X. [Google Scholar] [CrossRef]
- Jia, H.; Wang, C.; Zhang, C.; Haider, M.S.; Zhao, P.; Liu, Z.; Shangguan, L.; Pervaiz, T.; Fang, J. Functional Analysis of VvBG1 during Fruit Development and Ripening of Grape. J. Plant Growth Regul. 2016, 35, 987–999. [Google Scholar] [CrossRef]
- Jia, H.; Xu, J.; Orbović, V.; Zhang, Y.; Wang, N. Editing Citrus Genome via SaCas9/SgRNA System. Front. Plant Sci. 2017, 8, 2135. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, Y.; Irish, V.F. CENTRORADIALIS Maintains Shoot Meristem Indeterminacy by Antagonizing THORN IDENTITY1 in Citrus. Curr. Biol. 2021, 31, 2237–2242. [Google Scholar] [CrossRef]
- Alquézar, B.; Bennici, S.; Carmona, L.; Gentile, A.; Peña, L. Generation of Transfer-DNA-Free Base-Edited Citrus Plants. Front. Plant Sci. 2022, 13, 835282. [Google Scholar] [CrossRef]
- Poles, L.; Licciardello, C.; Distefano, G.; Nicolosi, E.; Gentile, A.; La Malfa, S. Recent Advances of in Vitro Culture for the Application of New Breeding Techniques in Citrus. Plants 2020, 9, 938. [Google Scholar] [CrossRef]
- Butelli, E.; Garcia-Lor, A.; Licciardello, C.; Las Casas, G.; Hill, L. Reforgiato Recupero, G. Changes in anthocyanin production during domestication of Citrus. Plant Physiol. 2017, 173, 2225–2242. [Google Scholar] [CrossRef]
- Butelli, E.; Licciardello, C.; Ramadugu, C.; Durand-Hulak, M.; Celant, A.; Recupero, G.R.; Froelicher, Y.; Martin, C. Noemi Controls Production of Flavonoid Pigments and Fruit Acidity and Illustrates the Domestication Routes of Modern Citrus Varieties. Curr. Biol. 2019, 29, 158–164. [Google Scholar] [CrossRef]
- Dasgupta, K.; Thilmony, R.; Stover, E.; Oliveira, M.L.; Thomson, J. Novel R2R3-MYB Transcription Factors from Prunus Americana Regulate Differential Patterns of Anthocyanin Accumulation in Tobacco and Citrus. GM Crop. Food 2017, 8, 85–105. [Google Scholar] [CrossRef] [PubMed]
- Dutt, M.; Stanton, D.; Grosser, J.W. Ornacitrus: Development of Genetically Modified Anthocyanin-Expressing Citrus with Both Ornamental and Fresh Fruit Potential. J. Am. Soc. Hortic. Sci. 2016, 141, 54–61. [Google Scholar] [CrossRef]
- Ciacciulli, A.; Pappalardo, H.D.; Caruso, M.; Pindo, M.; Piazza, S.; Malnoy, M.; Licciardello, C. A Marker-Free Cisgenesis/Genome Editing System, a New Tool to Produce Fortified Citrus Fruits. Acta Hortic. 2023; in press. [Google Scholar]
- Endo, T.; Shimada, T.; Fujii, H.; Kobayashi, Y.; Araki, T.; Omura, M. Ectopic Expression of an FT Homolog from Citrus Confers an Early Flowering Phenotype on Trifoliate Orange (Poncirus trifoliata L. Raf.). Transgenic Res. 2005, 14, 703–712. [Google Scholar] [CrossRef]
- Li, R.; Li, R.; Li, X.; Fu, D.; Zhu, B.; Tian, H.; Luo, Y.; Zhu, H. Multiplexed CRISPR/Cas9-mediated Metabolic Engineering of Γ-aminobutyric Acid Levels in Solanum lycopersicum. Plant Biotechnol. J. 2018, 16, 415–427. [Google Scholar] [CrossRef]
- Natalini, A.; Acciarri, N.; Cardi, T. Breeding for Nutritional and Organoleptic Quality in Vegetable Crops: The Case of Tomato and Cauliflower. Agriculture 2021, 11, 606. [Google Scholar] [CrossRef]
- Kaur, N.; Alok, A.; Kumar, P.; Kaur, N.; Awasthi, P.; Chaturvedi, S.; Pandey, P.; Pandey, A.; Pandey, A.K.; Tiwari, S. CRISPR/Cas9 Directed Editing of Lycopene Epsilon-Cyclase Modulates Metabolic Flux for β-Carotene Biosynthesis in Banana Fruit. Metab. Eng. 2020, 59, 76–86. [Google Scholar] [CrossRef]
- Salonia, F.; Ciacciulli, A.; Pappalardo, H.D.; Poles, L.; Pindo, M.; Larger, S.; Caruso, P.; Caruso, M.; Licciardello, C. A Dual SgRNA-Directed CRISPR/Cas9 Construct Designed for Editing the Fruit-Specific β-Cyclase 2 Gene in Pigmented Citrus Fruits. Front. Plant Sci. 2022, 13, 975917. [Google Scholar] [CrossRef]
- Peng, A.; Zou, X.; Xu, L.; He, Y.; Lei, T.; Yao, L.; Li, Q.; Chen, S. Improved Protocol for the Transformation of Adult Citrus sinensis Osbeck ‘Tarocco’ Blood Orange Tissues. In Vitro Cell. Dev. Biol.-Plant 2019, 55, 659–667. [Google Scholar] [CrossRef]
- Jardak, R.; Boubakri, H.; Zemni, H.; Gandoura, S.; Mejri, S.; Mliki, A.; Ghorbel, A. Establishment of an in Vitro Regeneration System and Genetic Transformation of the Tunisian ‘Maltese Half-Blood’ (Citrus sinensis): An Agro-Economically Important Variety. 3 Biotech 2020, 10, 99. [Google Scholar] [CrossRef]
- Yamasaki, A.; Kitajima, A.; Ohara, N.; Tanaka, M.; Hasegawa, K. Histological Study of Expression of Seedlessness in Citrus kinokuni ‘Mukaku Kishu’ and Its Progenies. J. Am. Soc. Hortic. Sci. 2007, 132, 869–875. [Google Scholar] [CrossRef]
- Caruso, M.; Merelo, P.; Distefano, G.; La Malfa, S.; Lo Piero, A.R.; Tadeo, F.R.; Talon, M.; Gentile, A. Comparative Transcriptome Analysis of Stylar Canal Cells Identifies Novel Candidate Genes Implicated in the Self-Incompatibility Response of Citrus clementina. BMC Plant Biol. 2012, 12, 20. [Google Scholar] [CrossRef] [PubMed]
- Varoquaux, F.; Blanvillain, R.; Delseny, M.; Gallois, P. Less Is Better: New Approaches for Seedless Fruit Production. Trends Biotechnol. 2000, 18, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Vardi, A.; Levin, I.; Carmi, N. Induction of Seedlessness in Citrus: From Classical Techniques to Emerging Biotechnological Approaches. J. Am. Soc. Hortic. Sci. 2008, 133, 117–126. [Google Scholar] [CrossRef]
- Luo, M.; Dennis, E.S.; Berger, F.; Peacock, W.J.; Chaudhury, A. MINISEED3 (MINI3), a WRKY Family Gene, and HAIKU2 (IKU2), a Leucine-Rich Repeat (LRR) KINASE Gene, Are Regulators of Seed Size in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 17531–17536. [Google Scholar] [CrossRef]
- Wang, A.; Garcia, D.; Zhang, H.; Feng, K.; Chaudhury, A.; Berger, F.; Peacock, W.J.; Dennis, E.S.; Luo, M. The VQ Motif Protein IKU1 Regulates Endosperm Growth and Seed Size in Arabidopsis. Plant J. 2010, 63, 670–679. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, X.; Kang, X.; Zhao, X.; Zhang, X.; Ni, M. SHORT HYPOCOTYL UNDER BLUE1 Associates with MINISEED3 and HAIKU2 Promoters in Vivo to Regulate Arabidopsis Seed Development. Plant Cell 2009, 21, 106–117. [Google Scholar] [CrossRef]
- Poles, L.; Ciacciulli, A.; Pappalardo, D.H.; Salonia, F.; Distefano, G.; Gentile, A.; Caruso, M.; Larger, S.; Pindo, M.; La Malfa, S.; et al. Genome Editing of IKU1 to Obtain Citrus Seedless Fruits. Acta Hortic. 2023; in press. [Google Scholar]
- Li, N.; Xu, R.; Li, Y. Molecular Networks of Seed Size Control in Plants. Annu. Rev. Plant Biol. 2019, 70, 435–463. [Google Scholar] [CrossRef]
- Liang, M.; Cao, Z.; Zhu, A.; Liu, Y.; Tao, M.; Yang, H.; Xu, Q.; Wang, S.; Liu, J.; Li, Y. Evolution of Self-Compatibility by a Mutant Sm-RNase in Citrus. Nat. Plants 2020, 6, 131–142. [Google Scholar] [CrossRef]
- Vezzulli, S.; Gramaje, D.; Tello, J.; Gambino, G.; Bettinelli, P.; Pirrello, C.; Schwandner, A.; Barba, P.; Angelini, E.; Anfora, G. Genomic Designing for Biotic Stress Resistant Grapevine. In Genomic Designing for Biotic Stress Resistant Fruit Crops; Springer: Berlin/Heidelberg, Germany, 2022; pp. 87–255. [Google Scholar] [CrossRef]
- Foria, S.; Magris, G.; Jurman, I.; Schwope, R.; De Candido, M.; De Luca, E.; Ivanišević, D.; Morgante, M.; Di Gaspero, G. Extent of Wild–to–Crop Interspecific Introgression in Grapevine (Vitis vinifera) as a Consequence of Resistance Breeding and Implications for the Crop Species Definition. Hortic. Res. 2022, 9, uhab010. [Google Scholar] [CrossRef] [PubMed]
- Cesco, S.; Tolotti, A.; Nadalini, S.; Rizzi, S.; Valentinuzzi, F.; Mimmo, T.; Porfido, C.; Allegretta, I.; Giovannini, O.; Perazzolli, M. Plasmopara viticola Infection Affects Mineral Elements Allocation and Distribution in Vitis vinifera Leaves. Sci. Rep. 2020, 10, 18759. [Google Scholar] [CrossRef] [PubMed]
- Poni, S.; Chiari, G.; Caffi, T.; Bove, F.; Tombesi, S.; Moncalvo, A.; Gatti, M. Canopy Physiology, Vine Performance and Host-Pathogen Interaction in a Fungi Resistant Cv. Sangiovese x Bianca Accession vs. a Susceptible Clone. Sci. Rep. 2017, 7, 6092. [Google Scholar] [CrossRef] [PubMed]
- Pagliarani, C.; Vitali, M.; Ferrero, M.; Vitulo, N.; Incarbone, M.; Lovisolo, C.; Valle, G.; Schubert, A. The Accumulation of miRNAs Differentially Modulated by Drought Stress Is Affected by Grafting in Grapevine. Plant Physiol. 2017, 173, 2180–2195. [Google Scholar] [CrossRef]
- Corso, M.; Vannozzi, A.; Maza, E.; Vitulo, N.; Meggio, F.; Pitacco, A.; Telatin, A.; D’angelo, M.; Feltrin, E.; Negri, A.S. Comprehensive Transcript Profiling of Two Grapevine Rootstock Genotypes Contrasting in Drought Susceptibility Links the Phenylpropanoid Pathway to Enhanced Tolerance. J. Exp. Bot. 2015, 66, 5739–5752. [Google Scholar] [CrossRef]
- Bianchi, D.; Caramanico, L.; Grossi, D.; Brancadoro, L.; Lorenzis, G.D. How Do Novel M-Rootstock (Vitis spp.) Genotypes Cope with Drought? Plants 2020, 9, 1385. [Google Scholar] [CrossRef]
- Pessina, S.; Lenzi, L.; Perazzolli, M.; Campa, M.; Dalla Costa, L.; Urso, S.; Valè, G.; Salamini, F.; Velasco, R.; Malnoy, M. Knockdown of MLO Genes Reduces Susceptibility to Powdery Mildew in Grapevine. Hortic. Res. 2016, 3, 16016. [Google Scholar] [CrossRef]
- Liu, X.; Ao, K.; Yao, J.; Zhang, Y.; Li, X. Engineering Plant Disease Resistance against Biotrophic Pathogens. Curr. Opin. Plant Biol. 2021, 60, 101987. [Google Scholar] [CrossRef]
- Pirrello, C.; Malacarne, G.; Moretto, M.; Lenzi, L.; Perazzolli, M.; Zeilmaker, T.; Van den Ackerveken, G.; Pilati, S.; Moser, C.; Giacomelli, L. Grapevine DMR6-1 Is a Candidate Gene for Susceptibility to Downy Mildew. Biomolecules 2022, 12, 182. [Google Scholar] [CrossRef]
- Giacomelli, L.; Zeilmaker, T.; Scintilla, S.; Salvagnin, U.; van der Voort, J.R.; Moser, C. Vitis vinifera Plants Edited in DMR6 Genes Show Improved Resistance to Downy Mildew. bioRxiv 2022. [Google Scholar] [CrossRef]
- Foria, S.; Copetti, D.; Eisenmann, B.; Magris, G.; Vidotto, M.; Scalabrin, S.; Testolin, R.; Cipriani, G.; Wiedemann-Merdinoglu, S.; Bogs, J. Gene Duplication and Transposition of Mobile Elements Drive Evolution of the Rpv3 Resistance Locus in Grapevine. Plant J. 2020, 101, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Najafi, S.; Bertini, E.; D’Incà, E.; Fasoli, M.; Zenoni, S. DNA-Free Genome Editing in Grapevine Using CRISPR/Cas9 Ribonucleoprotein Complexes Followed by Protoplast Regeneration. Hortic. Res. 2022. [Google Scholar] [CrossRef]
- Jiang, N.; Meng, J.; Cui, J.; Sun, G.; Luan, Y. Function Identification of MIR482b, a Negative Regulator during Tomato Resistance to Phytophthora Infestans. Hortic. Res. 2018, 5, uhac240. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Jin, S.; Yuan, Y.; Liu, Q.; Zhang, X.; Wilson, I. CRISPR/Cas9-mediated Saturated Mutagenesis of the Cotton MIR482 Family for Dissecting the Functionality of Individual Members in Disease Response. Plant Direct 2022, 6, e410. [Google Scholar] [CrossRef] [PubMed]
- Forleo, L.R.; D’Amico, M.; Basile, T.; Marsico, A.D.; Cardone, M.F.; Maggiolini, F.A.M.; Velasco, R.; Bergamini, C. Somatic Embryogenesis in Vitis for Genome Editing: Optimization of Protocols for Recalcitrant Genotypes. Horticulturae 2021, 7, 511. [Google Scholar] [CrossRef]
- Mejia, N.; Soto, B.; Guerrero, M.; Casanueva, X.; Houel, C.; de los Angeles Miccono, M.; Ramos, R.; Le Cunff, L.; Boursiquot, J.-M.; Hinrichsen, P. Molecular, Genetic and Transcriptional Evidence for a Role of VvAGL11 in Stenospermocarpic Seedlessness in Grapevine. BMC Plant Biol. 2011, 11, 57. [Google Scholar] [CrossRef]
- Royo, C.; Torres-Pérez, R.; Mauri, N.; Diestro, N.; Cabezas, J.A.; Marchal, C.; Lacombe, T.; Ibáñez, J.; Tornel, M.; Carreño, J. The Major Origin of Seedless Grapes Is Associated with a Missense Mutation in the MADS-Box Gene VviAGL11. Plant Physiol. 2018, 177, 1234–1253. [Google Scholar] [CrossRef]
- Amato, A.; Cardone, M.F.; Ocarez, N.; Alagna, F.; Ruperti, B.; Fattorini, C.; Velasco, R.; Mejía, N.; Zenoni, S.; Bergamini, C. VviAGL11 Self-Regulates and Targets Hormone- and Secondary Metabolism-Related Genes during Seed Development. Hortic. Res. 2022, 9, uhac133. [Google Scholar] [CrossRef]
- Torregrosa, L.; Rienth, M.; Romieu, C.; Pellegrino, A. The Microvine, a Model for Studies in Grapevine Physiology and Genetics. OENO One 2019, 53, 373–391. [Google Scholar] [CrossRef]
- Nerva, L.; Guaschino, M.; Pagliarani, C.; De Rosso, M.; Lovisolo, C.; Chitarra, W. Spray-induced Gene Silencing Targeting a Glutathione S-transferase Gene Improves Resilience to Drought in Grapevine. Plant Cell Environ. 2022, 45, 347–361. [Google Scholar] [CrossRef]
- Clemens, M.; Faralli, M.; Lagreze, J.; Bontempo, L.; Piazza, S.; Varotto, C.; Malnoy, M.; Oechel, W.; Rizzoli, A.; Dalla Costa, L. VvEPFL9-1 Knock-Out via CRISPR/Cas9 Reduces Stomatal Density in Grapevine. Front. Plant Sci. 2022, 13, 878001. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, P.; Machado, H.; Silva, M.C.; Costa, R.L. A Histopathological Study Reveals New Insights Into Responses of Chestnut (Castanea spp.) to Root Infection by Phytophthora cinnamomi. Phytopathology 2021, 111, 345–355. [Google Scholar] [CrossRef]
- Beccaro, G.; Alma, A.; Bounous, G.; Gomes-Laranjo, J. The Chestnut Handbook: Crop & Forest Management; CRC Press: Boca Raton, FL, USA, 2019; ISBN 0-429-81919-6. [Google Scholar]
- Serrazina, S.; Santos, C.; Machado, H.; Pesquita, C.; Vicentini, R.; Pais, M.S.; Sebastiana, M.; Costa, R. Castanea Root Transcriptome in Response to Phytophthora cinnamomi Challenge. Tree Genet. Genomes 2015, 11, 6. [Google Scholar] [CrossRef]
- Santos, C.; Machado, H.; Correia, I.; Gomes, F.; Gomes-Laranjo, J.; Costa, R. Phenotyping Castanea Hybrids for Phytophthora cinnamomi Resistance. Plant Pathol. 2015, 64, 901–910. [Google Scholar] [CrossRef]
- Acquadro, A.; Torello Marinoni, D.; Sartor, C.; Dini, F.; Macchio, M.; Botta, R. Transcriptome Characterization and Expression Profiling in Chestnut Cultivars Resistant or Susceptible to the Gall Wasp Dryocosmus kuriphilus. Mol. Genet. Genom. 2020, 295, 107–120. [Google Scholar] [CrossRef]
- Pavese, V.; Moglia, A.; Gonthier, P.; Torello Marinoni, D.; Cavalet-Giorsa, E.; Botta, R. Identification of Susceptibility Genes in Castanea Sativa and Their Transcription Dynamics Following Pathogen Infection. Plants 2021, 10, 913. [Google Scholar] [CrossRef]
- Aebi, A.; Schönrogge, K.; Melika, G.; Alma, A.; Bosio, G.; Quacchia, A.; Picciau, L.; Abe, Y.; Moriya, S.; Yara, K. Parasitoid Recruitment to the Globally Invasive Chestnut Gall Wasp Dryocosmus kuriphilus. In Galling Arthropods and Their Associates; Springer: Tokyo, Japan, 2006; pp. 103–121. [Google Scholar]
- Lione, G.; Giordano, L.; Turina, M.; Gonthier, P. Hail-Induced Infections of the Chestnut Blight Pathogen Cryphonectria parasitica Depend on Wound Size and May Lead to Severe Diebacks. Phytopathology 2020, 110, 1280–1293. [Google Scholar] [CrossRef]
- Pavese, V.; Moglia, A.; Corredoira, E.; Martínez, M.T.; Torello Marinoni, D.; Botta, R. First Report of CRISPR/Cas9 Gene Editing in Castanea sativa Mill. Front. Plant Sci. 2021, 12, 728516. [Google Scholar] [CrossRef]
- Pan, C.; Ye, L.; Qin, L.; Liu, X.; He, Y.; Wang, J.; Chen, L.; Lu, G. CRISPR/Cas9-mediated Efficient and Heritable Targeted Mutagenesis in Tomato Plants in the First and Later Generations. Sci. Rep. 2016, 6, 24765. [Google Scholar] [CrossRef]
- Qin, G.; Gu, H.; Ma, L.; Peng, Y.; Deng, X.W.; Chen, Z.; Qu, L.-J. Disruption of Phytoene Desaturase Gene Results in Albino and Dwarf Phenotypes in Arabidopsis by Impairing Chlorophyll, Carotenoid, and Gibberellin Biosynthesis. Cell Res. 2007, 17, 471–482. [Google Scholar] [CrossRef]
- Wilson, F.M.; Harrison, K.; Armitage, A.D.; Simkin, A.J.; Harrison, R.J. CRISPR/Cas9-Mediated Mutagenesis of Phytoene Desaturase in Diploid and Octoploid Strawberry. Plant Methods 2019, 15, 45. [Google Scholar] [CrossRef] [PubMed]
- Walawage, S.L.; Zaini, P.A.; Mubarik, M.S.; Martinelli, F.; Balan, B.; Caruso, T.; Leslie, C.A.; Dandekar, A.M. Deploying Genome Editing Tools for Dissecting the Biology of Nut Trees. Front. Sustain. Food Syst. 2019, 3, 100. [Google Scholar] [CrossRef]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalán, C.; Cho, S.W.; Kim, H.; Kim, S.-G.; Kim, S.-T.; Choe, S.; Kim, J.-S. DNA-Free Genome Editing in Plants with Preassembled CRISPR-Cas9 Ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1164. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Abdeen, A.A.; Wang, Y.; Shahi, P.K.; Robertson, S.; Xie, R.; Suzuki, M.; Pattnaik, B.R.; Saha, K.; Gong, S. A Biodegradable Nanocapsule Delivers a Cas9 Ribonucleoprotein Complex for in Vivo Genome Editing. Nat. Nanotechnol. 2019, 14, 974–980. [Google Scholar] [CrossRef]
- Osakabe, Y.; Liang, Z.; Ren, C.; Nishitani, C.; Osakabe, K.; Wada, M.; Komori, S.; Malnoy, M.; Velasco, R.; Poli, M. CRISPR–Cas9-Mediated Genome Editing in Apple and Grapevine. Nat. Protoc. 2018, 13, 2844–2863. [Google Scholar] [CrossRef]
- Pavese, V.; Moglia, A.; Abbà, S.; Milani, A.M.; Torello Marinoni, D.; Corredoira, E.; Martínez, M.T.; Botta, R. First Report on Genome Editing via Ribonucleoprotein (RNP) in Castanea Sativa Mill. Int. J. Mol. Sci. 2022, 23, 5762. [Google Scholar] [CrossRef]
- Malnoy, M.; Viola, R.; Jung, M.-H.; Koo, O.-J.; Kim, S.; Kim, J.-S.; Velasco, R.; Nagamangala Kanchiswamy, C. DNA-Free Genetically Edited Grapevine and Apple Protoplast Using CRISPR/Cas9 Ribonucleoproteins. Front. Plant Sci. 2016, 7, 1904. [Google Scholar] [CrossRef]
- Bertini, E.; Tornielli, G.B.; Pezzotti, M.; Zenoni, S. Regeneration of Plants from Embryogenic Callus-Derived Protoplasts of Garganega and Sangiovese Grapevine (Vitis vinifera L.) Cultivars. Plant Cell Tissue Organ. Cult. PCTOC 2019, 138, 239–246. [Google Scholar] [CrossRef]
- Kuzminsky, E.; Meschini, R.; Terzoli, S.; Pavani, L.; Silvestri, C.; Choury, Z.; Scarascia-Mugnozza, G. Isolation of Mesophyll Protoplasts from Mediterranean Woody Plants for the Study of DNA Integrity under Abiotic Stress. Front. Plant Sci. 2016, 7, 1168. [Google Scholar] [CrossRef]
- Gessler, C.; Patocchi, A.; Sansavini, S.; Tartarini, S.; Gianfranceschi, L. Venturia Inaequalis Resistance in Apple. Crit. Rev. Plant Sci. 2006, 25, 473–503. [Google Scholar] [CrossRef]
- Kellerhals, M.; Baumgartner, I.; Leumann, L.; Lussi, L.; Schütz, S.; Patocchi, A. Breeding High Quality Apples with Fire Blight Resistance. Acta Hortic. 2013, 1056, 225–230. [Google Scholar] [CrossRef]
- Dall’Agata, M.; Pagliarani, G.; Padmarasu, S.; Troggio, M.; Bianco, L.; Dapena, E.; Miñarro, M.; Aubourg, S.; Lespinasse, Y.; Durel, C.-E. Identification of Candidate Genes at the Dp-Fl Locus Conferring Resistance against the Rosy Apple Aphid Dysaphis Plantaginea. Tree Genet. Genomes 2018, 14, 12. [Google Scholar] [CrossRef]
- De Franceschi, P.; Dondini, L. Molecular Mapping of Major Genes and QTLs in Pear. In The Pear Genome; Springer: Cham, Switzerland, 2019; pp. 113–131. [Google Scholar]
- Peil, A.; Emeriewen, O.F.; Khan, A.; Kostick, S.; Malnoy, M. Status of Fire Blight Resistance Breeding in Malus. J. Plant Pathol. 2021, 103, 3–12. [Google Scholar] [CrossRef]
- Predieri, S. Mutation Induction and Tissue Culture in Improving Fruits. Plant Cell Tissue Organ. Cult. 2001, 64, 185–210. [Google Scholar] [CrossRef]
- Vanblaere, T.; Szankowski, I.; Schaart, J.; Schouten, H.; Flachowsky, H.; Broggini, G.A.; Gessler, C. The Development of a Cisgenic Apple Plant. J. Biotechnol. 2011, 154, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Krens, F.A.; Schaart, J.G.; Van der Burgh, A.M.; Tinnenbroek-Capel, I.E.; Groenwold, R.; Kodde, L.P.; Broggini, G.A.; Gessler, C.; Schouten, H.J. Cisgenic Apple Trees; Development, Characterization, and Performance. Front. Plant Sci. 2015, 6, 286. [Google Scholar] [CrossRef][Green Version]
- Kost, T.D.; Gessler, C.; Jänsch, M.; Flachowsky, H.; Patocchi, A.; Broggini, G.A. Development of the First Cisgenic Apple with Increased Resistance to Fire Blight. PLoS ONE 2015, 10, e0143980. [Google Scholar] [CrossRef]
- Fischer, T.; Malnoy, M.; Hofmann, T.; Schwab, W.; Palmieri, L.; Wehrens, R.; Schuch, L.; Müller, M.; Schimmelpfeng, H.; Velasco, R. F1 Hybrid of Cultivated Apple (Malus × domestica) and European Pear (Pyrus communis) with Fertile F2 Offspring. Mol. Breed. 2014, 34, 817–828. [Google Scholar] [CrossRef]
- Perchepied, L.; Chevreau, E.; Ravon, E.; Gaillard, S.; Pelletier, S.; Bahut, M.; Berthelot, P.; Cournol, R.; Schouten, H.; Vergne, E. Successful Intergeneric Transfer of a Major Apple Scab Resistance Gene (Rvi6) from Apple to Pear and Precise Comparison of the Downstream Molecular Mechanisms of This Resistance in Both Species. BMC Genom. 2021, 22, 843. [Google Scholar] [CrossRef]
- Yousaf, A.; Baldi, P.; Piazza, S.; Patocch, A.; Malnoy, M. A Perspective to Durable Apple Scab Resistance by Attributing Functions of Rvi12 Gene. In Proceedings of the 10th Rosaceae Genomics Conference, Barcelona, Spain, 1–30 July 2020. [Google Scholar]
- Domenichini, C.; Negri, P.; Defrancesco, M.; Alessandri, S.; Bergonzoni, L.; Verde, I.; Malnoy, M.; Broggini, G.; Patocchi, A.; Peil, A.; et al. New Breeding Technology Approaches to Improve Apple and Pear Varieties. Acta Hortic. 2023; in press. [Google Scholar]
- Emeriewen, O.F.; Richter, K.; Piazza, S.; Micheletti, D.; Broggini, G.A.; Berner, T.; Keilwagen, J.; Hanke, M.-V.; Malnoy, M.; Peil, A. Towards Map-Based Cloning of FB_Mfu10: Identification of a Receptor-like Kinase Candidate Gene Underlying the Malus Fusca Fire Blight Resistance Locus on Linkage Group 10. Mol. Breed. 2018, 38, 106. [Google Scholar] [CrossRef]
- Emeriewen, O.F.; Flachowsky, H.; Peil, A. Characterization of Genomic DNA Sequence of the Candidate Gene for FB_Mfu10 Associated with Fire Blight Resistance in Malus Species. BMC Res. Notes 2021, 14, 291. [Google Scholar] [CrossRef] [PubMed]
- Nishitani, C.; Hirai, N.; Komori, S.; Wada, M.; Okada, K.; Osakabe, K.; Yamamoto, T.; Osakabe, Y. Efficient Genome Editing in Apple Using a CRISPR/Cas9 System. Sci. Rep. 2016, 6, 31481. [Google Scholar] [CrossRef] [PubMed]
- Sarker, N.C.; Ray, P.; Pfau, C.; Kalavacharla, V.; Hossain, K.; Quadir, M. Development of Functional Nanomaterials from Wheat Bran Derived Arabinoxylan for Nucleic Acid Delivery. J. Agric. Food Chem. 2020, 68, 4367–4373. [Google Scholar] [CrossRef] [PubMed]
- Whitaker, V.M.; Knapp, S.J.; Hardigan, M.A.; Edger, P.P.; Slovin, J.P.; Bassil, N.V.; Hytönen, T.; Mackenzie, K.K.; Lee, S.; Jung, S. A Roadmap for Research in Octoploid Strawberry. Hortic. Res. 2020, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, R.F.; Carvalho, S.D.; O’Grady, K.; Folta, K.M. Agroinfiltration of Strawberry Fruit—A Powerful Transient Expression System for Gene Validation. Curr. Plant Biol. 2016, 6, 19–37. [Google Scholar] [CrossRef]
- Gaston, A.; Potier, A.; Alonso, M.; Sabbadini, S.; Delmas, F.; Tenreira, T.; Cochetel, N.; Labadie, M.; Prévost, P.; Folta, K.M. The FveFT2 Florigen/FveTFL1 Antiflorigen Balance Is Critical for the Control of Seasonal Flowering in Strawberry While FveFT3 Modulates Axillary Meristem Fate and Yield. New Phytol. 2021, 232, 372–387. [Google Scholar] [CrossRef]
- Carvalho, R.F.; Folta, K.M. Assessment of Promoters and a Selectable Marker for Development of Strawberry Intragenic Vectors. Plant Cell Tissue Organ. Cult. PCTOC 2017, 128, 259–271. [Google Scholar] [CrossRef]
- Kashtwari, M.; Mansoor, S.; Wani, A.A.; Najar, M.A.; Deshmukh, R.K.; Baloch, F.S.; Abidi, I.; Zargar, S.M. Random Mutagenesis in Vegetatively Propagated Crops: Opportunities, Challenges and Genome Editing Prospects. Mol. Biol. Rep. 2022, 49, 5729–5749. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, G.; Liu, Z. Efficient Genome Editing of Wild Strawberry Genes, Vector Development and Validation. Plant Biotechnol. J. 2018, 16, 1868–1877. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Gao, C. Genome Editing in Rice and Wheat Using the CRISPR/Cas System. Nat. Protoc. 2014, 9, 2395–2410. [Google Scholar] [CrossRef] [PubMed]
- Gou, Y.-J.; Li, Y.-L.; Bi, P.-P.; Wang, D.-J.; Ma, Y.-Y.; Hu, Y.; Zhou, H.-C.; Wen, Y.-Q.; Feng, J.-Y. Optimization of the Protoplast Transient Expression System for Gene Functional Studies in Strawberry (Fragaria vesca). Plant Cell Tissue Organ. Cult. PCTOC 2020, 141, 41–53. [Google Scholar] [CrossRef]
- Martín-Pizarro, C.; Triviño, J.C.; Posé, D. Functional Analysis of the TM6 MADS-Box Gene in the Octoploid Strawberry by CRISPR/Cas9-Directed Mutagenesis. J. Exp. Bot. 2019, 70, 885–895. [Google Scholar] [CrossRef]
- Cappelletti, R.; Sabbadini, S.; Mezzetti, B. Strawberry (Fragaria × ananassa). In Agrobacterium Protocols; Springer: New York, NY, USA, 2015; pp. 217–227. [Google Scholar]
- Dirlewanger, E.; Kleinhentz, M.; Voisin, R.; Claverie, M.; Lecouls, A.; Esmenjaud, D.; Poessel, J.; Faurobert, M.; Arús, P.; Gómez-Aparisi, J.; et al. Breeding for a New Generation of Prunus Rootstocks: An Example of MAS. Acta Hortic. 2004, 658, 581–590. [Google Scholar] [CrossRef]
- Verde, I.; Bassil, N.; Scalabrin, S.; Gilmore, B.; Lawley, C.T.; Gasic, K.; Micheletti, D.; Rosyara, U.R.; Cattonaro, F.; Vendramin, E.; et al. Development and Evaluation of a 9K SNP Array for Peach by Internationally Coordinated SNP Detection and Validation in Breeding Germplasm. PLoS ONE 2012, 7, e35668. [Google Scholar] [CrossRef]
- Peace, C.; Bassil, N.; Main, D.; Ficklin, S.; Rosyara, U.R.; Stegmeir, T.; Sebolt, A.; Gilmore, B.; Lawley, C.; Mockler, T.C. Development and Evaluation of a Genome-Wide 6K SNP Array for Diploid Sweet Cherry and Tetraploid Sour Cherry. PLoS ONE 2012, 7, e48305. [Google Scholar] [CrossRef]
- Aranzana, M.J.; Decroocq, V.; Dirlewanger, E.; Eduardo, I.; Gao, Z.S.; Gasic, K.; Iezzoni, A.; Jung, S.; Peace, C.; Prieto, H. Prunus Genetics and Applications after de Novo Genome Sequencing: Achievements and Prospects. Hortic. Res. 2019, 6, 58. [Google Scholar] [CrossRef]
- Dardick, C.; Callahan, A.; Horn, R.; Ruiz, K.B.; Zhebentyayeva, T.; Hollender, C.; Whitaker, M.; Abbott, A.; Scorza, R. P Pe TAC 1 Promotes the Horizontal Growth of Branches in Peach Trees and Is a Member of a Functionally Conserved Gene Family Found in Diverse Plants Species. Plant J. 2013, 75, 618–630. [Google Scholar] [CrossRef]
- Claverie, M.; Dirlewanger, E.; Bosselut, N.; Van Ghelder, C.; Voisin, R.; Kleinhentz, M.; Lafargue, B.; Abad, P.; Rosso, M.-N.; Chalhoub, B.; et al. The Ma Gene for Complete-Spectrum Resistance to Meloidogyne Species in Prunus Is a TNL with a Huge Repeated C-Terminal Post-LRR Region. Plant Physiol. 2011, 156, 779–792. [Google Scholar] [CrossRef]
- Van Ghelder, C.; Esmenjaud, D.; Callot, C.; Dubois, E.; Mazier, M.; Duval, H. Ma Orthologous Genes in Prunus spp. Shed Light on a Noteworthy NBS-LRR Cluster Conferring Differential Resistance to Root-Knot Nematodes. Front. Plant Sci. 2018, 9, 1269. [Google Scholar] [CrossRef]
- Claverie, M.; Bosselut, N.; Lecouls, A.; Voisin, R.; Lafargue, B.; Poizat, C.; Kleinhentz, M.; Laigret, F.; Dirlewanger, E.; Esmenjaud, D. Location of Independent Root-Knot Nematode Resistance Genes in Plum and Peach. Theor. Appl. Genet. 2004, 108, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Handoo, Z.; Nyczepir, A.; Esmenjaud, D.; Van der Beek, J.; Castagnone-Sereno, P.; Carta, L.; Skantar, A.; Higgins, J. Morphological, Molecular, and Differential-Host Characterization of Meloidogyne floridensis n. Sp. (Nematoda: Meloidogynidae), a Root-Knot Nematode Parasitizing Peach in Florida. J. Nematol. 2004, 36, 20. [Google Scholar] [PubMed]
- Clemente-Moreno, M.J.; Hernández, J.A.; Diaz-Vivancos, P. Sharka: How Do Plants Respond to Plum pox virus Infection? J. Exp. Bot. 2015, 66, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Robaglia, C.; Caranta, C. Translation Initiation Factors: A Weak Link in Plant RNA Virus Infection. Trends Plant Sci. 2006, 11, 40–45. [Google Scholar] [CrossRef]
- Rodamilans, B.; Valli, A.; García, J.A. Molecular Plant-Plum pox virus Interactions. Mol. Plant. Microbe Interact. 2020, 33, 6–17. [Google Scholar] [CrossRef]
- Rubio, J.; Sánchez, E.; Tricon, D.; Montes, C.; Eyquard, J.-P.; Chague, A.; Aguirre, C.; Prieto, H.; Decroocq, V. Silencing of One Copy of the Translation Initiation Factor EIFiso4G in Japanese Plum (Prunus salicina) Impacts Susceptibility to Plum pox virus (PPV) and Small RNA Production. BMC Plant Biol. 2019, 19, 440. [Google Scholar] [CrossRef]
- Miccoli, C.; Gambacorta, G.; Urbinati, G.; Santiago-Reyes, M.; Gentile, A.; Monticelli, S.; Caboni, E.; Prieto, H.; Verde, I.; Decroocq, V.; et al. Novel Breeding Strategies for Tackling Present and Future Challenges in Prunus Species. Acta Hortic. 2022, 1352, 419–426. [Google Scholar] [CrossRef]
- Miccoli, C.; Gambacorta, G.; Urbinati, G.; Santiago Reyes, M.; Gentile, A.; Vona, S.; Monticelli, S.; Caboni, E.; Verde, I.; Vendramin, E.; et al. Application of New Breeding Techniques to Improve Important Agronomical Traits in Prunus Species. In LXIV SIGA Annual Congress “Plant Genetic Innovation for Food Security in a Climate Change Scenario”; Italian Society of Agricultural Genetics: Portici, Italy, 2021; ISBN 978-88-944843-2-8. Available online: http://www.geneticagraria.it/congress_abstract.asp?a_pag=4&id=68#key_M (accessed on 20 December 2022).
- Glenn, D.; Bassett, C.; Tworkoski, T.; Scorza, R.; Miller, S. Tree Architecture of Pillar and Standard Peach Affect Canopy Transpiration and Water Use Efficiency. Sci. Hortic. 2015, 187, 30–34. [Google Scholar] [CrossRef]
- Hollender, C.A.; Pascal, T.; Tabb, A.; Hadiarto, T.; Srinivasan, C.; Wang, W.; Liu, Z.; Scorza, R.; Dardick, C. Loss of a Highly Conserved Sterile Alpha Motif Domain Gene (WEEP) Results in Pendulous Branch Growth in Peach Trees. Proc. Natl. Acad. Sci. USA 2018, 115, E4690–E4699. [Google Scholar] [CrossRef]
- Ku, L.; Wei, X.; Zhang, S.; Zhang, J.; Guo, S.; Chen, Y. Cloning and Characterization of a Putative TAC1 Ortholog Associated with Leaf Angle in Maize (Zea mays L.). PLoS ONE 2011, 6, e20621. [Google Scholar] [CrossRef]
- Li, H.; Zhang, L.; Hu, J.; Zhang, F.; Chen, B.; Xu, K.; Gao, G.; Li, H.; Zhang, T.; Li, Z. Genome-Wide Association Mapping Reveals the Genetic Control Underlying Branch Angle in Rapeseed (Brassica napus L.). Front. Plant Sci. 2017, 8, 1054. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Huai, Z.; Xiao, Y.; Wang, X.; Yu, J.; Ding, G.; Peng, J. Natural Variation and Genetic Analysis of the Tiller Angle Gene MsTAC1 in Miscanthus sinensis. Planta 2014, 240, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Qi, X.; Li, J.; Han, X.; Wang, J.; Jiang, Y.; Tian, Y.; Wang, Y. PzTAC and PzLAZY from a Narrow-Crown Poplar Contribute to Regulation of Branch Angles. Plant Physiol. Biochem. 2017, 118, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Flachowsky, H.; Le Roux, P.; Peil, A.; Patocchi, A.; Richter, K.; Hanke, M. Application of a High-speed Breeding Technology to Apple (Malus × domestica) Based on Transgenic Early Flowering Plants and Marker-assisted Selection. New Phytol. 2011, 192, 364–377. [Google Scholar] [CrossRef]
- Zhu, Y.; Klasfeld, S.; Jeong, C.W.; Jin, R.; Goto, K.; Yamaguchi, N.; Wagner, D. TERMINAL FLOWER 1-FD Complex Target Genes and Competition with FLOWERING LOCUS T. Nat. Commun. 2020, 11, 5118. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Nasim, Z.; Susila, H.; Ahn, J.H. Evolution and Functional Diversification of FLOWERING LOCUS T/TERMINAL FLOWER 1 Family Genes in Plants. Semin. Cell Dev. Biol. 2020, 109, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, C.; Dardick, C.; Callahan, A.; Scorza, R. Plum (Prunus domestica) Trees Transformed with Poplar FT1 Result in Altered Architecture, Dormancy Requirement, and Continuous Flowering. PLoS ONE 2012, 7, e40715. [Google Scholar] [CrossRef]
- Ferguson, A.R. Botanical Description. In The Kiwifruit Genome; Springer: Cham, Vietnam, 2016; pp. 1–13. [Google Scholar] [CrossRef]
- Datson, P.; Ferguson, A. Actinidia. In Wild Crop Relatives: Genomic and Breeding Resources; Springer: Berlin/Heidelberg, Germany, 2011; pp. 1–20. [Google Scholar]
- Ferguson, A.R.; Huang, H. Genetic Resources of Kiwifruit: Domestication and Breeding. Hortic. Rev. 2007, 33, 1–121. [Google Scholar]
- Ferrante, P.; Scortichini, M. Identification of Pseudomonas Syringae Pv. Actinidiae as Causal Agent of Bacterial Canker of Yellow Kiwifruit (Actinidia chinensis Planchon) in Central Italy. J. Phytopathol. 2009, 157, 768–770. [Google Scholar] [CrossRef]
- Donati, I.; Cellini, A.; Sangiorgio, D.; Vanneste, J.L.; Scortichini, M.; Balestra, G.M.; Spinelli, F. Pseudomonas syringae Pv. Actinidiae: Ecology, Infection Dynamics and Disease Epidemiology. Microb. Ecol. 2020, 80, 81–102. [Google Scholar] [CrossRef]
- Vanneste, J.L. The Scientific, Economic, and Social Impacts of the New Zealand Outbreak of Bacterial Canker of Kiwifruit (Pseudomonas syringae Pv. Actinidiae). Annu. Rev. Phytopathol. 2017, 55, 377–399. [Google Scholar] [CrossRef] [PubMed]
- Cellini, A.; Donati, I.; Farneti, B.; Khomenko, I.; Buriani, G.; Biasioli, F.; Cristescu, S.M.; Spinelli, F. A Breach in Plant Defences: Pseudomonas syringae Pv. Actinidiae Targets Ethylene Signalling to Overcome Actinidia chinensis Pathogen Responses. Int. J. Mol. Sci. 2021, 22, 4375. [Google Scholar] [CrossRef] [PubMed]
- Michelotti, V.; Lamontanara, A.; Buriani, G.; Orrù, L.; Cellini, A.; Donati, I.; Vanneste, J.L.; Cattivelli, L.; Tacconi, G.; Spinelli, F. Comparative Transcriptome Analysis of the Interaction between Actinidia chinensis Var. Chinensis and Pseudomonas syringae Pv. Actinidiae in Absence and Presence of Acibenzolar-S-Methyl. BMC Genom. 2018, 19, 585. [Google Scholar] [CrossRef] [PubMed]
- Cellini, A.; Fiorentini, L.; Buriani, G.; Yu, J.; Donati, I.; Cornish, D.; Novak, B.; Costa, G.; Vanneste, J.; Spinelli, F. Elicitors of the Salicylic Acid Pathway Reduce Incidence of Bacterial Canker of Kiwifruit Caused by Pseudomonas syringae Pv. Actinidae. Ann. Appl. Biol. 2014, 165, 441–453. [Google Scholar] [CrossRef]
- Collina, M.; Donati, I.; Bertacchini, E.; Brunelli, A.; Spinelli, F. Greenhouse Assays on the Control of the Bacterial Canker of Kiwifruit (Pseudomonas syringae Pv. Actinidiae). J. Berry Res. 2016, 6, 407–415. [Google Scholar] [CrossRef]
- Wu, X.; Kriz, A.J.; Sharp, P.A. Target Specificity of the CRISPR-Cas9 System. Quant. Biol. 2014, 2, 59–70. [Google Scholar] [CrossRef]
- Caboni, E.; Biasi, R.; Delia, G.; Tonelli, M. Effect of CPPU on in Vitro Axillary Shoot Proliferation and Adventitious Shoot Regeneration in Kiwifruit. Plant Biosyst. 2009, 143, 456–461. [Google Scholar] [CrossRef]
- Prado, M.; Gonzalez, M.; Romo, S.; Herrera, M. Adventitious Plant Regeneration on Leaf Explants from Adult Male Kiwifruit and AFLP Analysis of Genetic Variation. Plant Cell Tissue Organ. Cult. 2007, 88, 1–10. [Google Scholar] [CrossRef]
- Michelotti, V.; Urbinati, G.; Gentile, A.; Lucioli, S.; Caboni, E.; Tacconi, G. Preliminary Results on the Development of a Genome Editing Protocol in Actinidia chinensis Var. Chinensis as Psa Resistance Approach. Acta Hortic. 2022, 1332, 111–116. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Mbaya, H.; Lillico, S.; Kemp, S.; Simm, G.; Raybould, A. Regulatory Frameworks Can Facilitate or Hinder the Potential for Genome Editing to Contribute to Sustainable Agricultural Development. Front. Bioeng. Biotechnol. 2022, 10, 959236. [Google Scholar] [CrossRef] [PubMed]
- Weidner, C.; Edelmann, S.; Moor, D.; Lieske, K.; Savini, C.; Jacchia, S.; Sacco, M.G.; Mazzara, M.; Lämke, J.; Eckermann, K.N. Assessment of the Real-Time PCR Method Claiming to Be Specific for Detection and Quantification of the First Commercialised Genome-Edited Plant. Food Anal. Methods 2022, 15, 2107–2125. [Google Scholar] [CrossRef]
- Gallinella, C.; Cillis, G.; L’abbate, M.; Manca, A.; Pignatone, D.C.G. AC 3393. Deputy Chamber of Italy. 2021. Available online: https://documenti.camera.it/leg18/pdl/pdf/leg.18.pdl.camera.3310.18PDL0162050.pdf (accessed on 20 December 2022).
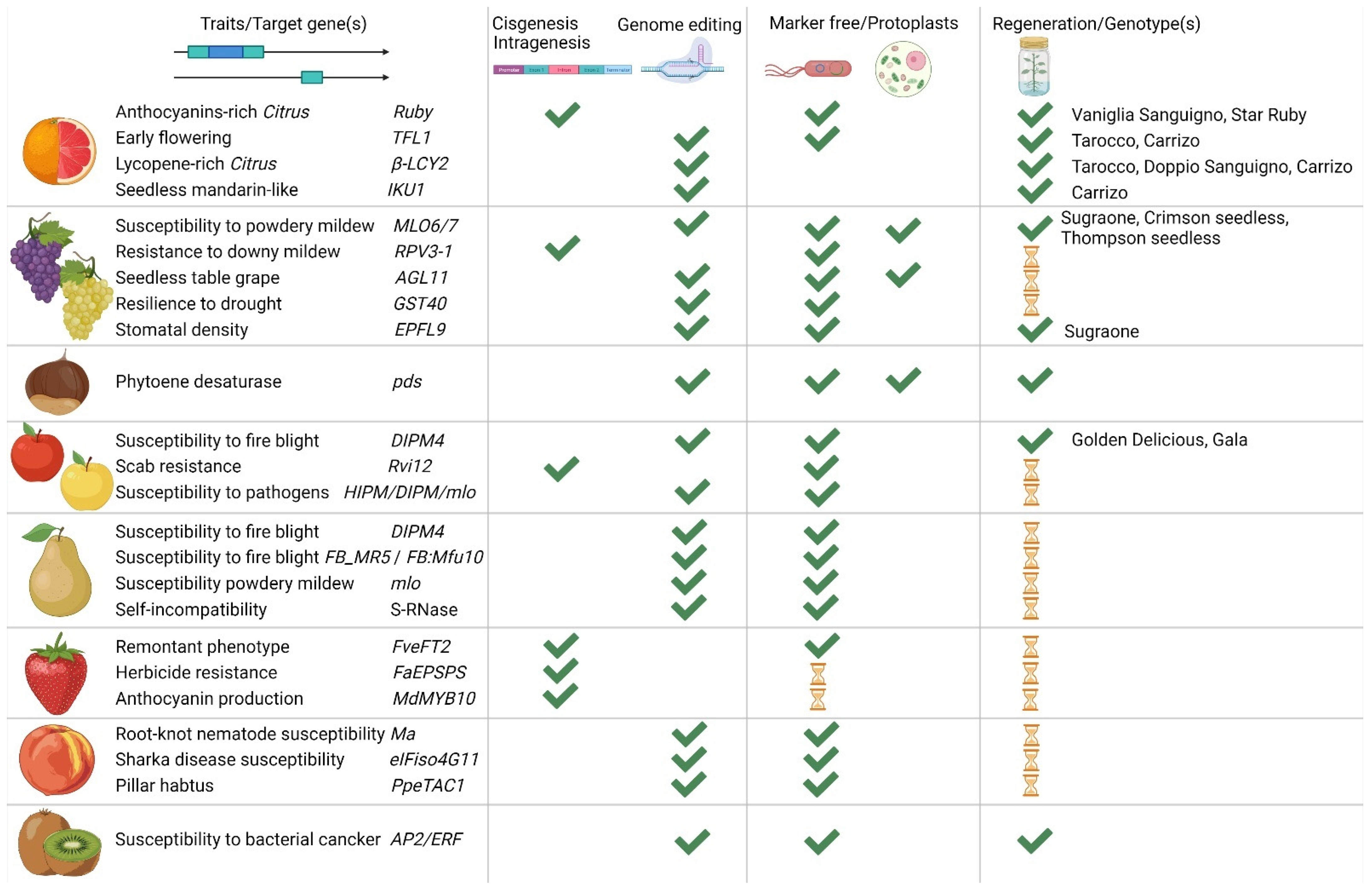
| Genus | Species | Trait | Modified Gene(s) | Approach | Ref. |
|---|---|---|---|---|---|
| Citrus | C. sinensis | Resistance to Citrus canker disease | Loss of function of CsNPR3 that represses NPR1 | CRISPR/Cas9; Protoplast transfection with Lipofectamine | [20] |
| C. paradisi | Mutation of an EBE in the promoter of LOB1 | CRISPR/Cas9; A. tumefaciens infection of grapefruit epicotyls | [21] | ||
| C. sinensis | Mutation of an EBE in the promoter of LOB1 | CRISPR/Cas9; A. tumefaciens infection of epicotyls and protoplast transfection. Improved binary vector | [21] | ||
| C. paradisi C. sinensis × Poncirus trifoliata | Loss of function of DMR6 | CRISPR/Cas9; A. tumefaciens infection of epicotyls | [22] | ||
| Vitis | V. vinifera | Resistance to Botrytis cinerea | Loss of function of VvWRKY52 | CRISPR/Cas9; A. tumefaciens infection of embryogenic callus | [23] |
| V. vinifera | Tolerance to downy mildew caused by Plasmopara viticola | Loss of function of PR4 | CRISPR/Cas9; A. tumefaciens infection of embryogenic callus | [24] | |
| V. vinifera | Resistance to powdery mildew caused by Erysiphe necator | Loss of function of VvMLO3 and VvMLO4 | CRISPR/Cas9; A. tumefaciens infection of embryogenic callus | [25] | |
| V. riparia × V. rupestris | Tolerance to Pierce’s disease and Red Blotch Disease | Disruption of the miRNA gene TAS4a/b | CRISPR/Cas9; A. tumefaciens infection of embryogenic callus | [26] | |
| V. vinifera × V. berlandieri | Control of grapevine shoot branching | Loss of function of CCD7 and CCD8 | CRISPR/Cas9; A. tumefaciens infection of embryogenic callus | [27] | |
| V. amurensis | Response to cold stress | Loss of function of PAT1 | CRISPR/Cas9; A. tumefaciens infection of embryogenic callus | [28] | |
| Castanea | C. sativa | Tolerance to Chestnut blight | Overexpression of the CsCh3 | Agrobacterium-mediated transformation of somatic embryos | [29] |
| C. dentata | Overexpression of the wheat OxO | Agrobacterium-mediated transformation of somatic embryos | [30] | ||
| Malus | Malus × domestica | Tolerance to Botryosphaeria dothidea | Loss of function of CNGC | CRISPR/Cas9; A. tumefaciens infection of leaf explants | [31] |
| Malus × domestica | Early flowering | Loss of function of TFL1 | CRISPR/Cas9; A. tumefaciens infection of leaf explants | [32] | |
| Malus × domestica | Proof of concept of base editing application | Base editing of ALS and PDS | CRISPR/Cas9; A. tumefaciens infection of leaf explants | [33] | |
| M. sieversii | Knockout of PDS | CRISPR/Cas9; A. tumefaciens infection of leaf explants | [34] | ||
| Pyrus | P. communis | Early flowering | Loss of function of TFL1 | CRISPR/Cas9; A. tumefaciens infection of leaf explants | [32] |
| P. communis | Proof of concept of base editing application | Base editing of ALS and PDS | CRISPR/Cas9; A. tumefaciens infection of leaf explants | [33] | |
| Fragaria | F. vesca | Investigation of the auxin synthesis sites during fruit and root development | Loss of function of FveYUC10 | CRISPR/Cas9; A. tumefaciens infection of leaf strips | [35] |
| Actinidia | A. chinensis | Compactness of growth habit, early flowering, and fruit development | Loss of function of AcCEN4 and AcCEN | CRISPR/Cas9; A. tumefaciens-mediated transformation of leaf explants | [36] |
| A. chinensis | Self-pollination and fast-flowering offspring. | Loss of function of SyGl and CEN-like genes | CRISPR/Cas9; A. tumefaciens-mediated transformation of leaf explants | [37] | |
| A. chinensis | Evergrowing but not early flowering phenotype | Genome editing of AcBFT2 | CRISPR/Cas9; A. tumefaciens-mediated transformation of leaf explants | [38] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nerva, L.; Dalla Costa, L.; Ciacciulli, A.; Sabbadini, S.; Pavese, V.; Dondini, L.; Vendramin, E.; Caboni, E.; Perrone, I.; Moglia, A.; et al. The Role of Italy in the Use of Advanced Plant Genomic Techniques on Fruit Trees: State of the Art and Future Perspectives. Int. J. Mol. Sci. 2023, 24, 977. https://doi.org/10.3390/ijms24020977
Nerva L, Dalla Costa L, Ciacciulli A, Sabbadini S, Pavese V, Dondini L, Vendramin E, Caboni E, Perrone I, Moglia A, et al. The Role of Italy in the Use of Advanced Plant Genomic Techniques on Fruit Trees: State of the Art and Future Perspectives. International Journal of Molecular Sciences. 2023; 24(2):977. https://doi.org/10.3390/ijms24020977
Chicago/Turabian StyleNerva, Luca, Lorenza Dalla Costa, Angelo Ciacciulli, Silvia Sabbadini, Vera Pavese, Luca Dondini, Elisa Vendramin, Emilia Caboni, Irene Perrone, Andrea Moglia, and et al. 2023. "The Role of Italy in the Use of Advanced Plant Genomic Techniques on Fruit Trees: State of the Art and Future Perspectives" International Journal of Molecular Sciences 24, no. 2: 977. https://doi.org/10.3390/ijms24020977
APA StyleNerva, L., Dalla Costa, L., Ciacciulli, A., Sabbadini, S., Pavese, V., Dondini, L., Vendramin, E., Caboni, E., Perrone, I., Moglia, A., Zenoni, S., Michelotti, V., Micali, S., La Malfa, S., Gentile, A., Tartarini, S., Mezzetti, B., Botta, R., Verde, I., ... Licciardello, C. (2023). The Role of Italy in the Use of Advanced Plant Genomic Techniques on Fruit Trees: State of the Art and Future Perspectives. International Journal of Molecular Sciences, 24(2), 977. https://doi.org/10.3390/ijms24020977















