Genome-Wide Identification and Analysis of FKBP Gene Family in Wheat (Triticum asetivum)
Abstract
:1. Introduction
2. Results
2.1. Identification of TaFKBP Gene Family in T. aestivum
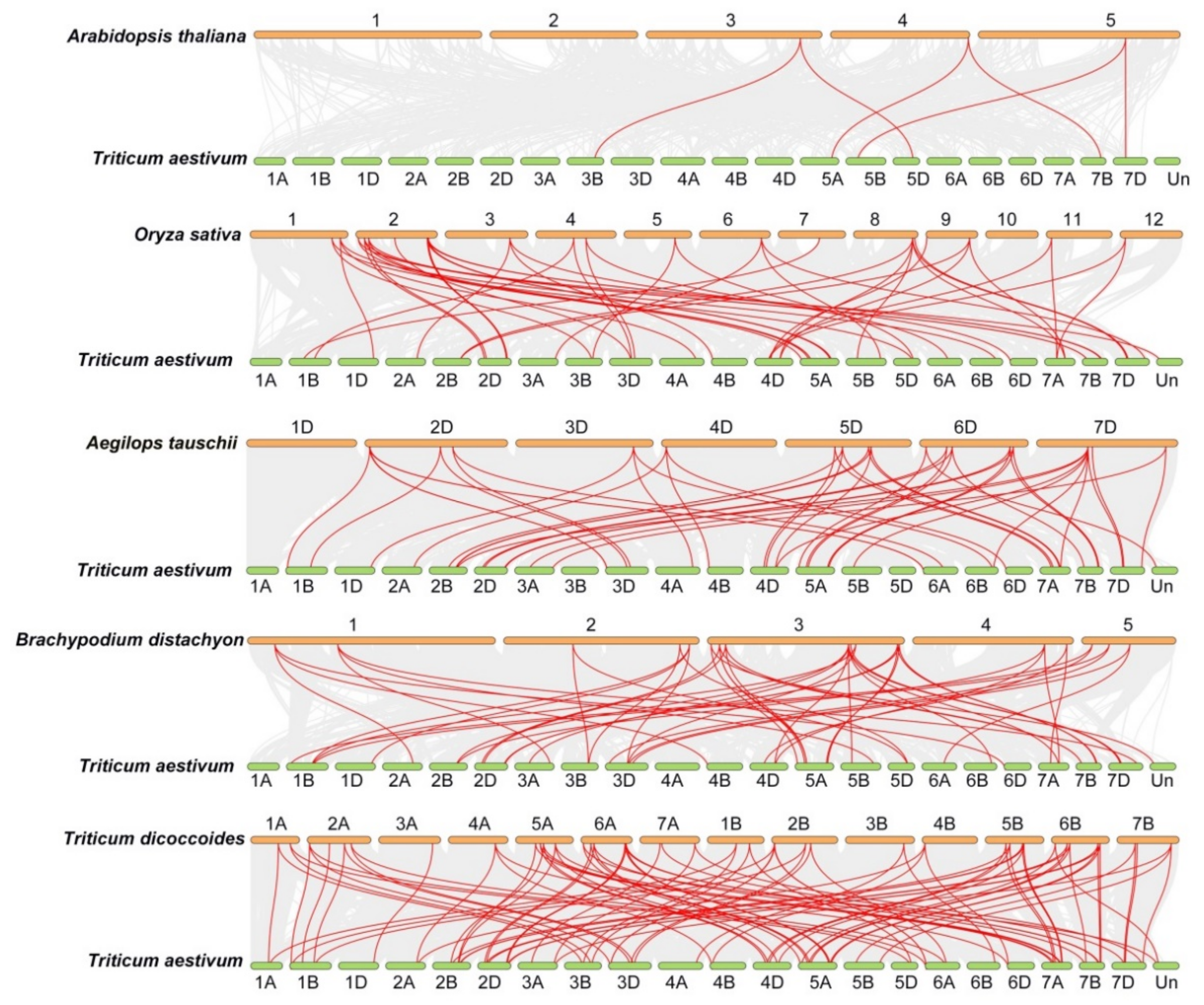
2.2. Chromosomal Distribution of TaFKBP Genes
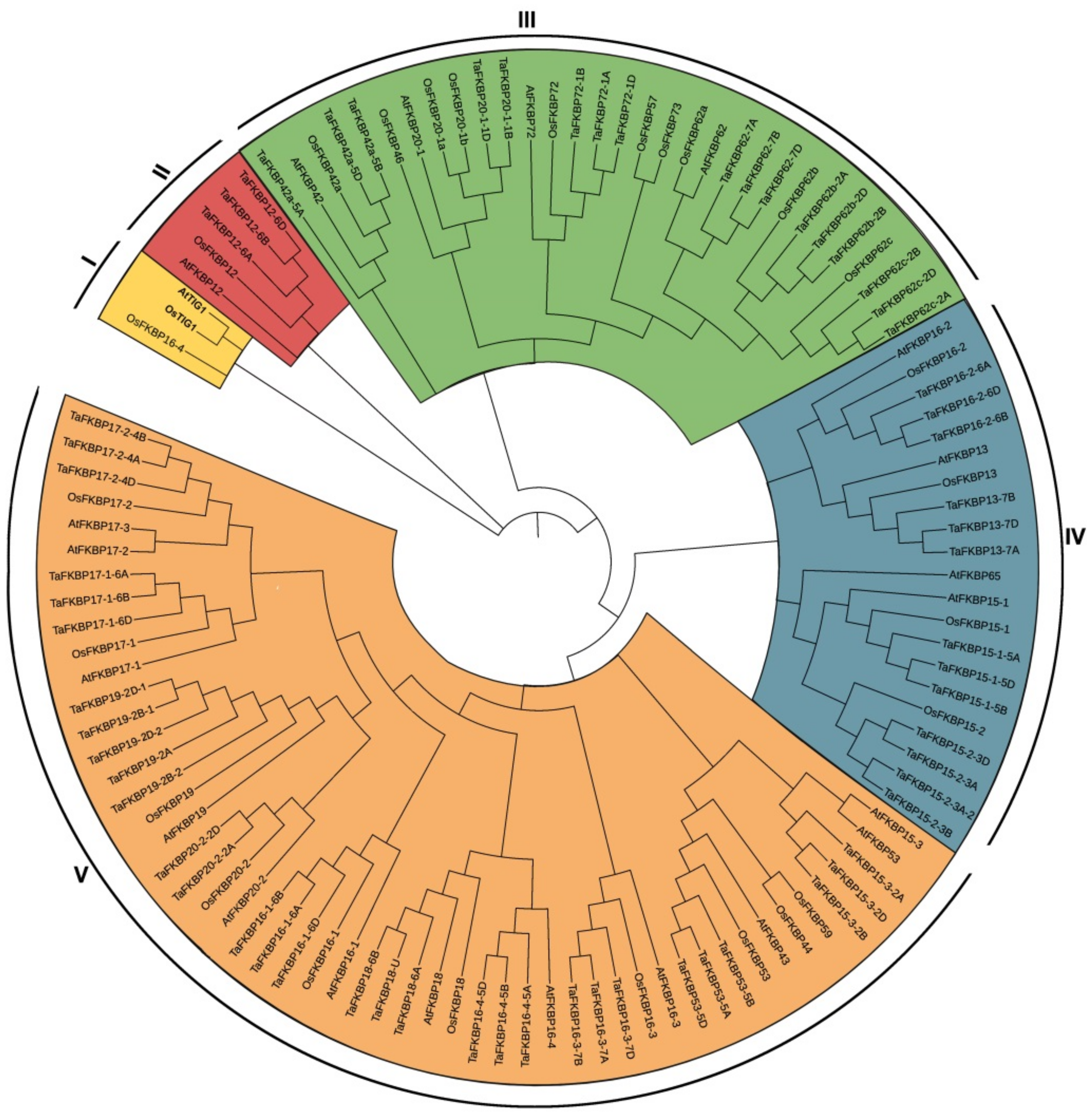
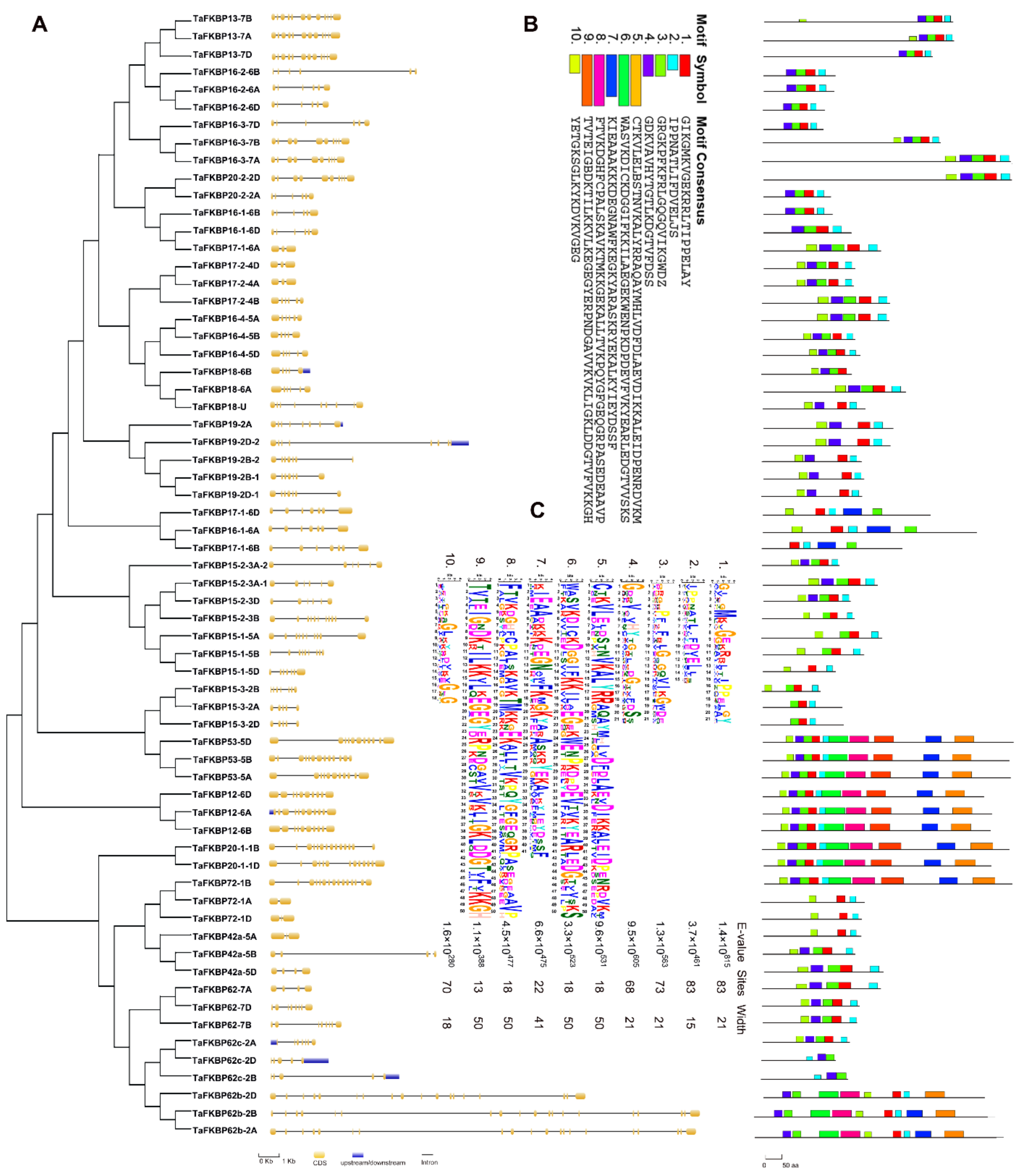
2.3. Gene Structure and Conserved Motif Analysis of TaFKBP Genes
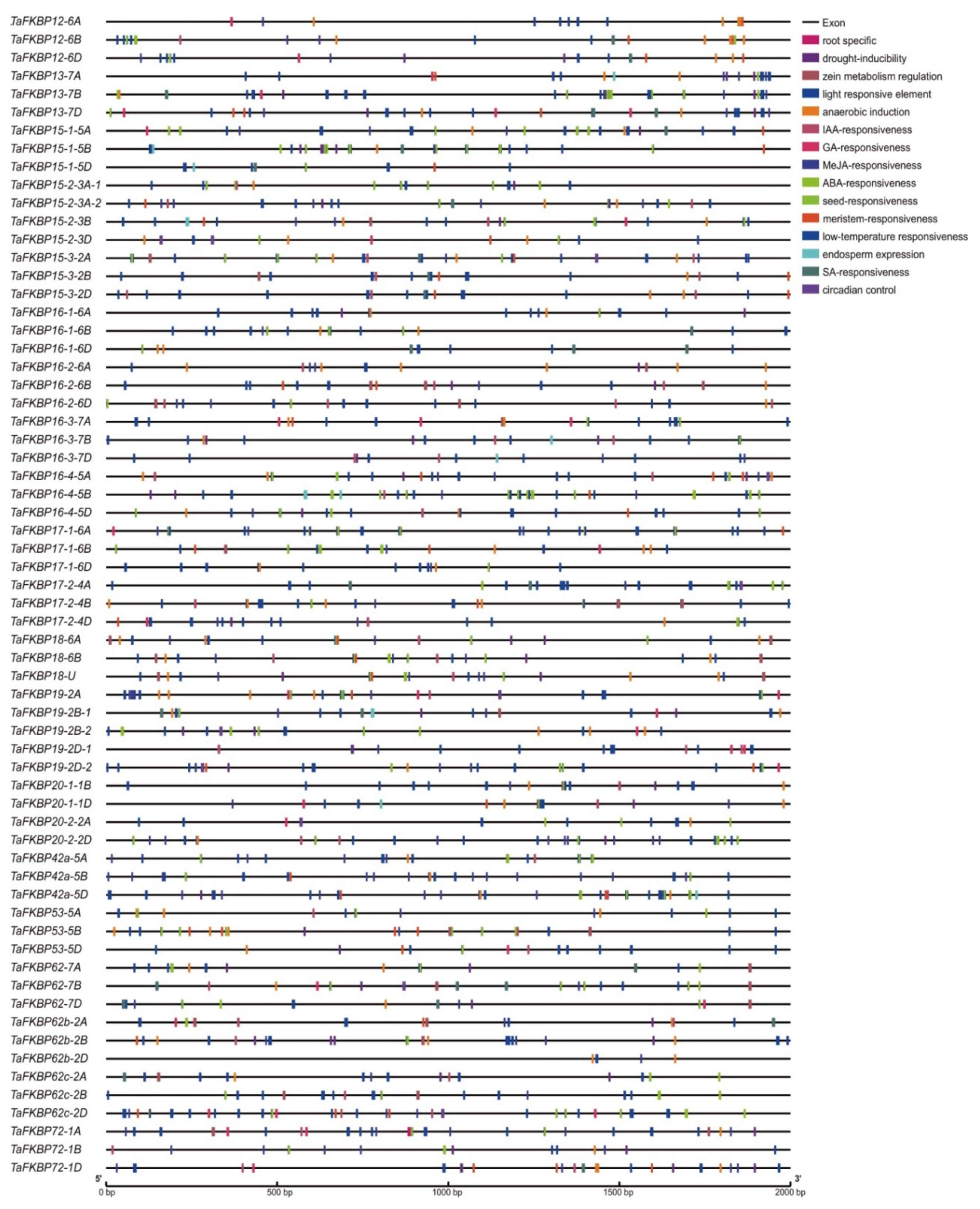
2.4. Cis-Acting Regulatory Elements Analysis of TaFKBP Genes
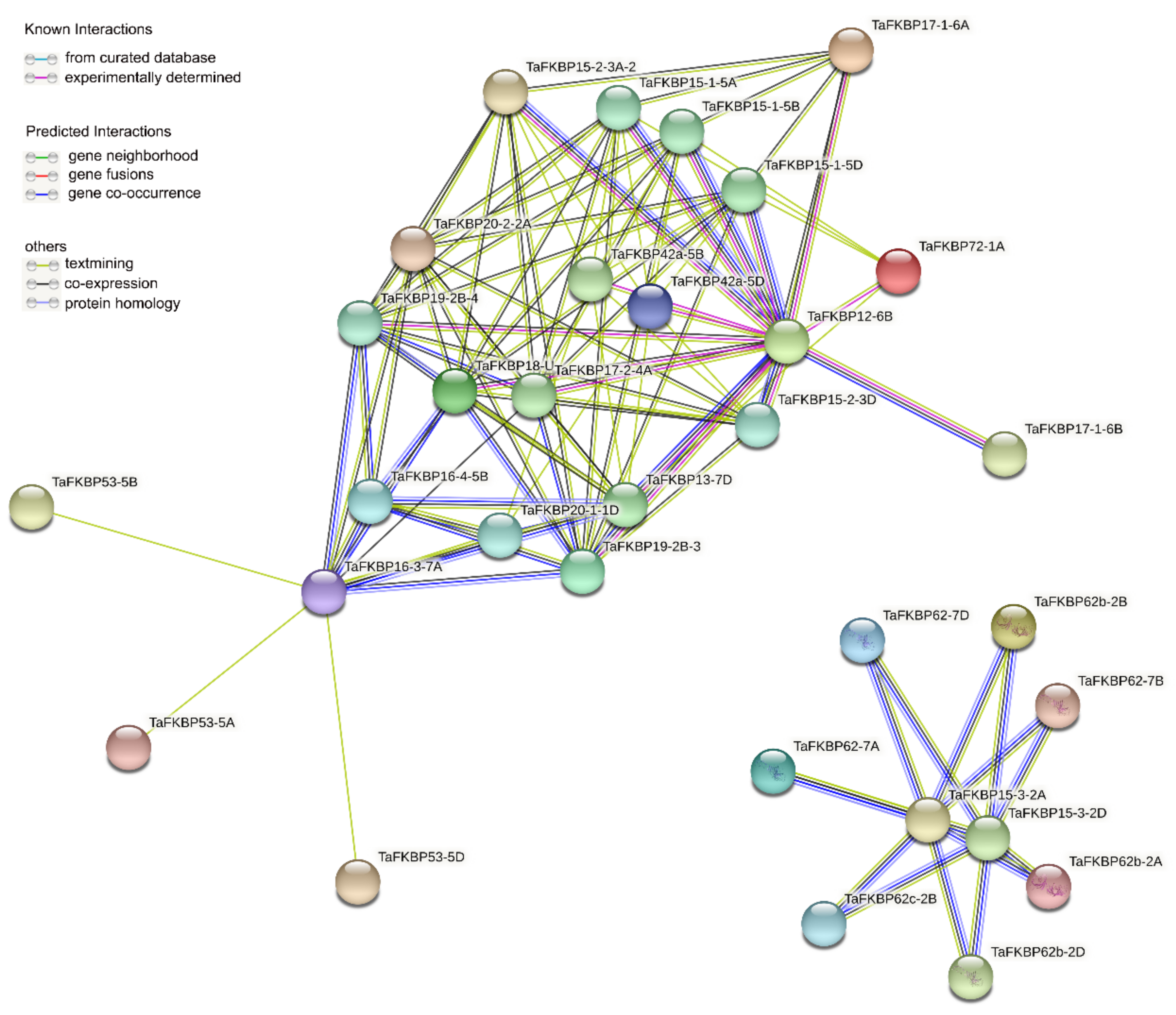
2.5. Gene Ontology (GO) Enrichment and Protein–Protein Network Analysis of TaFKBP Genes or Proteins
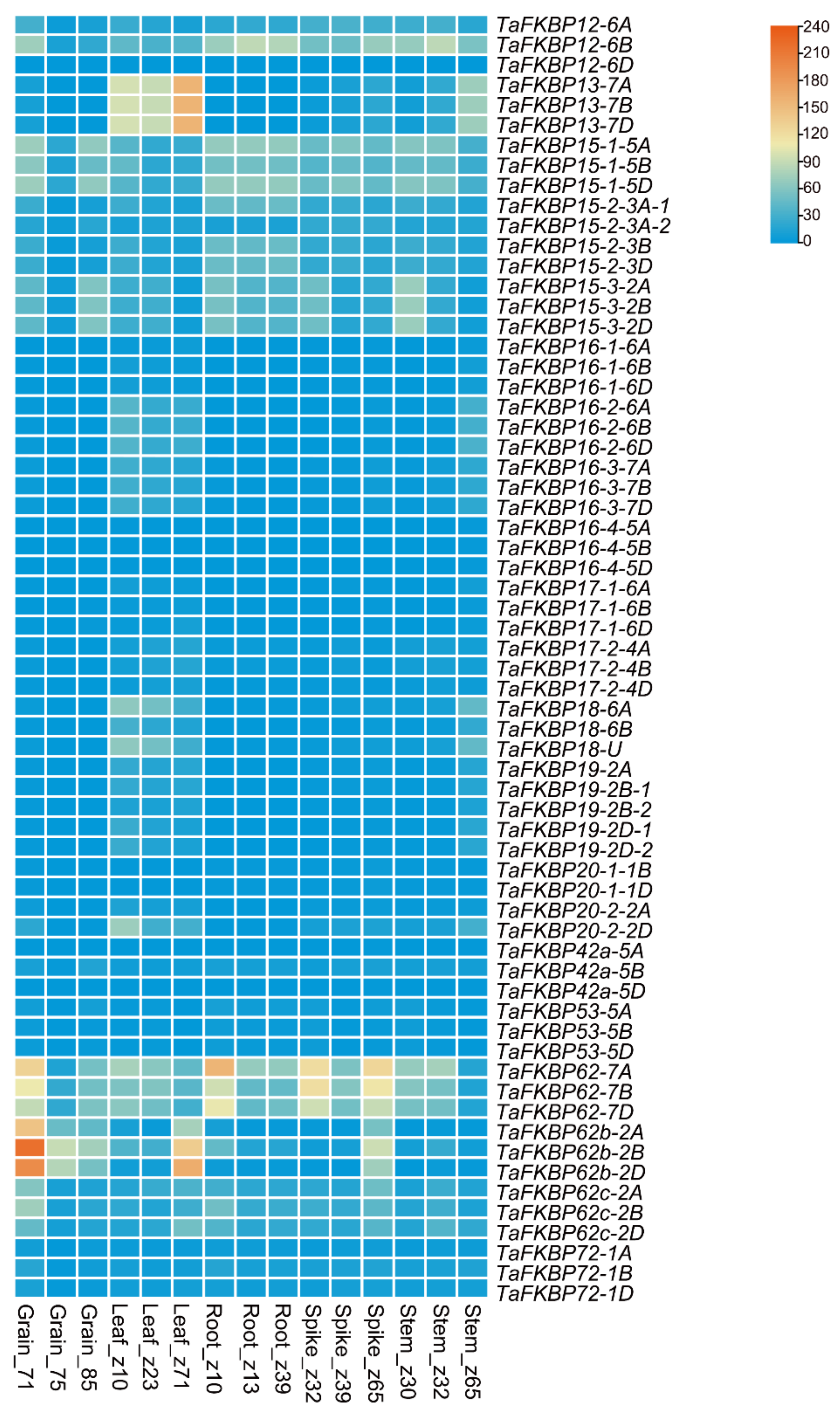
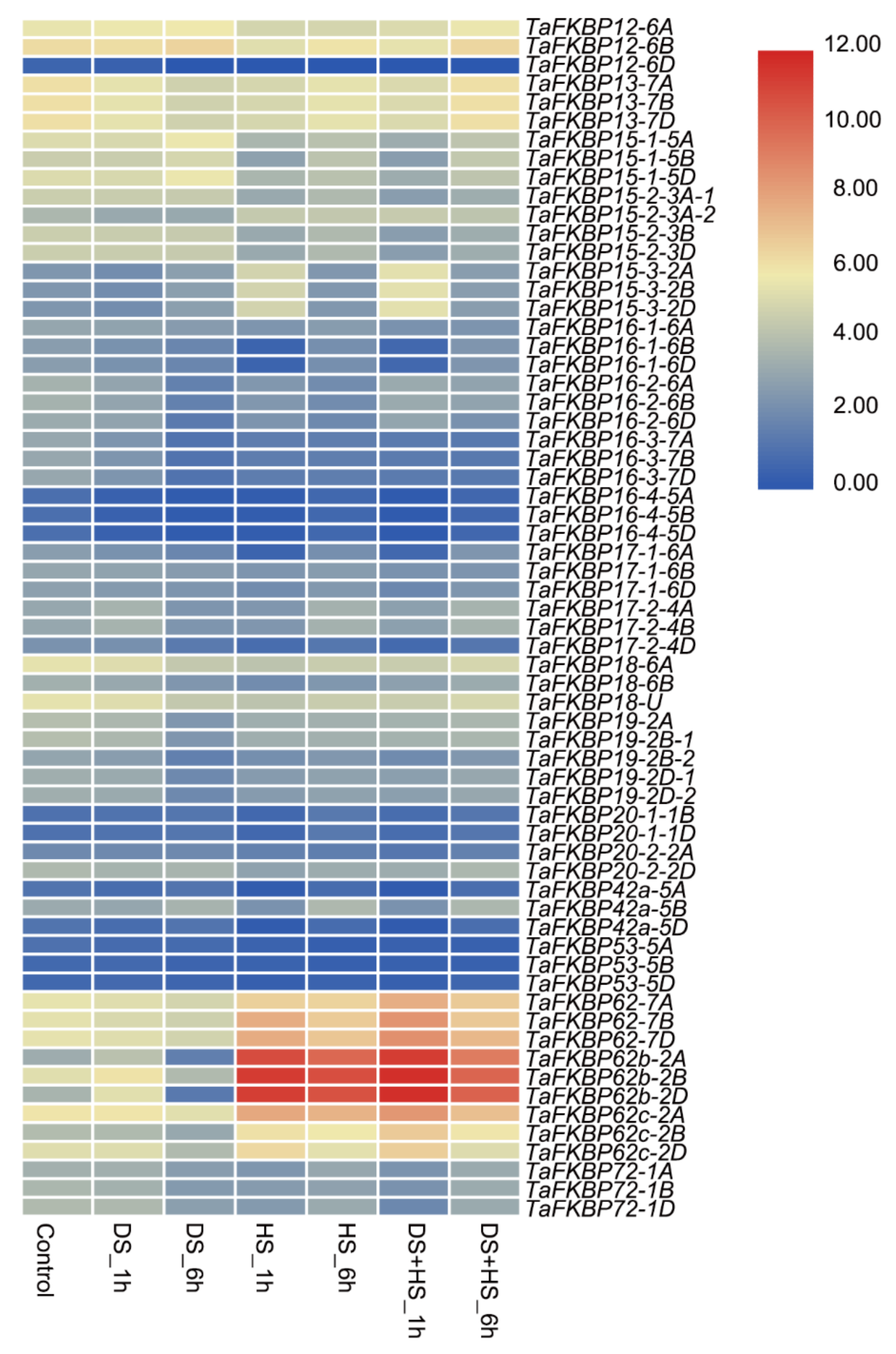
2.6. Expression Profiling of TaFKBP Genes under Different Developmental Stages and Stresses
3. Discussion
4. Materials and Methods
4.1. Identification of FKBP Genes in the Wheat Genome
4.2. Physico-Chemical Characteristics, Subcellular Localization, Gene Structure, Multiple Sequence Alignment and Construction of Phylogenetic Tree
4.3. Chromosome Localization, Motif Analysis and Gene Ontology
4.4. Cis-Acting Regulatory Elements (CAREs) Analysis and Protein Interaction Network
4.5. Expression Profiling of TaFKBP Genes
4.6. Plant Material and Growth Conditions
4.7. RNA Isolation and Real-Time PCR
4.8. Yeast Transformation
4.9. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dubcovsky, J.; Dvorak, J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 2007, 316, 1862–1866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Olejnik, P.; Madrzak, C.J.; Nuc, K. Cyclophilins and their functions in abiotic stress and plant-microbe interactions. Biomolecules 2021, 11, 1390. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Li, M.Z.; Wang, S.M.; Yin, H.J. Revisiting the role of plant transcription factors in the battle against abiotic stress. Int. J. Mol. Sci. 2018, 19, 1634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romano, P.; He, Z.; Luan, S. Introducing immunophilins. From organ transplantation to plant biology. Plant Physiol. 2004, 134, 1241–1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galat, A. Peptidylprolyl cis/trans isomerases (immunophilins): Biological diversity-targets-functions. Curr.Top. Med. Chem. 2003, 3, 1315–1347. [Google Scholar] [CrossRef]
- Vasudevan, D.; Gopalan, G.; Kumar, A.; Garcia, V.J.; Luan, S.; Swaminathan, K. Plant immunophilins: A review of their structure-function relationship. Biochim. Biophys. Acta 2015, 1850, 2145–2158. [Google Scholar] [CrossRef] [Green Version]
- Romano, P.; Gray, J.; Horton, P.; Luan, S. Plant immunophilins: Functional versatility beyond protein maturation. New Phytol. 2005, 166, 753–769. [Google Scholar] [CrossRef]
- Ghartey-Kwansah, G.; Li, Z.; Feng, R.; Wang, L.; Zhou, X.; Chen, F.Z.; Xu, M.M.; Jones, O.; Mu, Y.; Chen, S.; et al. Comparative analysis of FKBP family protein: Evaluation, structure, and function in mammals and Drosophila melanogaster. BMC Dev. Biol. 2018, 18, 7. [Google Scholar] [CrossRef] [Green Version]
- Gollan, P.J.; Bhave, M.; Aro, E.M. The FKBP families of higher plants: Exploring the structures and functions of protein interaction specialists. FEBS Lett. 2012, 586, 3539–3547. [Google Scholar] [CrossRef]
- Geisler, M.; Bailly, A. Tete-a-tete: The function of FKBPs in plant development. Trends Plant Sci. 2007, 12, 465–473. [Google Scholar] [CrossRef]
- Vallon, O. Chlamydomonas immunophilins and parvulins: Survey and critical assessment of gene models. Eukaryot Cell 2005, 4, 230–241. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, J.B.; Foor, F.; Siekierka, J.J.; Hsu, M.J.; Ramadan, N.; Morin, N.; Shafiee, A.; Dahl, A.M.; Brizuela, L.; Chrebet, G.; et al. Yeast FKBP-13 is a membrane-associated FK506-binding protein encoded by the nonessential gene FKB2. Proc. Natl. Acad. Sci. USA 1992, 89, 7471–7475. [Google Scholar] [CrossRef] [Green Version]
- Standaert, R.F.; Galat, A.; Verdine, G.L.; Schreiber, S.L. Molecular cloning and overexpression of the human FK506-binding protein FKBP. Nature 1990, 346, 671–674. [Google Scholar] [CrossRef]
- Fan, G.; Yang, Y.; Li, T.; Lu, W.; Du, Y.; Qiang, X.; Wen, Q.; Shan, W. A phytophthora capsici RXLR effector targets and inhibits a plant PPIase to suppress endoplasmic reticulum-mediated immunity. Mol. Plant 2018, 11, 1067–1083. [Google Scholar] [CrossRef] [Green Version]
- Pogorelko, G.V.; Mokryakova, M.; Fursova, O.V.; Abdeeva, I.; Piruzian, E.S.; Bruskin, S.A. Characterization of three Arabidopsis thaliana immunophilin genes involved in the plant defense response against Pseudomonas syringae. Gene 2014, 538, 12–22. [Google Scholar] [CrossRef]
- Meiri, D.; Tazat, K.; Cohen-Peer, R.; Farchi-Pisanty, O.; Aviezer-Hagai, K.; Avni, A.; Breiman, A. Involvement of Arabidopsis ROF2 (FKBP65) in thermotolerance. Plant Mol. Biol. 2010, 72, 191–203. [Google Scholar] [CrossRef]
- Meiri, D.; Breiman, A. Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J. 2009, 59, 387–399. [Google Scholar] [CrossRef]
- Charng, Y.Y.; Liu, H.C.; Liu, N.Y.; Chi, W.T.; Wang, C.N.; Chang, S.H.; Wang, T.T. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 2007, 143, 251–262. [Google Scholar] [CrossRef] [Green Version]
- Thirumalaikumar, V.P.; Gorka, M.; Schulz, K.; Masclaux-Daubresse, C.; Sampathkumar, A.; Skirycz, A.; Vierstra, R.D.; Balazadeh, S. Selective autophagy regulates heat stress memory in Arabidopsis by NBR1-mediated targeting of HSP90.1 and ROF1. Autophagy 2021, 17, 2184–2199. [Google Scholar] [CrossRef]
- Karali, D.; Oxley, D.; Runions, J.; Ktistakis, N.; Farmaki, T. The Arabidopsis thaliana immunophilin ROF1 directly interacts with PI(3)P and PI(3,5)P2 and affects germination under osmotic stress. PLoS ONE 2012, 7, e48241. [Google Scholar] [CrossRef] [Green Version]
- Alavilli, H.; Lee, H.; Park, M.; Yun, D.J.; Lee, B.H. Enhanced multiple stress tolerance in Arabidopsis by overexpression of the polar moss peptidyl prolyl isomerase FKBP12 gene. Plant Cell Rep. 2018, 37, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Nigam, N.; Singh, A.; Sahi, C.; Chandramouli, A.; Grover, A. SUMO-conjugating enzyme (Sce) and FK506-binding protein (FKBP) encoding rice (Oryza sativa L.) genes: Genome-wide analysis, expression studies and evidence for their involvement in abiotic stress response. Mol. Genet. Genom. 2008, 279, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Gollan, P.J.; Bhave, M. Genome-wide analysis of genes encoding FK506-binding proteins in rice. Plant Mol. Biol. 2010, 72, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Mao, K.; Duan, D.; Zhao, S.; Wang, Y.; Wang, Q.; Huang, D.; Li, C.; Liu, C.; Gong, X.; et al. Genome-wide analyses of genes encoding FK506-binding proteins reveal their involvement in abiotic stress responses in apple. BMC Genom. 2018, 19, 707. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Zhang, H.; Li, W.; Mu, C.; Zhang, F.; Wang, L.; Meng, Z. Genome-wide analysis and environmental response profiling of the FK506-binding protein gene family in maize (Zea mays L.). Gene 2012, 498, 212–222. [Google Scholar] [CrossRef]
- He, Z.; Li, L.; Luan, S. Immunophilins and parvulins. Superfamily of peptidyl prolyl isomerases in Arabidopsis. Plant Physiol. 2004, 134, 1248–1267. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.S.; Chen, Y.C.; Lu, C.H.; Hwang, J.K. Prediction of protein subcellular localization. Proteins 2006, 64, 643–651. [Google Scholar] [CrossRef]
- Rulten, S.; Thorpe, J.; Kay, J. Identification of eukaryotic parvulin homologues: A new subfamily of peptidylprolyl cis-trans isomerases. Biochem. Biophys. Res.Commun. 1999, 259, 557–562. [Google Scholar] [CrossRef]
- Abdeeva, I.A.; Pogorelko, G.V.; Maloshenok, L.G.; Mokrykova, M.V.; Fursova, O.V.; Bruskin, S.A. Search for partner proteins of A. thaliana immunophilins involved in the control of plant immunity. Molecules 2018, 23, 953. [Google Scholar] [CrossRef]
- Parada, C.A.; de Oliveira, I.P.; Gewehr, M.C.F.; Machado-Neto, J.A.; Lima, K.; Eichler, R.A.S.; Lopes, L.R.; Bechara, L.R.G.; Ferreira, J.C.B.; Festuccia, W.T.; et al. Effect of FKBP12-derived intracellular peptides on rapamycin-induced FKBP-FRB interaction and autophagy. Cells 2022, 11, 385. [Google Scholar] [CrossRef]
- Waseem, M.; Ahmad, F.; Habib, S.; Gao, Y.; Li, Z. Genome-wide identification of FK506-binding domain protein gene family, its characterization, and expression analysis in tomato (Solanum lycopersicum L.). Gene 2018, 678, 143–154. [Google Scholar] [CrossRef]
- Zhang, Y.; Han, J.; Liu, D.; Wen, X.; Li, Y.; Tao, R.; Peng, Y.; Fang, J.; Wang, C. Genome-wide identification and analysis of FK506-binding protein gene family in peach (Prunus persica). Gene 2014, 536, 416–424. [Google Scholar] [CrossRef]
- Leng, X.; Liu, D.; Zhao, M.; Sun, X.; Li, Y.; Mu, Q.; Zhu, X.; Li, P.; Fang, J. Genome-wide identification and analysis of FK506-binding protein family gene family in strawberry (Fragaria x ananassa). Gene 2014, 534, 390–399. [Google Scholar] [CrossRef]
- Wang, W.W.; Ma, Q.; Xiang, Y.; Zhu, S.W.; Cheng, B.J. Genome-wide analysis of immunophilin FKBP genes and expression patterns in Zea mays. Genet. Mol. Res. 2012, 11, 1690–1700. [Google Scholar] [CrossRef]
- Kesawat, M.S.; Kherawat, B.S.; Singh, A.; Dey, P.; Routray, S.; Mohapatra, C.; Saha, D.; Ram, C.; Siddique, K.H.M.; Kumar, A.; et al. Genome-wide analysis and characterization of the proline-rich extensin-like receptor kinases (PERKs) gene family reveals their role in different developmental stages and stress conditions in wheat (Triticum aestivum L.). Plants 2022, 11, 496. [Google Scholar] [CrossRef]
- Kumar, M.; Kherawat, B.S.; Dey, P.; Saha, D.; Singh, A.; Bhatia, S.K.; Ghodake, G.S.; Kadam, A.A.; Kim, H.U.; Manorama; et al. Genome-wide identification and characterization of PIN-FORMED (PIN) gene family reveals role in developmental and various stress conditions in Triticum aestivum L. Int. J. Mol. Sci. 2021, 22, 7396. [Google Scholar] [CrossRef]
- Kesawat, M.S.; Kherawat, B.S.; Singh, A.; Dey, P.; Kabi, M.; Debnath, D.; Saha, D.; Khandual, A.; Rout, S.; Manorama; et al. Genome-wide identification and characterization of the Brassinazole-resistant (BZR) gene family and its expression in the various developmental stage and stress conditions in wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2021, 22, 8743. [Google Scholar] [CrossRef]
- Zhu, T.; Liu, Y.; Ma, L.; Wang, X.; Zhang, D.; Han, Y.; Ding, Q.; Ma, L. Genome-wide identification, phylogeny and expression analysis of the SPL gene family in wheat. BMC Plant Biol. 2020, 20, 420. [Google Scholar] [CrossRef]
- Ye, H.; Qiao, L.; Guo, H.; Guo, L.; Ren, F.; Bai, J.; Wang, Y. Genome-wide identification of wheat WRKY gene family reveals that TaWRKY75-A is referred to drought and salt resistances. Front. Plant Sci. 2021, 12, 663118. [Google Scholar] [CrossRef]
- Shao, W.; Chen, W.; Zhu, X.; Zhou, X.; Jin, Y.; Zhan, C.; Liu, G.; Liu, X.; Ma, D.; Qiao, Y. Genome-wide identification and characterization of wheat 14-3-3 genes unravels the role of TaGRF6-A in salt stress tolerance by binding MYB transcription factor. Int. J. Mol. Sci. 2021, 22, 1904. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Liu, Z.; Guo, L.; Li, H.; Nie, X.; Chai, S.; Zheng, W. Genome-wide identification of wheat ZIP gene family and functional characterization of the TaZIP13-B in plants. Front. Plant Sci. 2021, 12, 748146. [Google Scholar] [CrossRef] [PubMed]
- Rohr, M.; Ries, F.; Herkt, C.; Gotsmann, V.L.; Westrich, L.D.; Gries, K.; Trosch, R.; Christmann, J.; Chaux-Jukic, F.; Jung, M.; et al. The role of plastidic trigger factor serving protein biogenesis in green algae and land plants. Plant Physiol. 2019, 179, 1093–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, J.C.; Kim, D.W.; You, Y.N.; Seok, M.S.; Park, J.M.; Hwang, H.; Kim, B.G.; Luan, S.; Park, H.S.; Cho, H.S. Classification of rice (Oryza sativa L. Japonica nipponbare) immunophilins (FKBPs, CYPs) and expression patterns under water stress. BMC Plant Biol. 2010, 10, 253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serrano-Bueno, G.; Said, F.E.; de Los Reyes, P.; Lucas-Reina, E.I.; Ortiz-Marchena, M.I.; Romero, J.M.; Valverde, F. CONSTANS-FKBP12 interaction contributes to modulation of photoperiodic flowering in Arabidopsis. Plant J. 2020, 101, 1287–1302. [Google Scholar] [CrossRef] [PubMed]
- Deng, K.; Yu, L.; Zheng, X.; Zhang, K.; Wang, W.; Dong, P.; Zhang, J.; Ren, M. Target of rapamycin is a key player for auxin signaling transduction in Arabidopsis. Front. Plant Sci. 2016, 7, 291. [Google Scholar] [CrossRef] [Green Version]
- Chauve, C.; Doyon, J.P.; El-Mabrouk, N. Gene family evolution by duplication, speciation, and loss. J. Comput. Biol. 2008, 15, 1043–1062. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Gaut, B.S.; Vision, T.J. Gene duplication and evolution. Science 2001, 293, 1551. [Google Scholar]
- Wang, J.; Sun, W.; Kong, X.; Zhao, C.; Li, J.; Chen, Y.; Gao, Z.; Zuo, K. The peptidyl-prolyl isomerases FKBP15-1 and FKBP15-2 negatively affect lateral root development by repressing the vacuolar invertase VIN2 in Arabidopsis. Planta 2020, 252, 52. [Google Scholar] [CrossRef]
- Cheung, M.Y.; Auyeung, W.K.; Li, K.P.; Lam, H.M. A Rice immunophilin homolog, OsFKBP12, is a negative regulator of both biotic and abiotic stress responses. Int. J. Mol. Sci. 2020, 21, 8791. [Google Scholar] [CrossRef]
- Niu, X.W.; Zheng, Z.Y.; Feng, Y.G.; Guo, W.Z.; Wang, X.Y. The fusarium graminearum virulence factor FGL targets an FKBP12 immunophilin of wheat. Gene 2013, 525, 77–83. [Google Scholar] [CrossRef]
- Xiong, F.; Liu, M.; Zhuo, F.; Yin, H.; Deng, K.; Feng, S.; Liu, Y.; Luo, X.; Feng, L.; Zhang, S.; et al. Host-induced gene silencing of BcTOR in Botrytis cinerea enhances plant resistance to grey mould. Mol. Plant Pathol. 2019, 20, 1722–1739. [Google Scholar] [CrossRef] [Green Version]
- Seok, M.S.; You, Y.N.; Park, H.J.; Lee, S.S.; Aigen, F.; Luan, S.; Ahn, J.C.; Cho, H.S. AtFKBP16-1, a chloroplast lumenal immunophilin, mediates response to photosynthetic stress by regulating PsaL stability. Physiol. Plant 2014, 150, 620–631. [Google Scholar] [CrossRef] [Green Version]
- Kurek, I.; Harvey, A.J.; Lonsdale, D.M.; Breiman, A. Isolation and characterization of the wheat prolyl isomerase FK506-binding protein (FKBP) 73 promoter. Plant Mol. Biol. 2000, 42, 489–497. [Google Scholar] [CrossRef]
- Alok, A.; Kaur, J.; Tiwari, S. Functional characterization of wheat myo-inositol oxygenase promoter under different abiotic stress conditions in Arabidopsis thaliana. Biotechnol. Lett. 2020, 42, 2035–2047. [Google Scholar] [CrossRef]
- Liu, J.; Ghelli, R.; Cardarelli, M.; Geisler, M. Arabidopsis TWISTED DWARF1 regulates stamen elongation by differential activation of ABCB1,19-mediated auxin transport. J. Exp. Bot. 2022, 73, 4818–4831. [Google Scholar] [CrossRef]
- Bailly, A.; Wang, B.; Zwiewka, M.; Pollmann, S.; Schenck, D.; Luthen, H.; Schulz, A.; Friml, J.; Geisler, M. Expression of TWISTED DWARF1 lacking its in-plane membrane anchor leads to increased cell elongation and hypermorphic growth. Plant J. 2014, 77, 108–118. [Google Scholar] [CrossRef]
- Bissoli, G.; Ninoles, R.; Fresquet, S.; Palombieri, S.; Bueso, E.; Rubio, L.; Garcia-Sanchez, M.J.; Fernandez, J.A.; Mulet, J.M.; Serrano, R. Peptidyl-prolyl cis-trans isomerase ROF2 modulates intracellular pH homeostasis in Arabidopsis. Plant J. 2012, 70, 704–716. [Google Scholar] [CrossRef] [Green Version]
- Zannas, A.S.; Wiechmann, T.; Gassen, N.C.; Binder, E.B. Gene-stress-epigenetic regulation of FKBP5: Clinical and translational implications. Neuropsychopharmacology 2016, 41, 261–274. [Google Scholar] [CrossRef] [Green Version]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, 412–419. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, 458–460. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, 200–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [PubMed]
- Yu, C.S.; Lin, C.J.; Hwang, J.K. Predicting subcellular localization of proteins for gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004, 13, 1402–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, 585–587. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Jiangtao, C.; Yingzhen, K.; Qian, W.; Yuhe, S.; Daping, G.; Jing, L.; Guanshan, L. MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Yi Chuan 2015, 37, 91–97. [Google Scholar]
- Chen, Y.; Song, W.; Xie, X.; Wang, Z.; Guan, P.; Peng, H.; Jiao, Y.; Ni, Z.; Sun, Q.; Guo, W. A Collinearity-Incorporating Homology inference strategy for connecting emerging assemblies in the Triticeae Tribe as a pilot practice in the plant pangenomic era. Mol. Plant 2020, 13, 1694–1708. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernandez-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, 309–314. [Google Scholar] [CrossRef] [Green Version]
- Cantalapiedra, C.P.; Hernandez-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Rombauts, S.; Dehais, P.; Van Montagu, M.; Rouze, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative Toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, 447–452. [Google Scholar] [CrossRef]
- Pearce, S.; Vazquez-Gross, H.; Herin, S.Y.; Hane, D.; Wang, Y.; Gu, Y.Q.; Dubcovsky, J. WheatExp: An RNA-seq expression database for polyploid wheat. BMC Plant Biol. 2015, 15, 299. [Google Scholar] [CrossRef] [Green Version]
- Li, G.Z.; Li, H.X.; Xu, M.J.; Wang, P.F.; Xiao, X.H.; Kang, G.Z. Functional characterization and regulatory mechanism of wheat CPK34 kinase in response to drought stress. BMC Genom. 2020, 21, 577. [Google Scholar] [CrossRef]
- Gutierrez-Lomeli, M.; Torres-Guzman, J.C.; Gonzalez-Hernandez, G.A.; Cira-Chavez, L.A.; Pelayo-Ortiz, C.; Ramirez-Cordova Jde, J. Overexpression of ADH1 and HXT1 genes in the yeast Saccharomyces cerevisiae improves the fermentative efficiency during tequila elaboration. Antonie Leeuw. Int. J. G 2008, 93, 363–371. [Google Scholar] [CrossRef]


| Name | Gene ID | GRAVY | MW | Start | End | Chromosome | CDS Length (bp) | Protein Length (aa) | PI | Localization |
|---|---|---|---|---|---|---|---|---|---|---|
| TaFKBP72-1A | TraesCS1A02G184600 | 0.55 | 69.61 | 335,012,317 | 335,034,601 | 1A | 2332 | 624 | 5.75 | Nuclear, Cytoplasmic |
| TaFKBP72-1B | TraesCS1B02G192700 | 0.48 | 67.97 | 345,303,083 | 345,325,533 | 1B | 2055 | 607 | 5.52 | Chloroplast, Mitochondrial |
| TaFKBP20-1-1B | TraesCS1B02G292400 | 0.42 | 21.78 | 509,891,166 | 509,892,675 | 1B | 609 | 202 | 7.35 | Nuclear, Cytoplasmic |
| TaFKBP72-1D | TraesCS1D02G192100 | 0.51 | 63.98 | 267,788,383 | 267,804,883 | 1D | 2195 | 575 | 5.26 | Nuclear, Cytoplasmic |
| TaFKBP20-1-1D | TraesCS1D02G282500 | 0.41 | 21.74 | 379,796,335 | 379,797,841 | 1D | 609 | 202 | 7.35 | Nuclear, Cytoplasmic |
| TaFKBP62c-2A | TraesCS2A02G050600 | 0.52 | 71.81 | 19,645,993 | 19,652,506 | 2A | 2508 | 645 | 5.08 | Cytoplasmic, Endoplasmic reticulum |
| TaFKBP19-2A | TraesCS2A02G053200 | 0.49 | 29.04 | 21,261,065 | 21,267,439 | 2A | 1184 | 263 | 10.48 | Mitochondrial |
| TaFKBP20-2-2A | TraesCS2A02G226800 | 0.40 | 27.14 | 236,827,327 | 236,834,045 | 2A | 735 | 244 | 9.88 | Chloroplast, Mitochondrial |
| TaFKBP62b-2A | TraesCS2A02G277100 | 0.67 | 64.9 | 45,805,0841 | 45,8054,214 | 2A | 2289 | 582 | 4.94 | Cytoplasmic, Nuclear |
| TaFKBP15-3-2A | TraesCS2A02G314800 | 0.97 | 48.84 | 539,964,818 | 539,968,229 | 2A | 1829 | 444 | 6.23 | Nuclear |
| TaFKBP62c-2B | TraesCS2B02G063900 | 0.52 | 71.31 | 30,494,467 | 30,499,674 | 2B | 2404 | 638 | 5.24 | Cytoplasmic, Endoplasmic reticulum |
| TaFKBP19-2B-1 | TraesCS2B02G067100 | 0.36 | 24.28 | 33,848,383 | 33,851,216 | 2B | 663 | 220 | 9.59 | Chloroplast |
| TaFKBP19-2B-2 | TraesCS2B02G067200 | 0.48 | 16.83 | 33,907,018 | 33,908,399 | 2B | 506 | 151 | 5.31 | Cytoplasmic, Nuclear |
| TaFKBP62b-2B | TraesCS2B02G294500 | 0.69 | 65.61 | 411,140,309 | 411,143,734 | 2B | 2382 | 587 | 4.82 | Nuclear, Cytoplasmic |
| TaFKBP15-3-2B | TraesCS2B02G333300 | 1.00 | 53.47 | 476,557,369 | 476,560,999 | 2B | 1960 | 487 | 5.52 | Nuclear |
| TaFKBP62c-2D | TraesCS2D02G050300 | 0.51 | 72.08 | 18,642,151 | 18,646,485 | 2D | 2174 | 649 | 4.99 | Endoplasmic reticulum, Cytoplasmic |
| TaFKBP19-2D-1 | TraesCS2D02G053100 | 0.40 | 20.91 | 20,661,350 | 20,663,246 | 2D | 922 | 187 | 8.70 | Cytoplasmic, Nuclear |
| TaFKBP19-2D-2 | TraesCS2D02G053400 | 0.46 | 29.09 | 20,778,047 | 20,783,120 | 2D | 1307 | 264 | 10.40 | Mitochondrial, Chloroplast |
| TaFKBP20-2-2D | TraesCS2D02G239700 | 0.44 | 21.03 | 255,681,859 | 255,683,584 | 2D | 579 | 192 | 10.52 | Chloroplast |
| TaFKBP62b-2D | TraesCS2D02G276000 | 0.65 | 65.45 | 345,623,668 | 345,626,945 | 2D | 2095 | 550 | 6.59 | Cytoplasmic, Nuclear |
| TaFKBP15-3-2D | TraesCS2D02G313000 | 1.02 | 53.45 | 402,709,856 | 402,713,456 | 2D | 2102 | 487 | 5.28 | Nuclear |
| TaFKBP15-2-3A-1 | TraesCS3A02G421300 | 0.32 | 17.53 | 66,281,3307 | 66,282,0850 | 3A | 471 | 156 | 5.78 | Secreted |
| TaFKBP15-2-3A-2 | TraesCS3A02G422000 | 0.19 | 16.69 | 663,292,556 | 663,297,701 | 3A | 728 | 158 | 9.10 | Secreted |
| TaFKBP15-2-3B | TraesCS3B02G457500 | 0.18 | 16.72 | 699,693,748 | 699,696,737 | 3B | 792 | 158 | 9.46 | Cytoplasmic, Mitochondrial, |
| TaFKBP15-2-3D | TraesCS3D02G417400 | 0.18 | 16.52 | 529,138,315 | 529,141,328 | 3D | 786 | 156 | 8.47 | Secreted |
| TaFKBP17-2-4A | TraesCS4A02G270400 | 0.31 | 26.4 | 581,749,445 | 581,750,928 | 4A | 1318 | 251 | 5.12 | Chloroplast |
| TaFKBP17-2-4B | TraesCS4B02G043700 | 0.32 | 26.76 | 31,849,101 | 31,850,336 | 4B | 1092 | 255 | 5.40 | Chloroplast |
| TaFKBP17-2-4D | TraesCS4D02G041200 | 0.29 | 26.25 | 19,398,540 | 19,399,673 | 4D | 1038 | 250 | 6.02 | Chloroplast |
| TaFKBP42a-5A | TraesCS5A02G134500 | 0.58 | 36.28 | 303,282,052 | 303,287,233 | 5A | 1414 | 315 | 9.97 | Chloroplast |
| TaFKBP53-5A | TraesCS5A02G158900 | 0.90 | 47.59 | 338,893,087 | 338,896,927 | 5A | 1810 | 432 | 5.24 | Nuclear |
| TaFKBP16-4-5A | TraesCS5A02G264500 | 0.06 | 23.29 | 476,682,552 | 476,687,413 | 5A | 1069 | 223 | 10.66 | Chloroplast |
| TaFKBP15-1-5A | TraesCS5A02G279500 | 0.15 | 16.14 | 488,408,951 | 488,411,182 | 5A | 665 | 152 | 5.78 | Secreted |
| TaFKBP42a-5B | TraesCS5B02G131200 | 0.59 | 42.11 | 243,916,316 | 243,920,638 | 5B | 1624 | 370 | 6.61 | Cytoplasmic |
| TaFKBP53-5B | TraesCS5B02G156700 | 0.89 | 47.76 | 289,160,446 | 289,164,785 | 5B | 1796 | 433 | 5.09 | Nuclear |
| TaFKBP16-4-5B | TraesCS5B02G263800 | 0.11 | 23.43 | 448,220,159 | 448,223,932 | 5B | 1053 | 223 | 10.75 | Chloroplast |
| TaFKBP15-1-5B | TraesCS5B02G278800 | 0.12 | 16.02 | 464,587,956 | 464,590,390 | 5B | 865 | 152 | 6.26 | Cytoplasmic |
| TaFKBP42a-5D | TraesCS5D02G139000 | 0.58 | 41.93 | 221,310,848 | 221,315,012 | 5D | 1473 | 370 | 6.27 | Nuclear |
| TaFKBP53-5D | TraesCS5D02G164000 | 0.92 | 47.81 | 255,575,313 | 255,579,380 | 5D | 1835 | 433 | 5.35 | Nuclear |
| TaFKBP16-4-5D | TraesCS5D02G272100 | 0.05 | 23.24 | 375,377,111 | 375,387,488 | 5D | 1141 | 222 | 10.85 | Chloroplast |
| TaFKBP15-1-5D | TraesCS5D02G286500 | 0.11 | 16.06 | 386,732,844 | 386,735,285 | 5D | 787 | 152 | 5.78 | Secreted |
| TaFKBP18-6A | TraesCS6A02G050000 | 0.33 | 24.32 | 25,636,074 | 25,636,973 | 6A | 445 | 138 | 10.06 | Chloroplast |
| TaFKBP16-1-6A | TraesCS6A02G143200 | 0.03 | 21.86 | 119,619,077 | 119,621,219 | 6A | 903 | 207 | 10.06 | Chloroplast |
| TaFKBP17-1-6A | TraesCS6A02G162000 | 0.20 | 23.24 | 157,134,336 | 157,138,036 | 6A | 891 | 212 | 9.01 | Chloroplast |
| TaFKBP16-2-6A | TraesCS6A02G302500 | 0.11 | 22.46 | 535,990,059 | 535,991,654 | 6A | 1002 | 222 | 9.25 | Chloroplast |
| TaFKBP12-6A | TraesCS6A02G314100 | 0.23 | 20.08 | 550,732,566 | 550,738,491 | 6A | 864 | 193 | 8.94 | Chloroplast |
| TaFKBP18-6B | TraesCS6B02G066300 | 0.34 | 24.02 | 44,104,775 | 44,108,505 | 6B | 977 | 225 | 10.06 | Chloroplast |
| TaFKBP17-1-6B | TraesCS6B02G171400 | 0.09 | 21.89 | 183,753,733 | 183,762,420 | 6B | 624 | 207 | 10.26 | Chloroplast |
| TaFKBP16-1-6B | TraesCS6B02G189300 | 0.17 | 23.08 | 219,095,831 | 219,097,981 | 6B | 941 | 212 | 9.01 | Chloroplast |
| TaFKBP16-2-6B | TraesCS6B02G331700 | 0.18 | 22.47 | 583,050,670 | 583,052,365 | 6B | 943 | 221 | 9.25 | Chloroplast |
| TaFKBP12-6B | TraesCS6B02G344100 | 0.33 | 20.49 | 606,285,921 | 606,289,161 | 6B | 731 | 195 | 9.79 | Chloroplast |
| TaFKBP17-1-6D | TraesCS6D02G132400 | 0.04 | 21.85 | 99,667,030 | 99,66,9069 | 6D | 845 | 168 | 10.06 | Chloroplast |
| TaFKBP16-1-6D | TraesCS6D02G150200 | 0.19 | 22.66 | 123,966,526 | 1239,688,73 | 6D | 759 | 208 | 9.55 | Cytoplasmic |
| TaFKBP16-2-6D | TraesCS6D02G282000 | 0.13 | 22.49 | 389,596,958 | 389,598,490 | 6D | 1022 | 222 | 9.25 | Chloroplast |
| TaFKBP12-6D | TraesCS6D02G293400 | 0.28 | 20.45 | 404,219,105 | 404,222,466 | 6D | 914 | 195 | 9.79 | Chloroplast |
| TaFKBP62-7A | TraesCS7A02G257100 | 0.65 | 62.01 | 246,264,792 | 246,270,837 | 7A | 2394 | 559 | 5.03 | Peroxisome |
| TaFKBP16-3-7A | TraesCS7A02G266500 | 0.08 | 24.85 | 268,865,930 | 268,867,994 | 7A | 1119 | 247 | 7.66 | Chloroplast |
| TaFKBP13-7A | TraesCS7A02G485300 | 0.06 | 20.93 | 676,102,667 | 676,1039,60 | 7A | 1059 | 203 | 8.60 | Chloroplast |
| TaFKBP62-7B | TraesCS7B02G153100 | 0.65 | 62.02 | 203,740,576 | 203,746,113 | 7B | 2069 | 559 | 5.09 | Peroxisome |
| TaFKBP16-3-7B | TraesCS7B02G166000 | 0.04 | 21.94 | 228,914,871 | 228,916,908 | 7B | 760 | 220 | 7.60 | Chloroplast |
| TaFKBP13-7B | TraesCS7B02G388800 | 0.09 | 20.86 | 655,040,607 | 655,041,893 | 7B | 1065 | 203 | 8.37 | Chloroplast |
| TaFKBP62-7D | TraesCS7D02G257300 | 0.65 | 61.98 | 232,811,431 | 232,816,797 | 7D | 2334 | 559 | 5.03 | Peroxisome |
| TaFKBP16-3-7D | TraesCS7D02G268300 | 0.07 | 23.76 | 252,238,804 | 25,224,0730 | 7D | 1060 | 235 | 6.47 | Chloroplast |
| TaFKBP13-7D | TraesCS7D02G472600 | 0.07 | 20.9 | 585,650,878 | 585,652,169 | 7D | 1057 | 203 | 8.60 | Chloroplast |
| TaFKBP18-U | TraesCSU02G108900 | 0.34 | 23.59 | 94,822,583 | 94,826,891 | Un | 802 | 219 | 10.06 | Nuclear, Mitochondrial |
| Plant Species | Genome Size (Approx.) | Coding Genes | FKBP Genes |
|---|---|---|---|
| Triticum aestivum (6n) | 17 Gb | 107,891 | 64 |
| Arabidopsis thaliana (2n) | 135 MB | 27,655 | 23 |
| Oryza sativa (2n) | 500 MB | 37,960 | 29 |
| Zea mays (2n) | 2.4 Gb | 39,591 | 30 |
| Chlamydomonas (n) | 111 MB | 17,741 | 23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ge, Q.; Peng, P.; Cheng, M.; Meng, Y.; Cao, Y.; Zhang, S.; Long, Y.; Li, G.; Kang, G. Genome-Wide Identification and Analysis of FKBP Gene Family in Wheat (Triticum asetivum). Int. J. Mol. Sci. 2022, 23, 14501. https://doi.org/10.3390/ijms232314501
Ge Q, Peng P, Cheng M, Meng Y, Cao Y, Zhang S, Long Y, Li G, Kang G. Genome-Wide Identification and Analysis of FKBP Gene Family in Wheat (Triticum asetivum). International Journal of Molecular Sciences. 2022; 23(23):14501. https://doi.org/10.3390/ijms232314501
Chicago/Turabian StyleGe, Qiang, Peipei Peng, Mingyue Cheng, Yanjun Meng, Yuan Cao, Shuya Zhang, Yu Long, Gezi Li, and Guozhang Kang. 2022. "Genome-Wide Identification and Analysis of FKBP Gene Family in Wheat (Triticum asetivum)" International Journal of Molecular Sciences 23, no. 23: 14501. https://doi.org/10.3390/ijms232314501
APA StyleGe, Q., Peng, P., Cheng, M., Meng, Y., Cao, Y., Zhang, S., Long, Y., Li, G., & Kang, G. (2022). Genome-Wide Identification and Analysis of FKBP Gene Family in Wheat (Triticum asetivum). International Journal of Molecular Sciences, 23(23), 14501. https://doi.org/10.3390/ijms232314501






