Quantitative Transcriptome Analysis of Purified Equine Mast Cells Identifies a Dominant Mucosal Mast Cell Population with Possible Inflammatory Functions in Airways of Asthmatic Horses
Abstract
1. Introduction
2. Results
2.1. Method to Isolate MCs from BALF of Asthmatic Horses
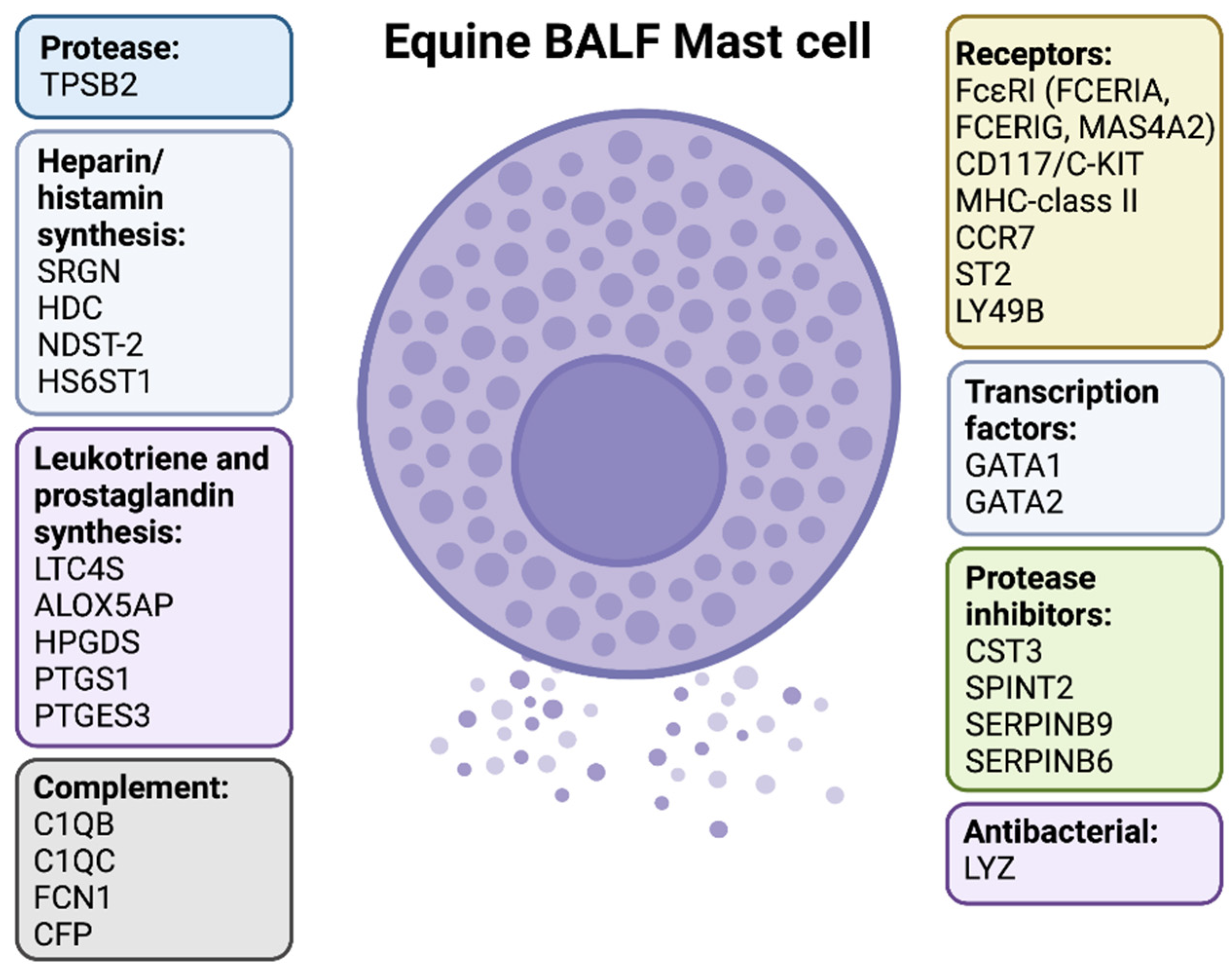
2.2. Transcriptome Analysis of Purified MCs and Total BALF Cells
2.3. Protease Profiles Reveal a Typical Mucosal MC Population in Horse BALF
2.4. Expression of FcεRI and Other MC Surface Markers
2.5. Low Expression of Toll-Like Receptors and Most Cytokine/Chemokine Receptors
2.6. Enzymes Related to Histamine and Arachidonic Acid Metabolites
2.7. Genes Related to Glycosaminoglycans
2.8. Expression of Transcription Factors
2.9. Markers Related to Antigen Presentation and other Immune Related Functions
3. Discussion
4. Materials and Methods
4.1. Horses and Collection of BALF
4.2. Mast Cell Isolation
4.3. Mast Cell Purity and Morphology
4.4. RNA Purification and Quantitative Transcriptome Analysis
4.5. RNA-Sequencing Data Analysis
4.6. Differential Gene Expression Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Couetil, L.L.; Cardwell, J.M.; Gerber, V.; Lavoie, J.P.; Leguillette, R.; Richard, E.A. Inflammatory Airway Disease of Horses--Revised Consensus Statement. J. Vet. Intern. Med. 2016, 30, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Bond, S.; Leguillette, R.; Richard, E.A.; Couetil, L.; Lavoie, J.P.; Martin, J.G.; Pirie, R.S. Equine asthma: Integrative biologic relevance of a recently proposed nomenclature. J. Vet. Intern. Med. 2018, 32, 2088–2098. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.L.; Newton, J.R.; Chanter, N.; Mumford, J.A. Inflammatory airway disease, nasal discharge and respiratory infections in young British racehorses. Equine Vet. J. 2005, 37, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Ivester, K.M.; Couetil, L.L.; Moore, G.E. An observational study of environmental exposures, airway cytology, and performance in racing thoroughbreds. J. Vet. Intern. Med. 2018, 32, 1754–1762. [Google Scholar] [CrossRef]
- Couetil, L.L.; Ward, M.P. Analysis of risk factors for recurrent airway obstruction in North American horses: 1,444 cases (1990–1999). J. Am. Vet. Med. Assoc. 2003, 223, 1645–1650. [Google Scholar] [CrossRef]
- Jean, D.; Vrins, A.; Beauchamp, G.; Lavoie, J.P. Evaluation of variations in bronchoalveolar lavage fluid in horses with recurrent airway obstruction. Am. J. Vet. Res. 2011, 72, 838–842. [Google Scholar] [CrossRef]
- Davis, K.U.; Sheats, M.K. Bronchoalveolar Lavage Cytology Characteristics and Seasonal Changes in a Herd of Pastured Teaching Horses. Front. Vet. Sci. 2019, 6, 74. [Google Scholar] [CrossRef]
- McGorum, B.; Dixon, P.; Halliwell, R. Comparison of cellular and molecular components of bronchoalveolar lavage fluid harvested from different segments of the equine lungs. Res. Vet. Sci. 1993, 55, 57–59. [Google Scholar] [CrossRef]
- Basano, I.; Romolo, A.; Iamone, G.; Memoli, G.; Riccio, B.; Lavoie, J.P.; Miniscalco, B.; Bullone, M. Giant Multinucleated Cells Are Associated with Mastocytic Inflammatory Signature Equine Asthma. Animals 2022, 12, 1070. [Google Scholar] [CrossRef]
- Wernersson, S.; Pejler, G. Mast cell secretory granules: Armed for battle. Nat. Rev. Immunol. 2014, 14, 478–494. [Google Scholar] [CrossRef]
- Moon, T.C.; Befus, A.D.; Kulka, M. Mast Cell Mediators: Their Differential Release and the Secretory Pathways Involved. Front. Immunol. 2014, 5, 569. [Google Scholar] [CrossRef]
- Krystel-Whittemore, M.; Dileepan, K.N.; Wood, J.G. Mast Cell: A Multi-Functional Master Cell. Front. Immunol. 2016, 6, 620. [Google Scholar] [CrossRef]
- Bradding, P.; Walls, A.F.; Holgate, S.T. The role of the mast cell in the pathophysiology of asthma. J. Allergy Clin. Immunol. 2006, 117, 1277–1284. [Google Scholar] [CrossRef] [PubMed]
- Galli, S.J.; Tsai, M. IgE and mast cells in allergic disease. Nat. Med. 2012, 18, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Enríquez, E.; Hallgren, J. Mast Cells and Their Progenitors in Allergic Asthma. Front. Immunol. 2019, 10, 821. [Google Scholar] [CrossRef] [PubMed]
- Brightling, C.E.; Bradding, P.; Symon, F.A.; Holgate, S.T.; Wardlaw, A.J.; Pavord, I.D. Mast-cell infiltration of airway smooth muscle in asthma. N. Engl. J. Med. 2002, 346, 1699–1705. [Google Scholar] [CrossRef]
- Dougherty, R.H.; Sidhu, S.S.; Raman, K.; Solon, M.; Solberg, O.D.; Caughey, G.H.; Woodruff, P.G.; Fahy, J.V. Accumulation of intraepithelial mast cells with a unique protease phenotype in T(H)2-high asthma. J. Allergy Clin. Immunol. 2010, 125, 1046–1053.e8. [Google Scholar] [CrossRef]
- Balzar, S.; Fajt, M.L.; Comhair, S.A.; Erzurum, S.C.; Bleecker, E.; Busse, W.W.; Castro, M.; Gaston, B.; Israel, E.; Schwartz, L.B.; et al. Mast cell phenotype, location, and activation in severe asthma. Data from the Severe Asthma Research Program. Am. J. Respir. Crit. Care Med. 2011, 183, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Andersson, C.K.; Bergqvist, A.; Mori, M.; Mauad, T.; Bjermer, L.; Erjefält, J.S. Mast cell-associated alveolar inflammation in patients with atopic uncontrolled asthma. J. Allergy Clin. Immunol. 2011, 127, 905–912.e1-7. [Google Scholar] [CrossRef]
- Yu, M.; Tsai, M.; Tam, S.Y.; Jones, C.; Zehnder, J.; Galli, S.J. Mast cells can promote the development of multiple features of chronic asthma in mice. J. Clin. Investig. 2006, 116, 1633–1641. [Google Scholar] [CrossRef]
- Cui, Y.; Dahlin, J.S.; Feinstein, R.; Bankova, L.G.; Xing, W.; Shin, K.; Gurish, M.F.; Hallgren, J. Mouse Mast Cell Protease-6 and MHC Are Involved in the Development of Experimental Asthma. J. Immunol. 2014, 193, 4783. [Google Scholar] [CrossRef]
- Waern, I.; Lundequist, A.; Pejler, G.; Wernersson, S. Mast cell chymase modulates IL-33 levels and controls allergic sensitization in dust-mite induced airway inflammation. Mucosal Immunol. 2013, 6, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Richard, E.A.; Fortier, G.D.; Denoix, J.M.; Art, T.; Lekeux, P.M.; Van Erck, E. Influence of subclinical inflammatory airway disease on equine respiratory function evaluated by impulse oscillometry. Equine Vet. J. 2009, 41, 384–389. [Google Scholar] [CrossRef]
- Enerback, L. Mast cells in rat gastrointestinal mucosa. I. Effects of fixation. Acta Pathol. Microbiol. Scand. 1966, 66, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, S.; Xu, J.; Zhang, X.; Han, D.; Liu, J.; Xia, M.; Yi, L.; Shen, Q.; Xu, S.; et al. Adult Connective Tissue-Resident Mast Cells Originate from Late Erythro-Myeloid Progenitors. Immunity 2018, 49, 640–653.e5. [Google Scholar] [CrossRef]
- Gentek, R.; Ghigo, C.; Hoeffel, G.; Bulle, M.J.; Msallam, R.; Gautier, G.; Launay, P.; Chen, J.; Ginhoux, F.; Bajenoff, M. Hemogenic Endothelial Fate Mapping Reveals Dual Developmental Origin of Mast Cells. Immunity 2018, 48, 1160–1171.e5. [Google Scholar] [CrossRef]
- Irani, A.A.; Schechter, N.M.; Craig, S.S.; DeBlois, G.; Schwartz, L.B. Two types of human mast cells that have distinct neutral protease compositions. Proc. Natl. Acad. Sci. USA 1986, 83, 4464–4468. [Google Scholar] [CrossRef]
- Cildir, G.; Yip, K.H.; Pant, H.; Tergaonkar, V.; Lopez, A.F.; Tumes, D.J. Understanding mast cell heterogeneity at single cell resolution. Trends Immunol. 2021, 42, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Pejler, G. The emerging role of mast cell proteases in asthma. Eur. Respir. J. 2019, 54, 1900685. [Google Scholar] [CrossRef] [PubMed]
- Waern, I.; Taha, S.; Lorenzo, J.; Montpeyó, D.; Covaleda-Cortés, G.; Avilés, F.X.; Wernersson, S. Carboxypeptidase inhibition by NvCI suppresses airway hyperreactivity in a mouse asthma model. Allergy 2021, 76, 2234–2237. [Google Scholar] [CrossRef] [PubMed]
- Hellman, L.T.; Akula, S.; Thorpe, M.; Fu, Z. Tracing the Origins of IgE, Mast Cells, and Allergies by Studies of Wild Animals. Front. Immunol. 2017, 8, 1749. [Google Scholar] [CrossRef] [PubMed]
- Galli, S.J.; Zsebo, K.M.; Geissler, E.N. The kit ligand, stem cell factor. Adv. Immunol. 1994, 55, 1–96. [Google Scholar] [PubMed]
- Wang, J.X.; Kaieda, S.; Ameri, S.; Fishgal, N.; Dwyer, D.; Dellinger, A.; Kepley, C.L.; Gurish, M.F.; Nigrovic, P.A. IL-33/ST2 axis promotes mast cell survival via BCLXL. Proc. Natl. Acad. Sci. USA 2014, 111, 10281–10286. [Google Scholar] [CrossRef]
- McNeil, B.D.; Pundir, P.; Meeker, S.; Han, L.; Undem, B.J.; Kulka, M.; Dong, X. Identification of a mast-cell-specific receptor crucial for pseudo-allergic drug reactions. Nature 2015, 519, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Akula, S.; Paivandy, A.; Fu, Z.; Thorpe, M.; Pejler, G.; Hellman, L. Quantitative In-Depth Analysis of the Mouse Mast Cell Transcriptome Reveals Organ-Specific Mast Cell Heterogeneity. Cells 2020, 9, 211. [Google Scholar] [CrossRef] [PubMed]
- Motakis, E.; Guhl, S.; Ishizu, Y.; Itoh, M.; Kawaji, H.; de Hoon, M.; Lassmann, T.; Carninci, P.; Hayashizaki, Y.; Zuberbier, T.; et al. Redefinition of the human mast cell transcriptome by deep-CAGE sequencing. Blood 2014, 123, e58–e67. [Google Scholar] [CrossRef]
- Varricchi, G.; Pecoraro, A.; Loffredo, S.; Poto, R.; Rivellese, F.; Genovese, A.; Marone, G.; Spadaro, G. Heterogeneity of Human Mast Cells with Respect to MRGPRX2 Receptor Expression and Function. Front. Cell Neurosci. 2019, 13, 299. [Google Scholar] [CrossRef]
- Nakamura, T. The roles of lipid mediators in type I hypersensitivity. J. Pharmacol. Sci. 2021, 147, 126–131. [Google Scholar] [CrossRef]
- Sterk, A.R.; Ishizaka, T. Binding properties of IgE receptors on normal mouse mast cells. J. Immunol. 1982, 128, 838–843. [Google Scholar]
- Ohneda, K.; Ohmori, S.; Yamamoto, M. Mouse Tryptase Gene Expression is Coordinately Regulated by GATA1 and GATA2 in Bone Marrow-Derived Mast Cells. Int. J. Mol. Sci. 2019, 20, 4603. [Google Scholar] [CrossRef]
- Baba, Y.; Maeda, K.; Yashiro, T.; Inage, E.; Kasakura, K.; Suzuki, R.; Niyonsaba, F.; Hara, M.; Tanabe, A.; Ogawa, H.; et al. GATA2 is a critical transactivator for the human IL1RL1/ST2 promoter in mast cells/basophils: Opposing roles for GATA2 and GATA1 in human IL1RL1/ST2 gene expression. J. Biol. Chem. 2012, 287, 32689–32696. [Google Scholar] [CrossRef]
- Siddhuraj, P.; Clausson, C.M.; Sanden, C.; Alyamani, M.; Kadivar, M.; Marsal, J.; Wallengren, J.; Bjermer, L.; Erjefalt, J.S. Lung Mast Cells Have a High Constitutive Expression of Carboxypeptidase A3 mRNA That Is Independent from Granule-Stored CPA3. Cells 2021, 10, 309. [Google Scholar] [CrossRef]
- Platzer, B.; Stout, M.; Fiebiger, E. Functions of dendritic-cell-bound IgE in allergy. Mol. Immunol. 2015, 68 Pt A, 116–119. [Google Scholar] [CrossRef]
- Paivandy, A.; Akula, S.; Lara, S.; Fu, Z.; Olsson, A.K.; Kleinau, S.; Pejler, G.; Hellman, L. Quantitative In-Depth Transcriptome Analysis Implicates Peritoneal Macrophages as Important Players in the Complement and Coagulation Systems. Int. J. Mol. Sci. 2022, 23, 1185. [Google Scholar] [CrossRef] [PubMed]
- Lara, S.; Akula, S.; Fu, Z.; Olsson, A.K.; Kleinau, S.; Hellman, L. The Human Monocyte-A Circulating Sensor of Infection and a Potent and Rapid Inducer of Inflammation. Int. J. Mol. Sci. 2022, 23, 3890. [Google Scholar] [CrossRef] [PubMed]
- Cvitas, I.; Oberhänsli, S.; Leeb, T.; Dettwiler, M.; Müller, E.; Burgmann, R.; Marti, E.I. Investigating the epithelial barrier and immune signatures in the pathogenesis of equine insect bite hypersensitivity. PLoS ONE 2020, 15, e0232189. [Google Scholar] [CrossRef] [PubMed]
- Wernersson, S.; Riihimäki, M.; Pejler, G.; Waern, I. Equine Airway Mast Cells are Sensitive to Cell Death Induced by Lysosomotropic Agents. Scand. J. Immunol. 2017, 85, 30–34. [Google Scholar] [CrossRef]
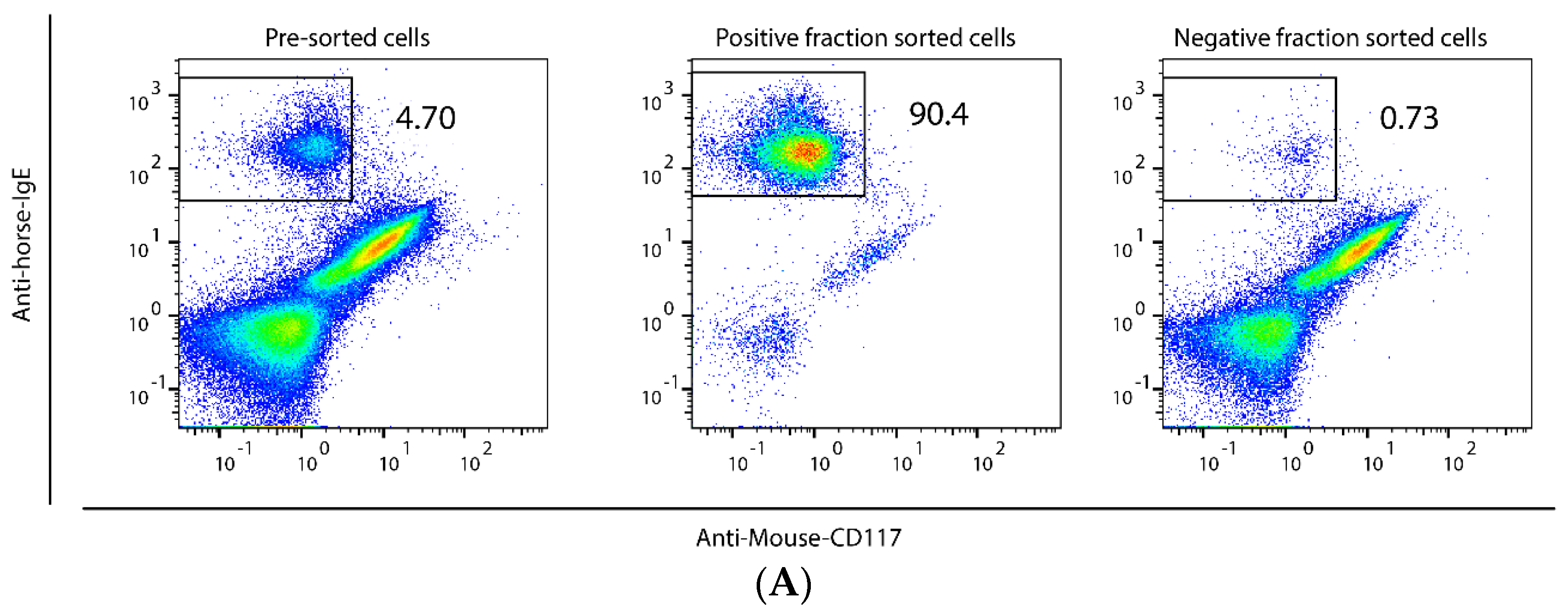
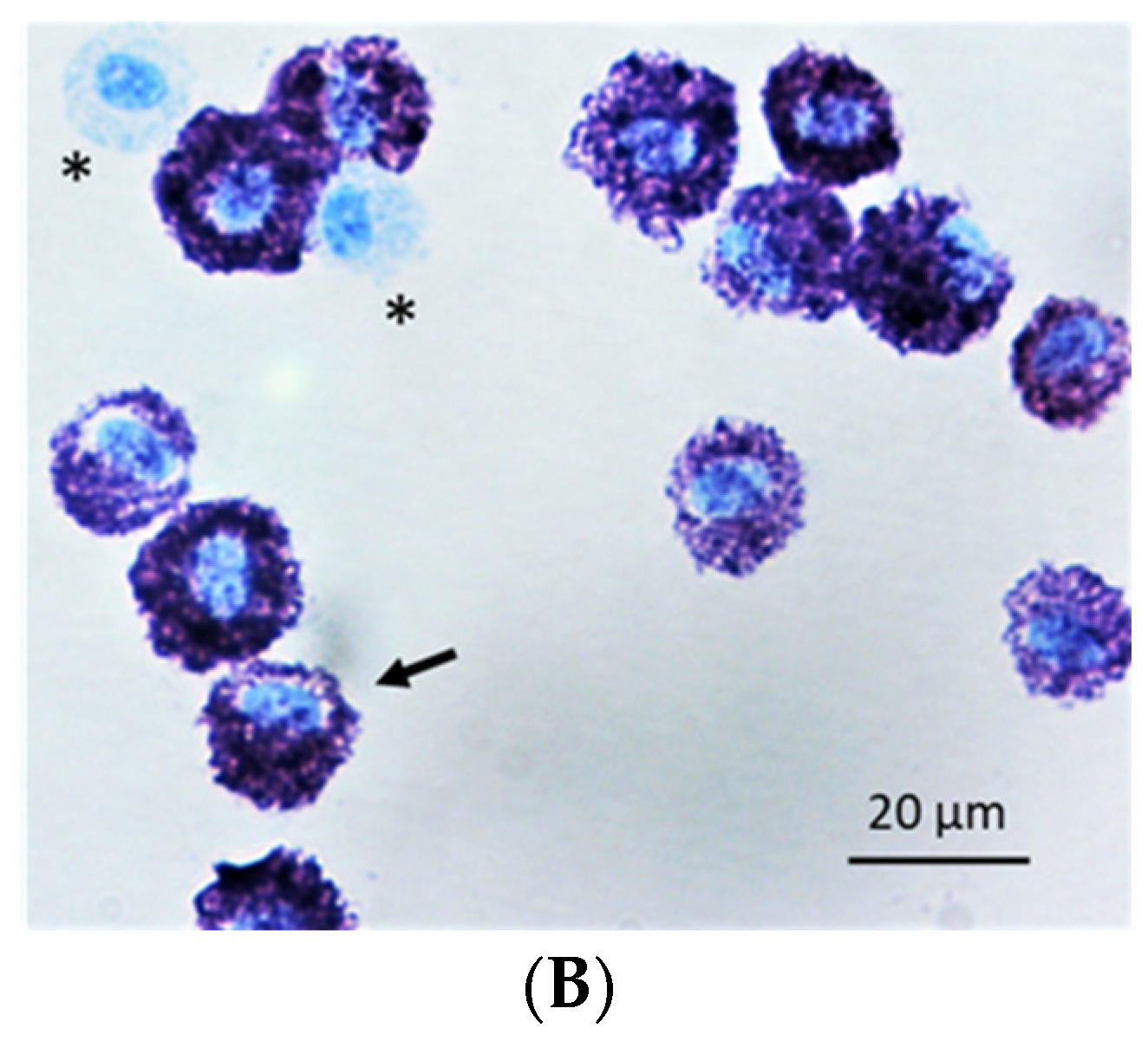
| Genes | BALF MCs | Total BALF Cells | |
|---|---|---|---|
| Proteases | CMA1 | 0 | 0.4 |
| CPA3 | 0 | 0 | |
| TPSB2 | 1530 | 43 | |
| TPSG1 | 16 | 0.3 | |
| CTSG | 0 | 0 | |
| GZMA | 68 | 166 | |
| GZMK | 63 | 109 | |
| GZMM | 2 | 2 | |
| ELANE | 0 | 0 | |
| AZU1 | 0 | 0 | |
| NSP4 | 0 | 0 | |
| MMP2 | 82 | 1.3 | |
| MMP9 | 28 | 3 | |
| CTSC | 275 | 100 | |
| CPM | 18 | 44 | |
| CPD | 11 | 5 | |
| CPE | 0 | 0 | |
| CPZ | 0 | 0 | |
| CPA4 | 0 | 0 | |
| CPA5 | 0 | 0 | |
| CPA1 | 0 | 0 | |
| CPB1 | 0 | 0 | |
| CPA6 | 0 | 0 | |
| CPB2 | 0 | 0 | |
| Protease inhibitors | CST3 | 6198 | 565 |
| SPINT2 | 244 | 16 | |
| SERPINB9 | 237 | 18 | |
| SERPINB6 | 230 | 27 |
| Genes | BALF MCs | Total BALF Cells | |
|---|---|---|---|
| Immunoglobulin Receptors | FCER1A | 1764 | 43 |
| FCER1G | 3387 | 4710 | |
| MS4A2 | 147 | 1 | |
| FCER2 | 5 | 0.6 | |
| FCMR | 2 | 0.8 | |
| Cytokine receptors | ST2 | 58 | 0.9 |
| C-KIT | 164 | 1.4 | |
| IL1R1 | 3 | 0.3 | |
| IL1R2 | 5 | 0.6 | |
| IL2RB | 60 | 20 | |
| IL4R | 78 | 48 | |
| IL9R | 6 | 0.1 | |
| IL10RA | 47 | 28 | |
| IL13RA1 | 54 | 18 | |
| IL18R1 | 8 | 1 | |
| IL21R | 53 | 4 | |
| IL27RA | 11 | 77 | |
| Chemokine receptors | CCR1 | 35 | 1.9 |
| CCR2 | 21 | 14 | |
| CCR3 | 5 | 6 | |
| CCR5 | 13 | 5 | |
| CCR7 | 650 | 26 | |
| Toll-like receptors | TLR1 | 18 | 4 |
| TLR2 | 16 | 47 | |
| TLR3 | 13 | 11 | |
| TLR4 | 0 | 0 | |
| TLR5 | 1.2 | 0.2 | |
| TLR6 | 8 | 5 | |
| TRL7 | 17 | 16 | |
| TLR8 | 13 | 32 | |
| TLR9 | 2 | 0.4 | |
| TLR10 | 13 | 0.2 | |
| Other receptors | LY49B | 55 | 0.9 |
| MILR1 | 13 | 4 | |
| HRH4 | 24 | 0.4 |
| Genes | BALF MCs | Total BALF Cells | |
|---|---|---|---|
| Heparin and Histamine synthesis | HDC | 166 | 2.5 |
| HS6ST1 | 69 | 8 | |
| HS3ST1 | 47 | 2 | |
| HS6ST2 | 2 | 0 | |
| NDST2 | 52 | 4 | |
| NDST1 | 1 | 3.5 | |
| SRGN | 5957 | 5935 | |
| Chondroitin sulfate synthesis | CSGALNACT1 | 1.2 | 2 |
| CSGALNACT2 | 21 | 7 | |
| CHST3 | 0.1 | 0.6 | |
| CHST7 | 45 | 5 | |
| CHST11 | 1.5 | 10 | |
| CHST13 | 32 | 2 | |
| CHST15 | 1.5 | 0.4 | |
| CHSY1 | 3 | 1.9 | |
| CHSY3 | 0.03 | 2.5 | |
| CHPF | 34 | 24 | |
| Leukotriene synthesis | LTC4S | 2413 | 109 |
| ALOX5 | 199 | 34 | |
| ALOX5AP | 2174 | 1887 | |
| Prostaglandin synthesis | HPGDS | 441 | 16 |
| PTGS1 | 297 | 5 | |
| PTGES3 | 216 | 211 | |
| PTGDS | 18 | 0.9 | |
| Transcription factors | GATA1 | 59 | 2 |
| GATA2 | 93 | 2.2 | |
| GATA3 | 20 | 5 |
| Genes | BALF MCs | Total BALF Cells | |
|---|---|---|---|
| MHC-II and CD1 | DRA | 17,906 | 2344 |
| DQA | 78 | 7 | |
| DQB | 2988 | 375 | |
| DRB | 1990 | 378 | |
| HLA-DMA | 950 | 460 | |
| HLA-DOB | 120 | 6 | |
| CD74 | 14,764 | 3531 | |
| CD1A3 | 426 | 44 | |
| CD1A7 | 210 | 36 | |
| CD1B1 | 157 | 5 | |
| CD1B2 | 19 | 0.7 | |
| CD1C | 44 | 0.9 | |
| CD1E1 | 195 | 6 | |
| CD1E2 | 537 | 18 | |
| Complement components | C1QB | 1274 | 3896 |
| C1QC | 1525 | 6168 | |
| FCN1 | 48 | 1480 | |
| CFP | 72 | 789 | |
| Lysozyme | LYZ | 479 | 42 |
| Genes | BALF MCs | Total BALF Cells | |
|---|---|---|---|
| Cytokine and Chemokines | TGFB1 | 460 | 434 |
| IL1B | 7 | 3 | |
| IL4 | 1 | 0.4 | |
| IL4L1 | 83 | 17 | |
| IL5 | 6 | 0.4 | |
| IL6 | 9 | 3 | |
| IL13 | 1 | 0 | |
| IL15 | 6 | 12 | |
| IL17 | 9 | 3 | |
| IL18 | 68 | 160 | |
| IL25 | 0 | 0 | |
| IL33 | 0 | 0 | |
| LIF | 46 | 1.3 | |
| MIF | 631 | 862 | |
| CCL5 | 163 | 483 | |
| CCL8 | 242 | 242 | |
| CCL22 | 59 | 2 | |
| CCL24 | 268 | 1211 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akula, S.; Riihimäki, M.; Waern, I.; Åbrink, M.; Raine, A.; Hellman, L.; Wernersson, S. Quantitative Transcriptome Analysis of Purified Equine Mast Cells Identifies a Dominant Mucosal Mast Cell Population with Possible Inflammatory Functions in Airways of Asthmatic Horses. Int. J. Mol. Sci. 2022, 23, 13976. https://doi.org/10.3390/ijms232213976
Akula S, Riihimäki M, Waern I, Åbrink M, Raine A, Hellman L, Wernersson S. Quantitative Transcriptome Analysis of Purified Equine Mast Cells Identifies a Dominant Mucosal Mast Cell Population with Possible Inflammatory Functions in Airways of Asthmatic Horses. International Journal of Molecular Sciences. 2022; 23(22):13976. https://doi.org/10.3390/ijms232213976
Chicago/Turabian StyleAkula, Srinivas, Miia Riihimäki, Ida Waern, Magnus Åbrink, Amanda Raine, Lars Hellman, and Sara Wernersson. 2022. "Quantitative Transcriptome Analysis of Purified Equine Mast Cells Identifies a Dominant Mucosal Mast Cell Population with Possible Inflammatory Functions in Airways of Asthmatic Horses" International Journal of Molecular Sciences 23, no. 22: 13976. https://doi.org/10.3390/ijms232213976
APA StyleAkula, S., Riihimäki, M., Waern, I., Åbrink, M., Raine, A., Hellman, L., & Wernersson, S. (2022). Quantitative Transcriptome Analysis of Purified Equine Mast Cells Identifies a Dominant Mucosal Mast Cell Population with Possible Inflammatory Functions in Airways of Asthmatic Horses. International Journal of Molecular Sciences, 23(22), 13976. https://doi.org/10.3390/ijms232213976






