Characterization of a Glycolipid Synthase Producing α-Galactosylceramide in Bacteroides fragilis
Abstract
1. Introduction
2. Results
2.1. Bioinformatics Search of GT Candidates in the B. fragilis Genome
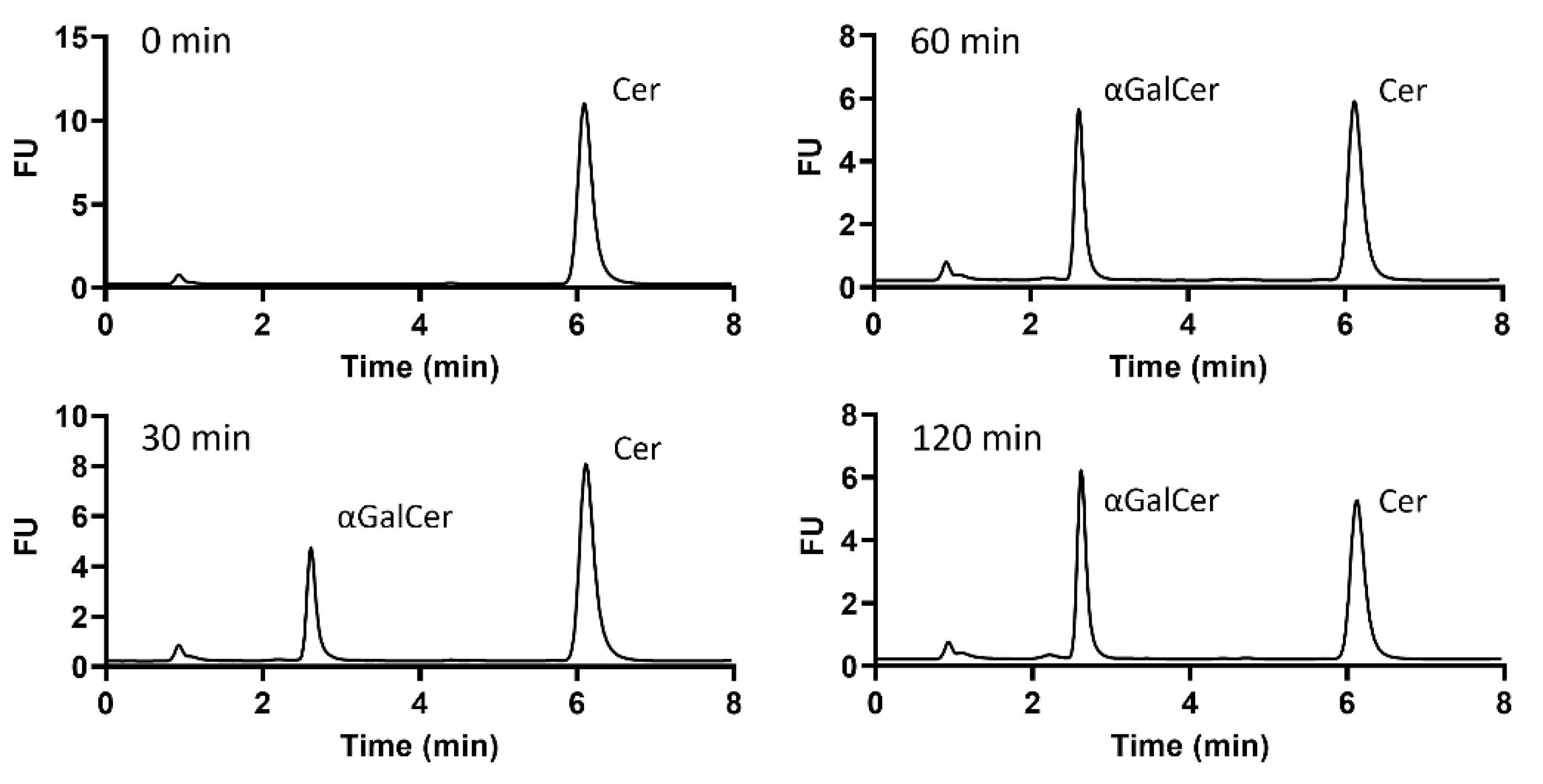
2.2. Identification of α-Galactosylceramide Transferase Activity in B. fragilis GT Candidates
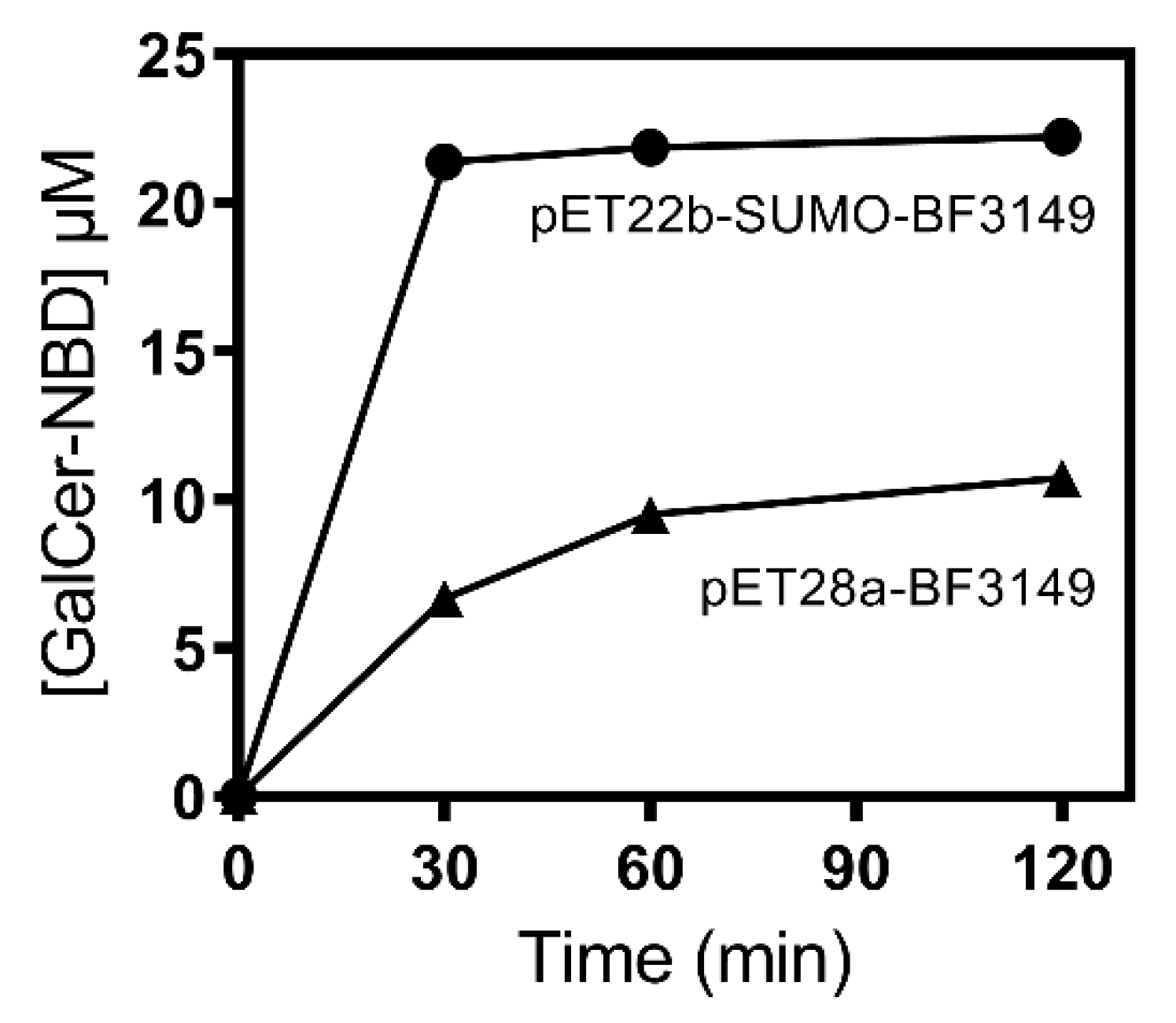
2.3. Expression and Purification of B. fragilis Glycosyltransferase
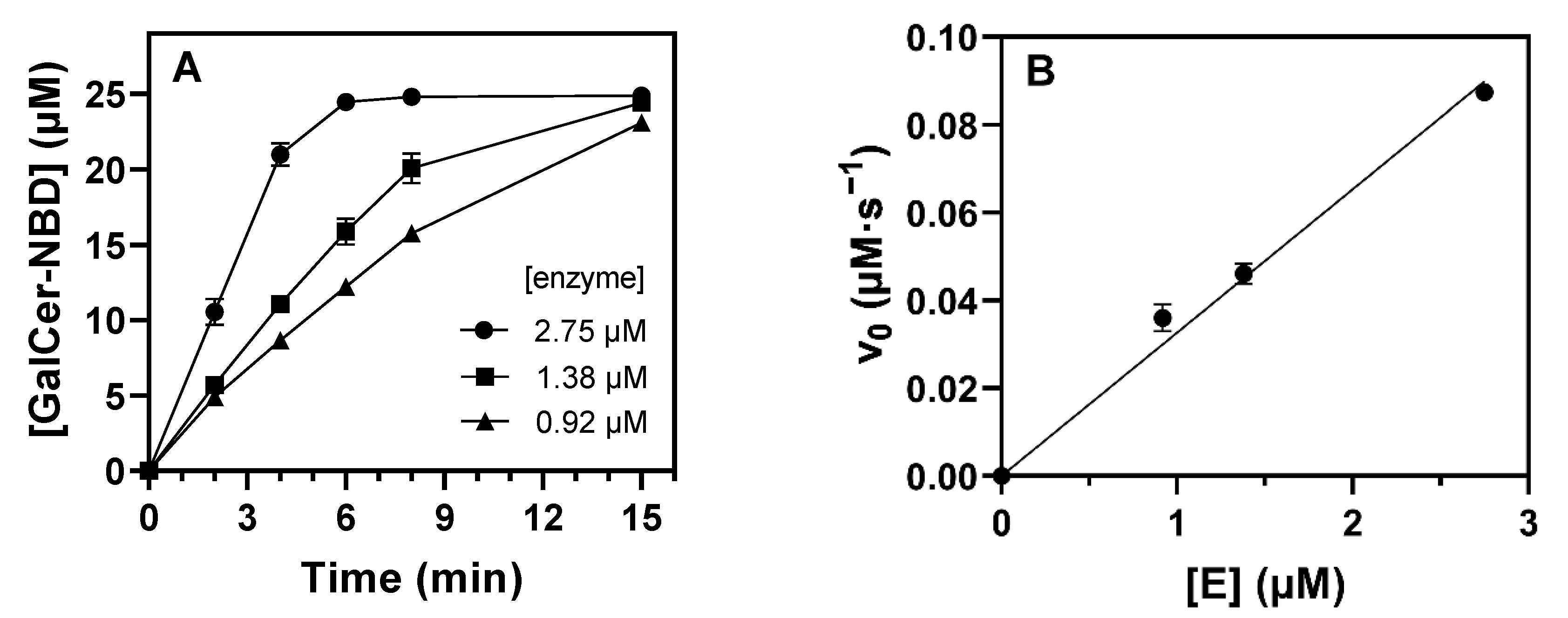
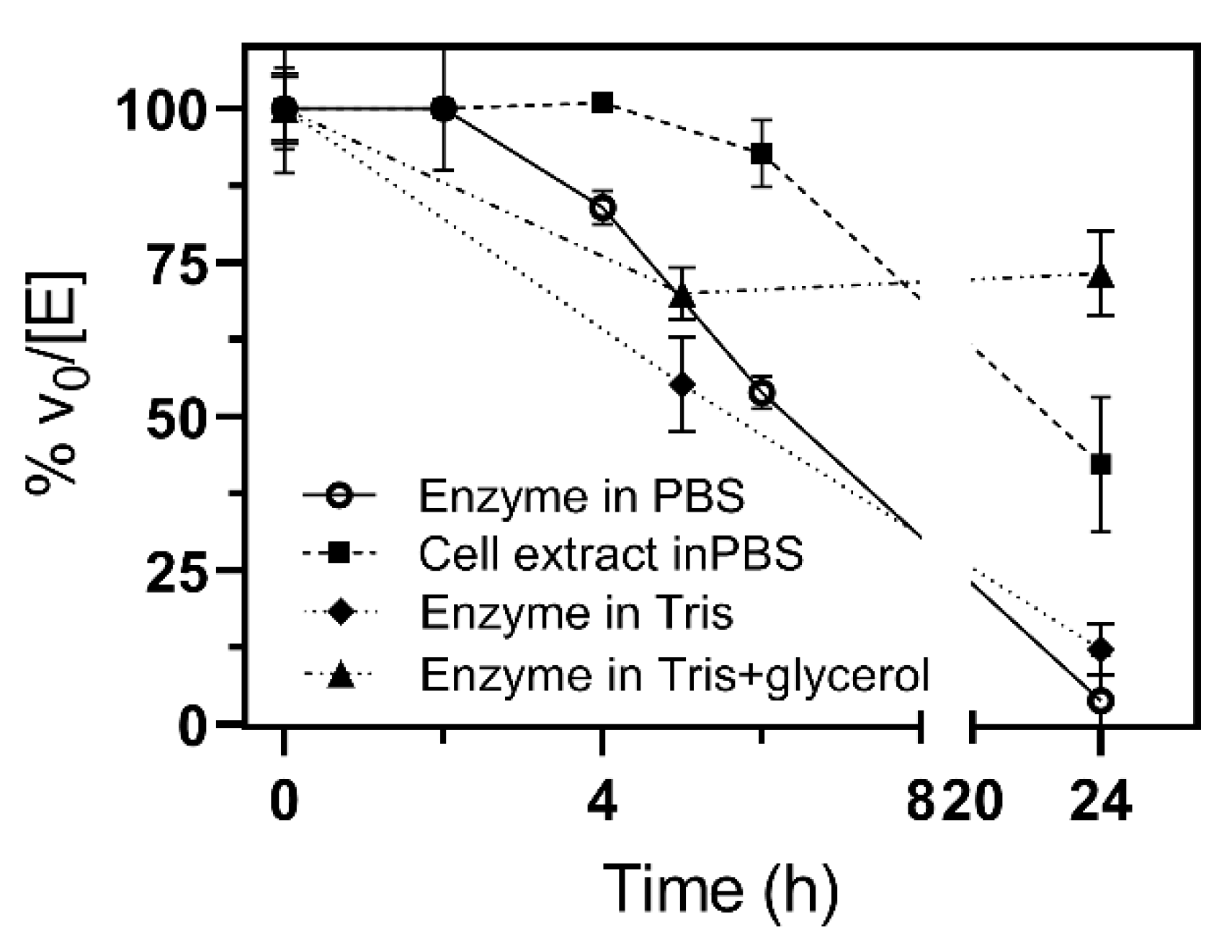
2.4. Kinetic Characterization
2.4.1. Specific Activity
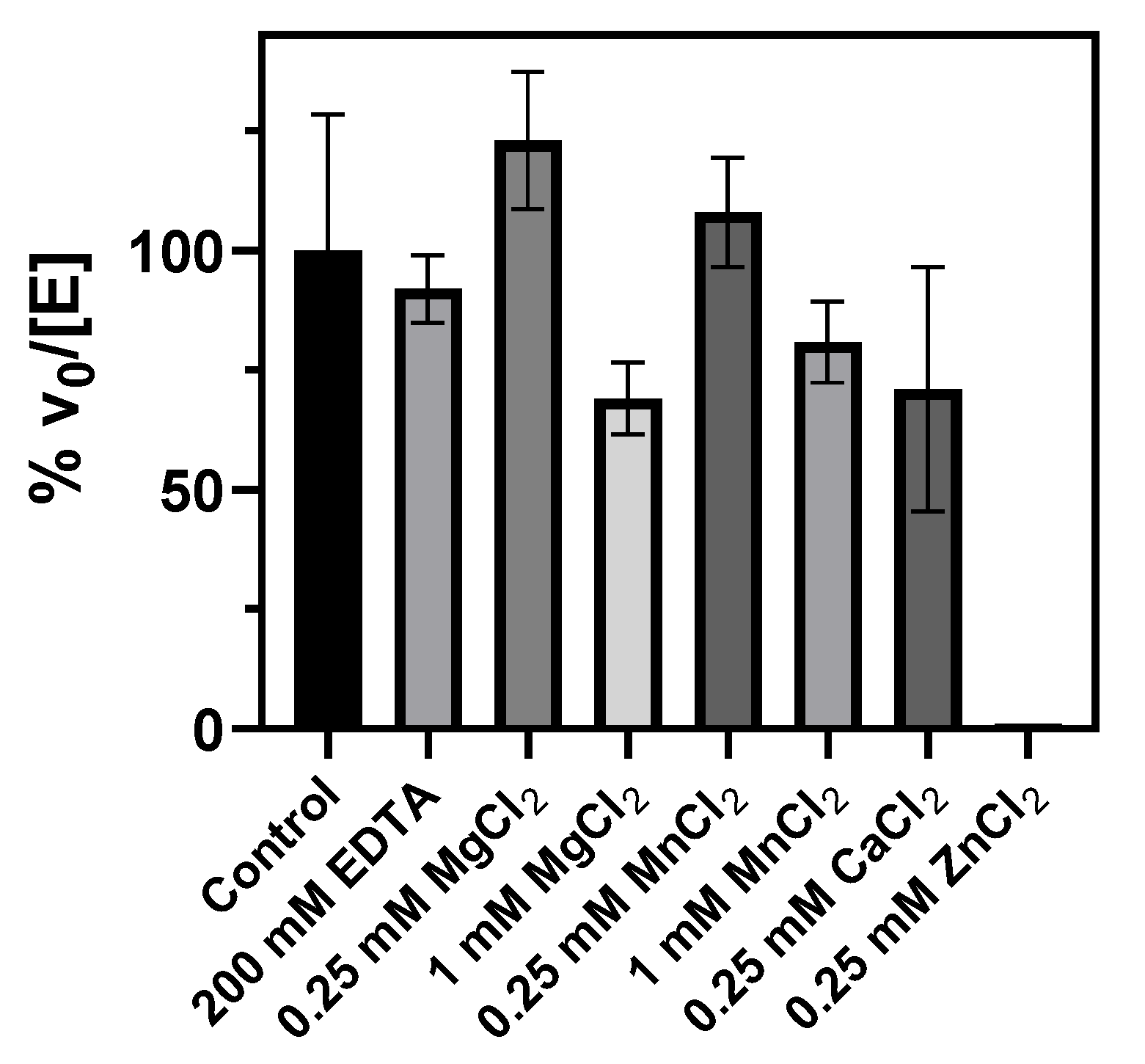
2.4.2. Metal Binding
2.4.3. Optimal pH and Temperature
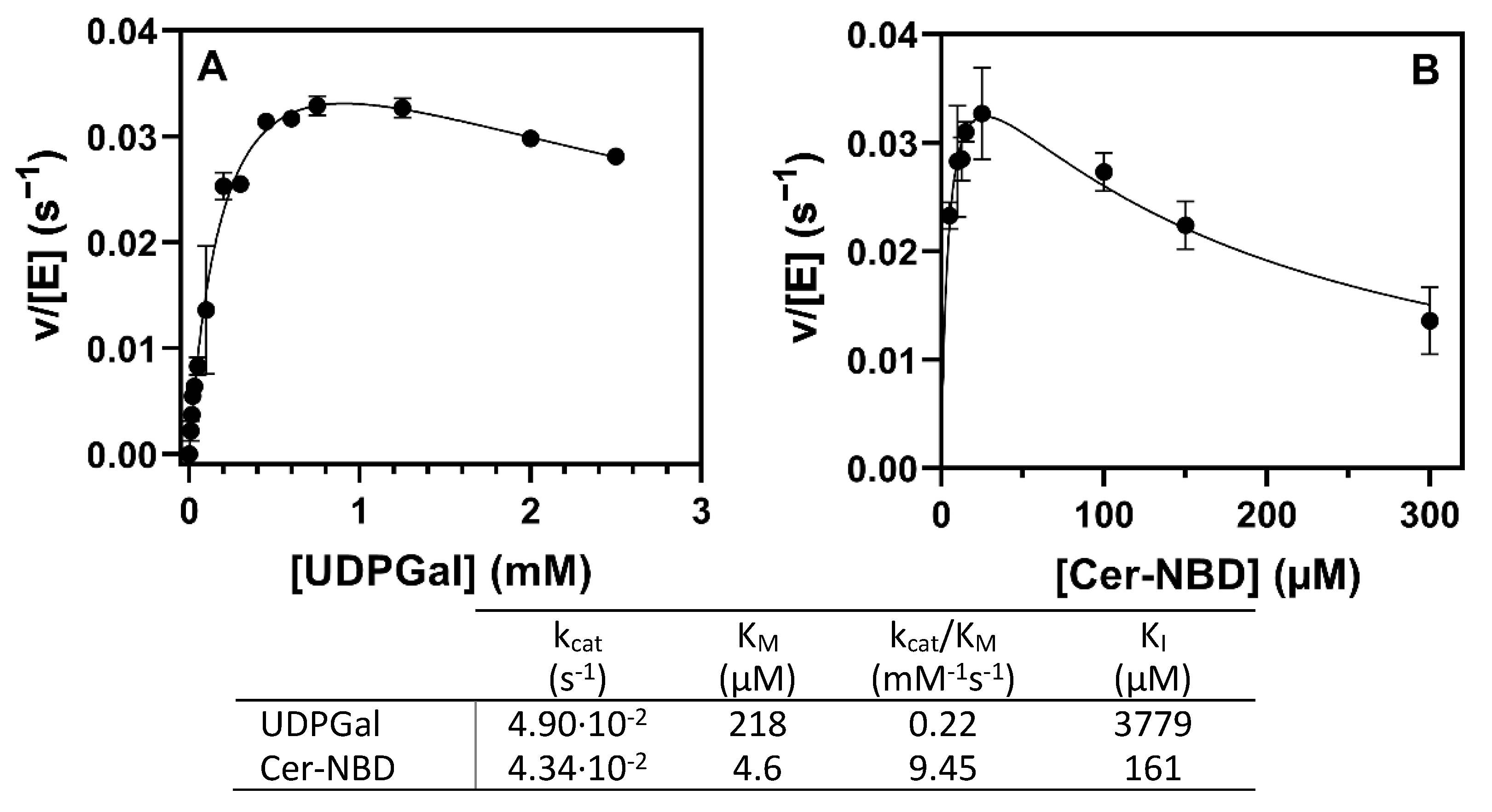
2.4.4. Kinetic Parameters for Donor and Acceptor Substrates
2.4.5. Activity in Mixed Micelles and in the Presence of Anionic Lipids
2.5. Modeled αGalCer_GT Structure and Ligand Binding
3. Discussion
4. Materials and Methods
4.1. In Silico Analysis
4.2. Strains, Plasmids and Reagents
4.3. Cloning of B. fragilis GT Sequences into E. coli Expression Vector
- (a)
- pET28a vector-Protein sequences were obtained from the UniProt database and then translated to DNA code optimized for E. coli. Synthetic sequences were obtained from GeneArt (ThermoFischer Scientific, Waltham, MA, USA) and cloned using restriction enzymes NcoI and XhoI into pET28a vector to obtain the expression vectors pET28a-HisTag-BF9343_XXXX. Positive transformants were selected on LB plates with 50 µg/mL of kanamycin. Successful plasmid constructions were verified by Sanger sequencing.
- (b)
- pET22b-Strep-SUMO vector-Sequence BF9343_3149 was later cloned into a pET22b vector containing a N-terminal fused Strep-tag and SUMO protein. Cloning was carried out by Circular Polymerase Extension cloning (CPEC), amplifying the linear vector and the insert containing BF9343_3149 with primers sharing homologous regions, and CPEC reaction to obtain pET22b-SUMO-BF9343_3149-StrepTag. Positive transformants were selected on LB plates with 100 µg/mL of ampicillin. Successful plasmid construction was verified by Sanger sequencing.
4.4. Expression of GT Candidates and αGalCer_GT in E. coli
4.5. Protein Purification by Strep-Tag Affinity Chromatography
4.6. Galactosylceramide Transferase Activity Assay
- (a)
- Ceramide-NBD with BSA: Reactions were performed with 25 µM of Cer-NBD, 25 µM of BSA, 1.25 mM of UDP-Gal, phosphate buffer (50 mM of Na2HPO4, 150 mM of NaCl, pH 7.5) and the appropriate amount of enzyme in a 200 µL final reaction volume. It was preincubated at 37 °C for five minutes and the reaction was started by adding UDP-Gal. Sampling time was adjusted depending on enzyme concentration. Aliquots were withdrawn and mixed with methanol (MeOH:sample 8:2) and centrifuged to eliminate debris. The samples were then analyzed by HPLC. Chromatographic peaks were assigned by co-injection with independent standards. Initial rates were obtained from the linear progress curve of product formation. Initial rates were determined from triplicate assays.
- (b)
- Ceramide-NBD in vesicles: Ceramide-NBD, DOPG and/or DOPG (matrix lipid) were dissolved in chloroform in a glass vial using the desired amounts for each experiment, as indicated. Chloroform was evaporated under a N2 stream and the lipid mixture dried under vacuum for 2 h. When the reactions were performed, the lipid film was solubilized to homogeneity in 80 µL of phosphate buffer solution (50 mM of Na2HPO4, 150 mM of NaCl, pH 7.5) by extensive vortexing and ultrasound bath for 5 min, and kept on ice for 1 min. This cycle was repeated 8 times. Vesicle formation was analyzed using ZetaSizer (Malvern Panalytical, Malvern, UK). For the enzymatic reaction, 25 µL of enzyme was added to 60 µL of freshly prepared vesicles solution. The mixture was kept for 30 min on ice and preincubated for 5 min at 37 °C. The reaction was started by adding 12.5 µL of UDP-Gal and incubated at 37 °C. The reaction conditions were: 1.25 mM of UDP-Gal, 25 µM of Cer-NBD, DOPC/DOPG vesicles (condition 1:1 mM of DOPC; 2:1 mM of total DOPC and DOPG (40 mol % DOPG); 3:1 mM of DOPG, phosphate buffer (50 mM of Na2HPO4, 150 mM of NaCl, pH 7.5), and enzyme (0.92–2.75 µM) in a 200 uL reaction volume. Aliquots were withdrawn every 30 min for 90 min, stopped as before and analyzed by HPLC.
4.7. Kinetic Parameters
4.8. Effect of Metal Cations
4.9. Modeled 3D Structure and Ligand Docking
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Quinville, B.M.; Deschenes, N.M.; Ryckman, A.E.; Walia, J.S. A Comprehensive Review: Sphingolipid Metabolism and Implications of Disruption in Sphingolipid Homeostasis. Int. J. Mol. Sci. 2021, 22, 5793. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Ikushiro, H.; Hayashi, H.; Kagamiyama, H. A Water-soluble Homodimeric Serine Palmitoyltransferase fromSphingomonas paucimobilis EY2395 T Strain complex from Chinese hamster ovary cells. J. Biol. Chem. 2001, 276, 18249–18256. [Google Scholar] [CrossRef] [PubMed]
- Stankeviciute, G.; Guan, Z.; Goldfine, H.; Klein, E.A. Caulobacter crescentus Adapts to Phosphate Starvation by Synthesizing Anionic Glycoglycerolipids and a Novel Glycosphingolipid. mBio 2019, 10, e00107-19. [Google Scholar] [CrossRef]
- Kawasaki, S.; Moriguchi, R.; Sekiya, K.; Nakai, T.; Ono, E.; Kume, K.; Kawahara, K. The cell envelope structure of the lipopolysaccharide-lacking gram-negative bacterium Sphingomonas paucimobilis. J. Bacteriol. 1994, 176, 284–290. [Google Scholar] [CrossRef]
- Health, R. Expression cloning of a cDNA for human ceramide glucosyltransferase that catalyzes the first glycosylation step of glycosphingolipid synthesis. Proc. Nat. Acad. Sci. USA 1996, 93, 4638–4643. [Google Scholar]
- Bendelac, A.; Savage, P.B.; Teyton, L. The Biology of NKT Cells. Annu. Rev. Immunol. 2007, 25, 297–336. [Google Scholar] [CrossRef]
- Natori, T.; Koezuka, Y.; Higa, T. Agelasphins, novel α-galactosylceramides from the marine sponge Agelas mauritianus. Tetrahedron Lett. 1993, 34, 5591–5592. [Google Scholar] [CrossRef]
- Natori, T.; Morita, M.; Akimoto, K.; Koezuka, Y. Agelasphins, novel antitumor and immunostimulatory cerebrosides from the marine sponge Agelas mauritianus. Tetrahedron 1994, 50, 2771–2784. [Google Scholar] [CrossRef]
- Morita, M.; Motoki, K.; Akimoto, K.; Natori, T.; Sakai, T.; Sawa, E.; Yamaji, K.; Koezuka, Y.; Kobayashi, E.; Fukushima, H. Structure-Activity Relationship of alpha-Galactosylceramides against B16-Bearing Mice. J. Med. Chem. 1995, 38, 2176–2187. [Google Scholar] [CrossRef]
- McEwen-Smith, R.M.; Salio, M.; Cerundolo, V. CD1d-dependent endogenous and exogenous lipid antigen presentation. Curr. Opin. Immunol. 2015, 34, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Kohlgruber, A.C.; Donado, C.A.; LaMarche, N.M.; Brenner, M.B.; Brennan, P.J. Activation strategies for invariant natural killer T cells. Immunogenetics 2016, 68, 649–663. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.C.W.; Penaranda, C.; Kashyap, P.C.; Williams, B.B.; Clardy, J.; Kronenberg, M.; Sonnenburg, J.L.; Comstock, L.E.; Bluestone, J.A.; Fischbach, M.A. Production of α-Galactosylceramide by a Prominent Member of the Human Gut Microbiota. PLoS Biol. 2013, 11, e1001610. [Google Scholar] [CrossRef]
- Okino, N.; Li, M.; Qu, Q.; Nakagawa, T.; Hayashi, Y.; Matsumoto, M.; Ishibashi, Y.; Ito, M. Two bacterial glycosphingolipid synthases responsible for the synthesis of glucuronosylceramide and α-galactosylceramide. J. Biol. Chem. 2020, 295, 10709–10725. [Google Scholar] [CrossRef] [PubMed]
- von Gerichten, J.; Schlosser, K.; Lamprecht, D.; Morace, I.; Eckhardt, M.; Wachten, D.; Jennemann, R.; Gröne, H.-J.; Mack, M.; Sandhoff, R. Diastereomer-specific quantification of bioactive hexosylceramides from bacteria and mammals. J. Lipid Res. 2017, 58, 1247–1258. [Google Scholar] [CrossRef] [PubMed]
- von Gerichten, J.; Lamprecht, D.; Opálka, L.; Soulard, D.; Marsching, C.; Pilz, R.; Sencio, V.; Herzer, S.; Galy, B.; Nordström, V.; et al. Bacterial immunogenic α-galactosylceramide identified in the murine large intestine: Dependency on diet and inflammation. J. Lipid Res. 2019, 60, 1892–1904. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, K.; Smiley, M.T.K.S.; Grusby, M.J.; Gui, M.; Burdin, M.N.; Brossay, L.; Koezuka, Y. Selective Ability of Mouse CD1 to Present Glycolipids: α-Galactosylceramide Specifically Stimulates Va14+ NK T Lymphocytes. J. Immunol. 1998, 161, 3271–3281. Available online: http://www.jimmunol.org/content/161/7/3271 (accessed on 18 September 2022).
- Shimizu, K.; Kurosawa, Y.; Taniguchi, M.; Steinman, R.M.; Fujii, S.-i. Cross-presentation of glycolipid from tumor cells loaded with α-galactosylceramide leads to potent and long-lived T cell–mediated immunity via dendritic cells. J. Exp. Med. 2007, 204, 2641–2653. [Google Scholar] [CrossRef]
- Mattarollo, S.R.; West, A.C.; Steegh, K.; Duret, H.; Paget, C.; Martin, B.; Matthews, G.M.; Shortt, J.; Chesi, M.; Bergsagel, P.L.; et al. NKT cell adjuvant-based tumor vaccine for treatment of myc oncogene-driven mouse B-cell lymphoma. Blood 2012, 120, 3019–3029. [Google Scholar] [CrossRef]
- Gableh, F.; Saeidi, M.; Hemati, S.; Hamdi, K.; Soleimanjahi, H.; Gorji, A.; Ghaemi, A. Combination of the toll like receptor agonist and α-Galactosylceramide as an efficient adjuvant for cancer vaccine. J. Biomed. Sci. 2016, 23, 1–11. [Google Scholar] [CrossRef]
- Plettenburg, O.; Bodmer-Narkevitch, A.V.; Wong, C.-H. Synthesis of α-Galactosyl Ceramide, a Potent Immunostimulatory Agent. J. Org. Chem. 2002, 67, 4559–4564. [Google Scholar] [CrossRef] [PubMed]
- Janssens, J.; Decruy, T.; Venken, K.; Seki, T.; Krols, S.; Van der Eycken, J.; Tsuji, M.; Elewaut, D.; Van Calenbergh, S. Efficient Divergent Synthesis of New Immunostimulant 4″-Modified α-Galactosylceramide Analogues. ACS Med. Chem. Lett. 2017, 8, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Drula, E.; Garron, M.-L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- Karlsson, O.P.; Dahlqvist, A.; Vikström, S.; Wieslander, Å. Lipid Dependence and Basic Kinetics of the Purified 1,2-Diacylglycerol 3-Glucosyltransferase from Membranes of Acholeplasma laidlawii. J. Biol. Chem. 1997, 272, 929–936. [Google Scholar] [CrossRef] [PubMed]
- Edman, M.; Berg, S.; Storm, P.; Wikström, M.; Vikström, S.; Öhman, A.; Wieslander, Å. Structural Features of Glycosyltransferases Synthesizing Major Bilayer and Nonbilayer-prone Membrane Lipids inAcholeplasma laidlawii and Streptococcus pneumoniae. J. Biol. Chem. 2003, 278, 8420–8428. [Google Scholar] [CrossRef] [PubMed]
- Sonnhammer, E.L.; Von Heijne, G.; Krogh, A. A hidden Markov model for predicting transmembrane helices in protein sequences. In Proceedings of the International Conference on Intelligent Systems for Molecular Biology, Montréal, QC, Canada, 28 June–1 July 1998; Volume 6, pp. 175–182. [Google Scholar]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Nishihara, K.; Kanemori, M.; Kitagawa, M.; Yanagi, H.; Yura, T. Chaperone Coexpression Plasmids: Differential and Synergistic Roles of DnaK-DnaJ-GrpE and GroEL-GroES in Assisting Folding of an Allergen of Japanese Cedar Pollen, Cryj2. Escherichia coli. Appl. Environ. Microbiol. 1998, 64, 1694–1699. [Google Scholar] [CrossRef]
- Malakhov, M.P.; Mattern, M.R.; Malakhova, O.A.; Drinker, M.; Weeks, S.; Butt, T.R. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J. Struct. Funct. Genom. 2004, 5, 75–86. [Google Scholar] [CrossRef]
- Thiyagarajan, N.; Pham, T.T.K.; Stinson, B.; Sundriyal, A.; Tumbale, P.; Lizotte-Waniewski, M.; Brew, K.; Acharya, K.R. Structure of a metal-independent bacterial glycosyltransferase that catalyzes the synthesis of histo-blood group A antigen. Sci. Rep. 2012, 2, srep00940. [Google Scholar] [CrossRef]
- Andrés, E.; Martínez, N.; Planas, A. Expression and Characterization of a Mycoplasma genitalium Glycosyltransferase in Membrane Glycolipid Biosynthesis: Potential Target against Mycoplasma Infections. J. Biol. Chem. 2011, 286, 35367–35379. [Google Scholar] [CrossRef] [PubMed]
- Dubots, E.; Audry, M.; Yamaryo, Y.; Bastien, O.; Ohta, H.; Breton, C.; Maréchal, E.; Block, M.A. Activation of the Chloroplast Monogalactosyldiacylglycerol Synthase MGD1 by Phosphatidic Acid and Phosphatidylglycerol. J. Biol. Chem. 2010, 285, 6003–6011. [Google Scholar] [CrossRef] [PubMed]
- Guerin, M.E.; Kordulakova, J.; Schaeffer, F.; Svetlikova, Z.; Buschiazzo, A.; Giganti, D.; Gicquel, B.; Mikusova, K.; Jackson, M.; Alzari, P.M. Molecular Recognition and Interfacial Catalysis by the Essential Phosphatidylinositol Mannosyltransferase PimA from Mycobacteria. J. Biol. Chem. 2007, 282, 20705–20714. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo-Unzueta, A.; Ghirardello, M.; Urresti, S.; Delso, I.; Giganti, D.; Anso, I.; Trastoy, B.; Comino, N.; Tersa, M.; D’Angelo, C.; et al. Dissecting the Structural and Chemical Determinants of the “Open-to-Closed” Motion in the Mannosyltransferase PimA from Mycobacteria. Biochemistry 2020, 59, 2934–2945. [Google Scholar] [CrossRef] [PubMed]
- Kocev, A.; Melamed, J.; Torgov, V.; Danilov, L.; Veselovsky, V.; Brockhausen, I. ThewclYgene ofEscherichia coliserotype O117 encodes an α1,4-glucosyltransferase with strict acceptor specificity but broad donor specificity. Glycobiology 2020, 30, 9003–9014. [Google Scholar] [CrossRef] [PubMed]
- Lairson, L.L.; Henrissat, B.; Davies, G.J.; Withers, S.G. Glycosyltransferases: Structures, Functions, and Mechanisms. Annu. Rev. Biochem. 2008, 77, 521–555. [Google Scholar] [CrossRef] [PubMed]
- Albesa-Jové, D.; Giganti, D.; Jackson, M.; Alzari, P.M.; Guerin, M.E. Structure-function relationships of membrane-associated GT-B glycosyltransferases. Glycobiology 2014, 24, 108–124. [Google Scholar] [CrossRef] [PubMed]
- Berg, S.; Edman, M.; Li, L.; Wikström, M.; Wieslander, Å. Wieslander, Sequence properties of the 1,2-diacylglycerol 3-glucosyltransferase from Acholeplasma laidlawii membranes. Recognition of a large group of lipid glycosyltransferases in eubacteria and archaea. J. Biol. Chem. 2001, 276, 22056–22063. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2–A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Orive-Milla, N.; Delmulle, T.; de Mey, M.; Faijes, M.; Planas, A. Metabolic engineering for glycoglycerolipids production in E. coli: Tuning phosphatidic acid and UDP-glucose pathways. Metab. Eng. 2020, 61, 106–119. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Campanella, J.J.; Bitincka, L.; Smalley, J. MatGAT: An application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinform. 2003, 4, 1–4. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caballé, M.; Faijes, M.; Planas, A. Characterization of a Glycolipid Synthase Producing α-Galactosylceramide in Bacteroides fragilis. Int. J. Mol. Sci. 2022, 23, 13975. https://doi.org/10.3390/ijms232213975
Caballé M, Faijes M, Planas A. Characterization of a Glycolipid Synthase Producing α-Galactosylceramide in Bacteroides fragilis. International Journal of Molecular Sciences. 2022; 23(22):13975. https://doi.org/10.3390/ijms232213975
Chicago/Turabian StyleCaballé, Marc, Magda Faijes, and Antoni Planas. 2022. "Characterization of a Glycolipid Synthase Producing α-Galactosylceramide in Bacteroides fragilis" International Journal of Molecular Sciences 23, no. 22: 13975. https://doi.org/10.3390/ijms232213975
APA StyleCaballé, M., Faijes, M., & Planas, A. (2022). Characterization of a Glycolipid Synthase Producing α-Galactosylceramide in Bacteroides fragilis. International Journal of Molecular Sciences, 23(22), 13975. https://doi.org/10.3390/ijms232213975





