Urinary Measurement of Epigenetic DNA Modifications and 8-oxodG as Possible Noninvasive Markers of Colon Cancer Evolution
Abstract
1. Introduction
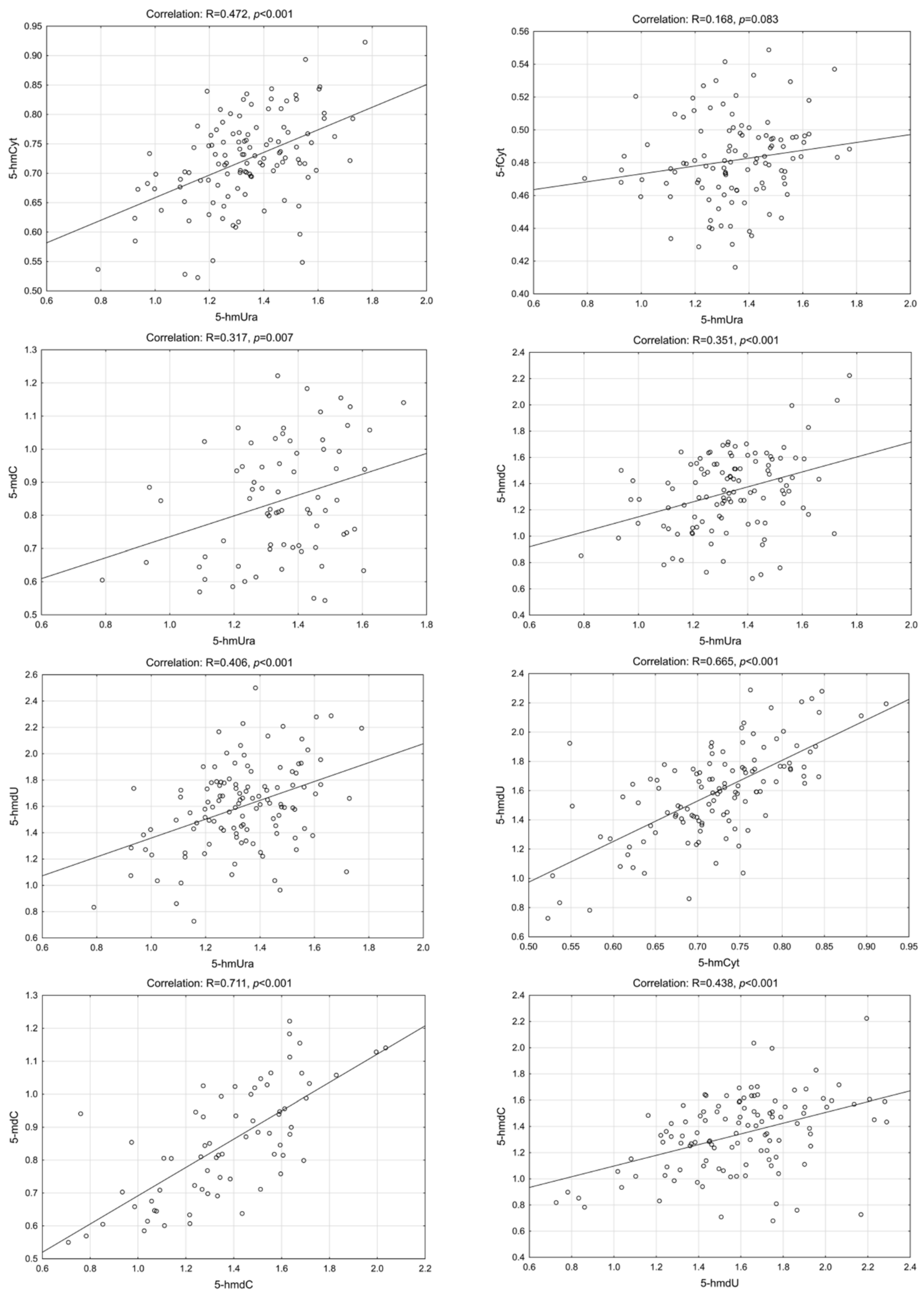
2. Results
3. Discussion
4. Materials and Methods
4.1. The Determination of the Epigenetic Modifications and 8-oxodG Levels in Urine [28]
4.2. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Pineros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer. 2021, 149, 778–789. [Google Scholar] [CrossRef]
- Effendi-Ys, R. Colonoscopy, Biomarkers, and Targeted Therapy in Colorectal Cancer. Acta Med. Indones. 2022, 54, 476–486. [Google Scholar] [PubMed]
- Chia, N.; Wang, L.; Lu, X.; Senut, M.C.; Brenner, C.; Ruden, D.M. Hypothesis: Environmental regulation of 5-hydroxymethylcytosine by oxidative stress. Epigenetics 2011, 6, 853–856. [Google Scholar] [CrossRef] [PubMed]
- Marsit, C.J. Influence of environmental exposure on human epigenetic regulation. J. Exp. Biol. 2015, 218, 71–79. [Google Scholar] [CrossRef] [PubMed]
- MacLennan, R. Diet and colorectal cancer. Int. J. Cancer 1997, 71 (Suppl. 10), 10–12. [Google Scholar] [CrossRef]
- Feng, S.; Jacobsen, S.E.; Reik, W. Epigenetic reprogramming in plant and animal development. Science 2010, 330, 622–627. [Google Scholar] [CrossRef]
- Bhutani, N.; Burns, D.M.; Blau, H.M. DNA Demethylation Dynamics. Cell 2011, 146, 866–872. [Google Scholar] [CrossRef]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef]
- Pfaffeneder, T.; Spada, F.; Wagner, M.; Brandmayr, C.; Laube, S.K.; Eisen, D.; Truss, M.; Steinbacher, J.; Hackner, B.; Kotljarova, O.; et al. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat. Chem. Biol. 2014, 10, 574–581. [Google Scholar] [CrossRef]
- Guo, J.U.; Su, Y.J.; Zhong, C.; Ming, G.L.; Song, H.J. Emerging roles of TET proteins and 5-Hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle 2011, 10, 2662–2668. [Google Scholar] [CrossRef]
- Branco, M.R.; Ficz, G.; Reik, W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat. Rev. Genet. 2012, 13, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Dziaman, T.; Gackowski, D.; Guz, J.; Linowiecka, K.; Bodnar, M.; Starczak, M.; Zarakowska, E.; Modrzejewska, M.; Szpila, A.; Szpotan, J.; et al. Characteristic profiles of DNA epigenetic modifications in colon cancer and its predisposing conditions-benign adenomas and inflammatory bowel disease. Clin. Epigenetics 2018, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kraus, S.; Arber, N. Inflammation and colorectal cancer. Curr. Opin. Pharmacol. 2009, 9, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Jess, T.; Gamborg, M.; Matzen, P.; Munkholm, P.; Sorensen, T.I. Increased risk of intestinal cancer in Crohn’s disease: A meta-analysis of population-based cohort studies. Am. J. Gastroenterol. 2005, 100, 2724–2729. [Google Scholar] [CrossRef] [PubMed]
- MacFarlane, A.J.; Stover, P.J. Convergence of genetic, nutritional and inflammatory factors in gastrointestinal cancers. Nutr. Rev. 2007, 65, S157–S166. [Google Scholar] [CrossRef]
- Munkholm, P. Review article: The incidence and prevalence of colorectal cancer in inflammatory bowel disease. Aliment. Pharmacol. Ther. 2003, 18 (Suppl. 2), 1–5. [Google Scholar] [CrossRef]
- Schnekenburger, M.; Diederich, M. Epigenetics Offer New Horizons for Colorectal Cancer Prevention. Curr. Color. Cancer Rep. 2012, 8, 66–81. [Google Scholar] [CrossRef]
- Carmona, F.J.; Esteller, M. Epigenomics of human colon cancer. Mutat. Res. 2010, 693, 53–60. [Google Scholar] [CrossRef]
- Rozalski, R.; Gackowski, D.; Siomek-Gorecka, A.; Banaszkiewicz, Z.; Olinski, R. Urinary Measurement of Epigenetic DNA Modifications: A Non-Invasive Assessment of the Whole-Body Epigenetic Status in Healthy Subjects and Colorectal Cancer Patients. Chemistryopen 2016, 5, 550–553. [Google Scholar] [CrossRef]
- Cooke, M.S.; Olinski, R.; Loft, S. Escula Measurement and meaning of oxidatively modified DNA lesions in urine. Cancer Epidemiol. Biomark. Prev. 2008, 17, 3–14. [Google Scholar] [CrossRef]
- Cooke, M.S.; Olinski, R.; Evans, M.D. Does measurement of oxidative damage to DNA have clinical significance? Clin. Chim. Acta 2006, 365, 30–49. [Google Scholar] [CrossRef] [PubMed]
- Fugger, K.; Bajrami, I.; Silva Dos Santos, M.; Young, S.J.; Kunzelmann, S.; Kelly, G.; Hewitt, G.; Patel, H.; Goldstone, R.; Carell, T.; et al. Targeting the nucleotide salvage factor DNPH1 sensitizes BRCA-deficient cells to PARP inhibitors. Science 2021, 372, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Baker, D.; Liu, P.F.; Burdzy, A.; Sowers, L.C. Characterization of the substrate specificity of a human 5-hydroxymethyluracil glycosylase activity. Chem. Res. Toxicol. 2002, 15, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, H.; Hong, S.; Bhagwat, A.S.; Zhang, X.; Cheng, X.D. Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: Its structural basis and implications for active DNA demethylation. Nucleic Acids Res. 2012, 40, 10203–10214. [Google Scholar] [CrossRef]
- Kemmerich, K.; Dingler, F.A.; Rada, C.; Neuberger, M.S. Germline ablation of SMUG1 DNA glycosylase causes loss of 5-hydroxymethyluracil- and UNG-backup uracil-excision activities and increases cancer predisposition of Ung(−/−)Msh2(−/−) mice. Nucleic Acids Res. 2012, 40, 6016–6025. [Google Scholar] [CrossRef] [PubMed]
- Zauri, M.; Berridge, G.; Thezenas, M.L.; Pugh, K.M.; Goldin, R.; Kessler, B.M.; Kriaucionis, S. CDA directs metabolism of epigenetic nucleosides revealing a therapeutic window in cancer. Nature 2015, 524, 114–118. [Google Scholar] [CrossRef]
- Grin, I.; Ishchenko, A.A. An interplay of the base excision repair and mismatch repair pathways in active DNA demethylation. Nucleic Acids Res. 2016, 44, 3713–3727. [Google Scholar] [CrossRef]
- Rozalski, R.; Gackowski, D.; Skalska-Bugala, A.; Starczak, M.; Siomek-Gorecka, A.; Zarakowska, E.; Modrzejewska, M.; Dziaman, T.; Szpila, A.; Linowiecka, K.; et al. The urinary excretion of epigenetically modified DNA as a marker of pediatric ALL status and chemotherapy response. Sci. Rep. 2021, 11, 21345. [Google Scholar] [CrossRef]
- Rozalski, R.; Gackowski, D.; Siomek-Gorecka, A.; Starczak, M.; Modrzejewska, M.; Banaszkiewicz, Z.; Olinski, R. Urinary 5-hydroxymethyluracil and 8-oxo-7,8-dihydroguanine as potential biomarkers in patients with colorectal cancer. Biomarkers 2015, 20, 287–291. [Google Scholar] [CrossRef]
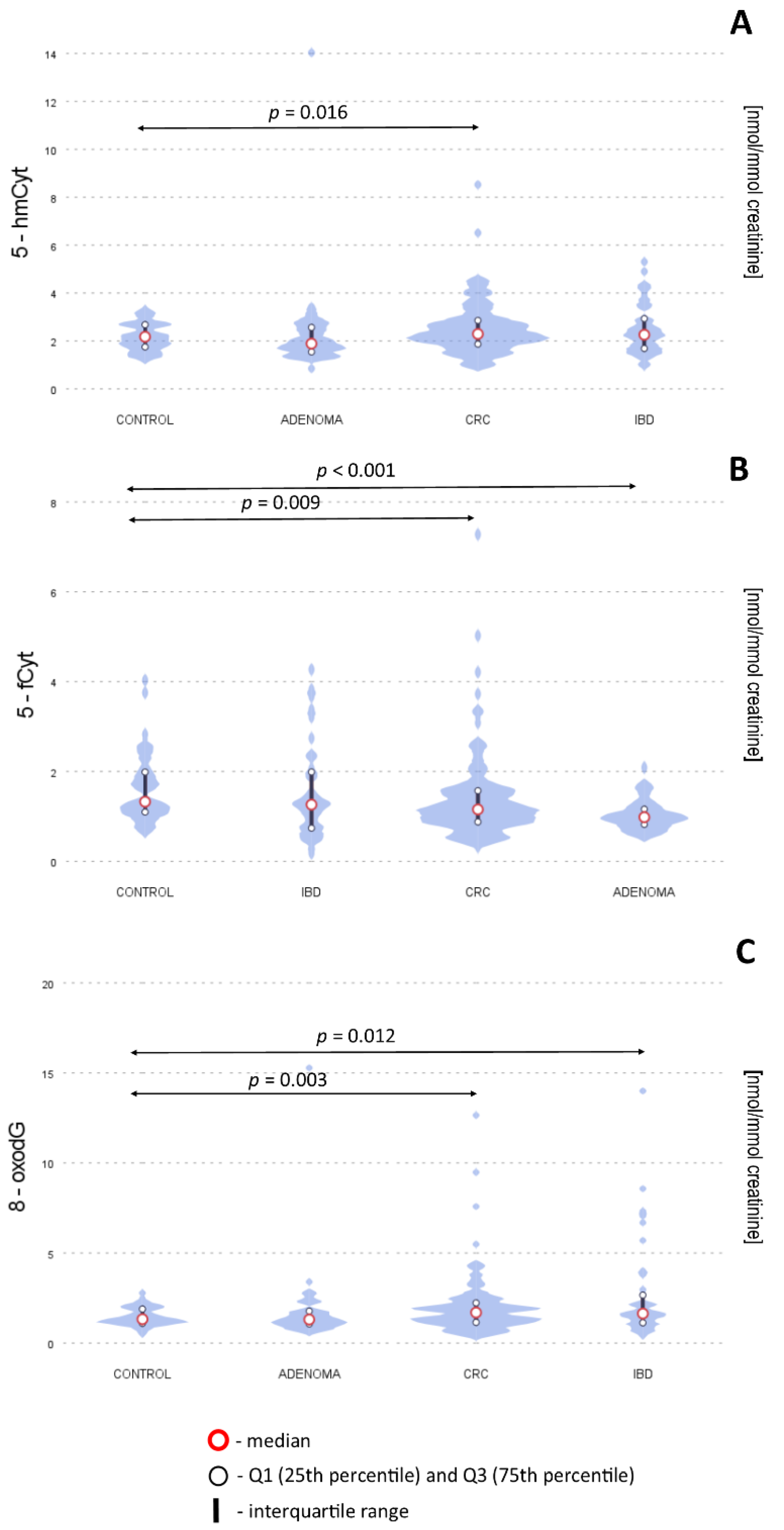



| [nmol/mmol Creatinine] | Control (N = 45) | CRC (N = 121) | AD (N = 65) | IBD (N = 42) | Control vs. CRC p Value | Control vs. AD p Value | Control vs. IBD p Value |
|---|---|---|---|---|---|---|---|
| 5-hmdU | 6.2 (4.6–7.4) | 6.4 (4.5–8.3) | 6.2 (4.4–7.7) | 6.5 (4.6–8.3) | 0.061 | 0.377 | 0.363 |
| 5-mdC | 1.8 (1.2–3.2) | 1.7 (0.82–4.0) | 2.0 (1.0–5.1) | 2.9 (1.0–4.0) | 0.729 | 0.426 | 0.500 |
| 5-hmCyt | 2.1 (1.7–2.6) | 2.3 (1.8–2.8) | 1.9 (1.5–2.4) | 2.3 (1.7–3.0) | 0.016 * | 0.735 | 0.091 |
| 5-fCyt | 1.3 (1.1–1.9) | 1.1 (0.8–1.5) | 1.0 (0.8–1.1) | 1.3 (0.7–2.1) | 0.009 * | <0.001 * | 0.750 |
| 5-hmdC | 2.1 (1.7–2.9) | 2.3 (1.3–3.2) | 1. 9 (1.1–2.8) | 2.1 (1.2–3.4) | 0.903 | 0.160 | 0.803 |
| 8-oxodG | 1.3 (1.1–1.8) | 1.7 (1.1–2.2) | 1.3 (1.0–1.8) | 1.6 (1.2–2.7) | 0.003 * | 0.368 | 0.012 * |
| 5-hmUra | 7.3 (5.8–8.2) | 6.2 (5.0–8.2) | 6.5 (6.1–8.3) | 7.9 (6.3–9.9) | 0.256 | 0.290 | 0.171 |
| Control (N = 45) | CRC (N = 121) | AD (N = 65) | IBD (N = 42) | |
|---|---|---|---|---|
| Male (%) | 44% | 56% | 59% | 34% |
| Age (year) | 56 | 65 | 64 | 34 |
| Weight (kg) | 77 | 75 | 80 | 61 |
| Height (cm) | 168 | 168 | 170 | 169 |
| Body Mass Index | 26.3 | 26.4 | 27.3 | 21.2 |
| Histological Grade (%) | Stage: A—8% B—47% C—29% D—7% Hagitt scale I–IV—8% | AT—85% ATV—15% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skalska-Bugala, A.; Siomek-Gorecka, A.; Banaszkiewicz, Z.; Olinski, R.; Rozalski, R. Urinary Measurement of Epigenetic DNA Modifications and 8-oxodG as Possible Noninvasive Markers of Colon Cancer Evolution. Int. J. Mol. Sci. 2022, 23, 13826. https://doi.org/10.3390/ijms232213826
Skalska-Bugala A, Siomek-Gorecka A, Banaszkiewicz Z, Olinski R, Rozalski R. Urinary Measurement of Epigenetic DNA Modifications and 8-oxodG as Possible Noninvasive Markers of Colon Cancer Evolution. International Journal of Molecular Sciences. 2022; 23(22):13826. https://doi.org/10.3390/ijms232213826
Chicago/Turabian StyleSkalska-Bugala, Aleksandra, Agnieszka Siomek-Gorecka, Zbigniew Banaszkiewicz, Ryszard Olinski, and Rafal Rozalski. 2022. "Urinary Measurement of Epigenetic DNA Modifications and 8-oxodG as Possible Noninvasive Markers of Colon Cancer Evolution" International Journal of Molecular Sciences 23, no. 22: 13826. https://doi.org/10.3390/ijms232213826
APA StyleSkalska-Bugala, A., Siomek-Gorecka, A., Banaszkiewicz, Z., Olinski, R., & Rozalski, R. (2022). Urinary Measurement of Epigenetic DNA Modifications and 8-oxodG as Possible Noninvasive Markers of Colon Cancer Evolution. International Journal of Molecular Sciences, 23(22), 13826. https://doi.org/10.3390/ijms232213826







