Mammalian Target of Rapamycin (mTOR) Signaling at the Crossroad of Muscle Fiber Fate in Sarcopenia
Abstract
1. Introduction
2. Supramolecular Organization of the Mammalian Target of Rapamycin
3. Muscle Protein Synthesis: The Mammalian Target of Rapamycin Complex 1 Axis
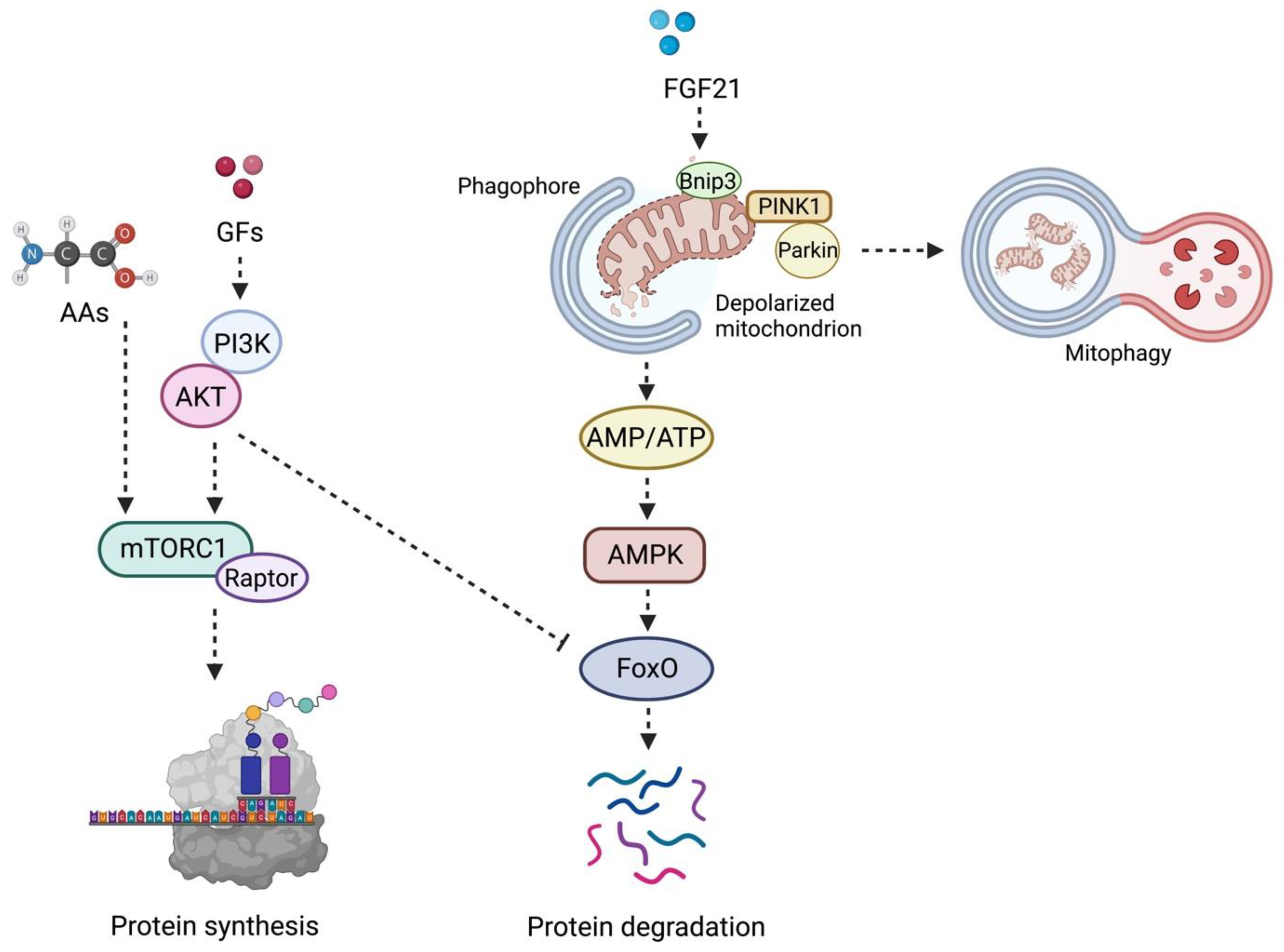
mTORC1 Axis: Different Stimuli for Muscle Protein Synthesis
4. Muscle Protein Degradation: The Other Face of the mTORC1 Axis during Aging
5. The mTORC2 Axis: Not a Twin Complex
6. mTOR Axis in Sarcopenia: All That Glitters Is Not Gold
7. Current Strategies Targeting mTOR to Manage Sarcopenia
| Strategy | Model | Reference | Main Findings |
|---|---|---|---|
| Lactococcus cremoris subsp. cremoris FC-fermented milk | Aged mouse | Aoi et al., 2022 [85] | Increase in mTOR phosphorylation and MPS |
| LRS-UNE-L peptide | C2C12 cells and aged mouse | Baek et al., 2022 [97] | Stimulation of mTORC1 axis and enhanced muscle fiber regeneration |
| Clinical trial on testosterone + resistance exercise | Men (65–75 years) | Gharahdaghi et al., 2019 [95] | Upregulation of mTOR signaling and increase in muscle mass and function |
| Collagen hydrolysate tripeptides (CTP) | Aged mouse | Kim et al., 2022 [96] | Stimulation of mTOR signaling and increase in muscle mass |
| Chrysanthemum extracts (CME) | Aged rat and mouse | Kwon et al., 2021 [100] | Increase in mTOR phosphorylation and amelioration of muscle mass and function |
| Rimonabant | C2C12 cells | Le Bacquer et al., 2021 [99] | Increase in mTOR phosphorylation and MPS |
| Magnesium | C2C12 cells and aged mouse | Liu et al., 2021 [84] | Increase in mTOR phosphorylation, enhanced muscle regeneration, preservation of muscle mass and strength |
| Fish proteins | Young rat | Morisasa et al., 2022 [83] | Promotion of muscle hypertrophy via AKT–mTOR signaling |
| Calorie restriction | Aged rat | Chen et al., 2019 [86] | Decrease in mTOR content/phosphorylation and preservation of muscle mass |
| Root of Maca | C2C12 cells | Yi et al., 2022 [98] | Promotion of muscle hypertrophy via AKT–mTOR signaling |
| Resistance exercise | Aged rat | Zeng et al., 2020 [82] | Decrease of mTOR signaling, stimulation of autophagy, preservation of muscle mass and function |
8. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marzetti, E.; Calvani, R.; Tosato, M.; Cesari, M.; Di Bari, M.; Cherubini, A.; Collamati, A.; D’Angelo, E.; Pahor, M.; Bernabei, R.; et al. Sarcopenia: An Overview. Aging Clin. Exp. Res. 2017, 29, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Bernabei, R.; Landi, F.; Calvani, R.; Cesari, M.; Del Signore, S.; Anker, S.D.; Bejuit, R.; Bordes, P.; Cherubini, A.; Cruz-Jentoft, A.J.; et al. Multicomponent Intervention to Prevent Mobility Disability in Frail Older Adults: Randomised Controlled Trial (SPRINTT Project). BMJ 2022, 377, e068788. [Google Scholar] [CrossRef] [PubMed]
- Sartori, R.; Romanello, V.; Sandri, M. Mechanisms of Muscle Atrophy and Hypertrophy: Implications in Health and Disease. Nat. Commun. 2021, 12, 330. [Google Scholar] [CrossRef] [PubMed]
- Fry, C.S.; Rasmussen, B.B. Skeletal Muscle Protein Balance and Metabolism in the Elderly. Curr. Aging Sci. 2011, 4, 260–268. [Google Scholar] [CrossRef]
- Yoon, M.S. mTOR as a Key Regulator in Maintaining Skeletal Muscle Mass. Front. Physiol. 2017, 8, 788. [Google Scholar] [CrossRef]
- Tang, H.; Shrager, J.B.; Goldman, D. Rapamycin Protects Aging Muscle. Aging 2019, 11, 5868–5870. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. MTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef]
- Aylett, C.H.S.; Sauer, E.; Imseng, S.; Boehringer, D.; Hall, M.N.; Ban, N.; Maier, T. Architecture of Human mTOR Complex 1. Science 2016, 351, 48–52. [Google Scholar] [CrossRef]
- Yu, Z.; Chen, J.; Takagi, E.; Wang, F.; Saha, B.; Liu, X.; Joubert, L.M.; Gleason, C.E.; Jin, M.; Li, C.; et al. Interactions between mTORC2 Core Subunits Rictor and MSin1 Dictate Selective and Context-Dependent Phosphorylation of Substrate Kinases SGK1 and Akt. J. Biol. Chem. 2022, 298, 102288. [Google Scholar] [CrossRef]
- Scaiola, A.; Mangia, F.; Imseng, S.; Boehringer, D.; Berneiser, K.; Shimobayashi, M.; Stuttfeld, E.; Hall, M.N.; Ban, N.; Maier, T. The 3.2-Å Resolution Structure of Human mTORC2. Sci. Adv. 2020, 6, eabc1251. [Google Scholar] [CrossRef]
- Baraldo, M.; Nogara, L.; Dumitras, G.A.; Dondjang, A.H.T.; Geremia, A.; Scalabrin, M.; Türk, C.; Telkamp, F.; Zentilin, L.; Giacca, M.; et al. Raptor Is Critical for Increasing the Mitochondrial Proteome and Skeletal Muscle Force during Hypertrophy. FASEB J. 2021, 35, e22031. [Google Scholar] [CrossRef] [PubMed]
- Baehr, L.M.; Hughes, D.C.; Waddell, D.S.; Bodine, S.C. SnapShot: Skeletal Muscle Atrophy. Cell 2022, 185, 1618–1618.e1. [Google Scholar] [CrossRef] [PubMed]
- Vainshtein, A.; Sandri, M. Signaling Pathways That Control Muscle Mass. Int. J. Mol. Sci. 2020, 21, 5759. [Google Scholar] [CrossRef] [PubMed]
- Ham, D.J.; Börsch, A.; Lin, S.; Thürkauf, M.; Weihrauch, M.; Reinhard, J.R.; Delezie, J.; Battilana, F.; Wang, X.; Kaiser, M.S.; et al. The Neuromuscular Junction Is a Focal Point of MTORC1 Signaling in Sarcopenia. Nat. Commun. 2020, 11, 4510. [Google Scholar] [CrossRef]
- Baraldo, M.; Geremia, A.; Pirazzini, M.; Nogara, L.; Solagna, F.; Türk, C.; Nolte, H.; Romanello, V.; Megighian, A.; Boncompagni, S.; et al. Skeletal Muscle mTORC1 Regulates Neuromuscular Junction Stability. J. Cachexia Sarcopenia Muscle 2020, 11, 208–225. [Google Scholar] [CrossRef]
- Ketilly, P.; Alves, N.; Cruz, A.; Silva, W.J.; Labeit, S.; Moriscot, A.S. MiR-29c Increases Protein Synthesis in Skeletal Muscle Independently of AKT/mTOR. Int. J. Mol. Sci. 2022, 23, 7198. [Google Scholar] [CrossRef]
- Jin, J.; Li, F.; Fan, C.; Wu, Y.; He, C. Elevated Mir-145-5p Is Associated with Skeletal Muscle Dysfunction and Triggers Apoptotic Cell Death in C2C12 Myotubes. J. Muscle Res. Cell Motil. 2022, 43, 135–145. [Google Scholar] [CrossRef]
- Picca, A.; Calvani, R.; Sirago, G.; Coelho-Junior, H.J.; Marzetti, E. Molecular Routes to Sarcopenia and Biomarker Development: Per Aspera Ad Astra. Curr. Opin. Pharmacol. 2021, 57, 140–147. [Google Scholar] [CrossRef]
- Liu, G.Y.; Sabatini, D.M. mTOR at the Nexus of Nutrition, Growth, Ageing and Disease. Nat. Rev. Mol. Cell Biol. 2020, 21, 183–203. [Google Scholar] [CrossRef]
- Nagao, H.; Cai, W.; Albrechtsen, N.J.W.; Steger, M.; Batista, T.M.; Pan, H.; Dreyfuss, J.M.; Mann, M.; Kahn, C.R. Distinct Signaling by Insulin and IGF-1 Receptors and Their Extra- And Intracellular Domains. Proc. Natl. Acad. Sci. USA 2021, 118, e2019474118. [Google Scholar] [CrossRef]
- Sarbassov, D.D.; Guertin, D.A.; Ali, S.M.; Sabatini, D.M. Phosphorylation and Regulation of Akt/PKB by the Rictor-mTOR Complex. Science 2005, 307, 1098–1101. [Google Scholar] [CrossRef] [PubMed]
- Philp, A.; Hamilton, D.L.; Baar, K. Highlighted Topic Signals Mediating Skeletal Muscle Remodeling by Activity Signals Mediating Skeletal Muscle Remodeling by Resistance Exercise: PI3-Kinase Independent Activation of mTORC1. J. Appl. Physiol. 2011, 110, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Frey, J.W.; Jacobs, B.L.; Goodman, C.A.; Hornberger, T.A. A Role for Raptor Phosphorylation in the Mechanical Activation of mTOR Signaling. Cell. Signal. 2014, 26, 313–322. [Google Scholar] [CrossRef] [PubMed]
- You, J.S.; Mcnally, R.M.; Jacobs, B.L.; Privett, R.E.; Gundermann, D.M.; Lin, K.H.; Steinert, N.D.; Goodman, C.A.; Hornberger, T.A. The Role of Raptor in the Mechanical Load-Induced Regulation of mTOR Signaling, Protein Synthesis, and Skeletal Muscle Hypertrophy. FASEB J. 2019, 33, 4021–4034. [Google Scholar] [CrossRef]
- Vary, T.C.; Jefferson, L.S.; Kimball, S.R. Amino Acid-Induced Stimulation of Translation Initiation in Rat Skeletal Muscle. Am. J. Physiol. 1999, 277, 1077–1086. [Google Scholar] [CrossRef]
- Jeyapalan, A.S.; Orellana, R.A.; Suryawan, A.; O’Connor, P.M.J.; Nguyen, H.V.; Escobar, J.; Frank, J.W.; Davis, T.A. Glucose Stimulates Protein Synthesis in Skeletal Muscle of Neonatal Pigs through an AMPK- and mTOR-Independent Process. Am. J. Physiol. Endocrinol. Metab. 2007, 293, 595–603. [Google Scholar] [CrossRef][Green Version]
- Sancak, Y.; Peterson, T.R.; Shaul, Y.D.; Lindquist, R.A.; Thoreen, C.C.; Bar-Peled, L.; Sabatini, D.M. The Rag GTPases Bind Raptor and Mediate Amino Acid Signaling to mTORC1. Science 2008, 320, 1496–1501. [Google Scholar] [CrossRef]
- Korzick, D.H.; Sharda, D.R.; Pruznak, A.M.; Lang, C.H. Aging Accentuates Alcohol-Induced Decrease in Protein Synthesis in Gastrocnemius. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013, 304, R887–R898. [Google Scholar] [CrossRef]
- Tang, H.; Inoki, K.; Brooks, S.V.; Okazawa, H.; Lee, M.; Wang, J.; Kim, M.; Kennedy, C.L.; Macpherson, P.C.D.; Ji, X.; et al. mTORC1 Underlies Age-Related Muscle Fiber Damage and Loss by Inducing Oxidative Stress and Catabolism. Aging Cell 2019, 18, e12943. [Google Scholar] [CrossRef]
- Cohen, S.; Zhai, B.; Gygi, S.P.; Goldberg, A.L. Ubiquitylation by Trim32 Causes Coupled Loss of Desmin, Z-Bands, and Thin Filaments in Muscle Atrophy. J. Cell Biol. 2012, 198, 575–589. [Google Scholar] [CrossRef]
- Mokhonova, E.I.; Avliyakulov, N.K.; Kramerova, I.; Kudryashova, E.; Haykinson, M.J.; Spencer, M.J. The E3 Ubiquitin Ligase TRIM32 Regulates Myoblast Proliferation by Controlling Turnover of NDRG2. Hum. Mol. Genet. 2015, 24, 2873–2883. [Google Scholar] [CrossRef] [PubMed]
- Skoglund, E.; Grönholdt-Klein, M.; Rullman, E.; Thornell, L.E.; Strömberg, A.; Hedman, A.; Cederholm, T.; Ulfhake, B.; Gustafsson, T. Longitudinal Muscle and Myocellular Changes in Community-Dwelling Men over Two Decades of Successful Aging–The ULSAM Cohort Revisited. J. Gerontol. A Biol. Sci. Med. Sci. 2020, 75, 654–663. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, C.J. The Genetics of Ageing. Nature 2010, 464, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Sirago, G.; Picca, A.; Giacomello, E.; Marzetti, E.; Toniolo, L. The Contribution of Genetics to Muscle Disuse, Retraining, and Aging. Genes 2022, 13, 1378. [Google Scholar] [CrossRef]
- Segalés, J.; Perdiguero, E.; Serrano, A.L.; Sousa-Victor, P.; Ortet, L.; Jardí, M.; Budanov, A.V.; Garcia-Prat, L.; Sandri, M.; Thomson, D.M.; et al. Sestrin Prevents Atrophy of Disused and Aging Muscles by Integrating Anabolic and Catabolic Signals. Nat. Commun. 2020, 11, 189. [Google Scholar] [CrossRef]
- Gu, X.; Jouandin, P.; Lalgudi, P.V.; Binari, R.; Valenstein, M.L.; Reid, M.A.; Allen, A.E.; Kamitaki, N.; Locasale, J.W.; Perrimon, N.; et al. Sestrin Mediates Detection of and Adaptation to Low-Leucine Diets in Drosophila. Nature 2022, 608, 209–216. [Google Scholar] [CrossRef]
- Xu, D.; Shimkus, K.L.; Lacko, H.A.; Kutzler, L.; Jefferson, L.S.; Kimball, S.R. Evidence for a Role for Sestrin1 in Mediating Leucine-Induced Activation of mTORC1 in Skeletal Muscle. Am. J. Physiol. Endocrinol. Metab. 2019, 316, E817–E828. [Google Scholar] [CrossRef]
- Bentzinger, C.F.; Romanino, K.; Cloëtta, D.; Lin, S.; Mascarenhas, J.B.; Oliveri, F.; Xia, J.; Casanova, E.; Costa, C.F.; Brink, M.; et al. Skeletal Muscle-Specific Ablation of Raptor, but Not of Rictor, Causes Metabolic Changes and Results in Muscle Dystrophy. Cell Metab. 2008, 8, 411–424. [Google Scholar] [CrossRef]
- Shaw, R.J. Raptor Swoops in on Metabolism. Cell Metab. 2008, 8, 343–344. [Google Scholar] [CrossRef][Green Version]
- Shu, L.; Houghton, P.J. The MTORC2 Complex Regulates Terminal Differentiation of C2C12 Myoblasts. Mol. Cell. Biol. 2009, 29, 4691–4700. [Google Scholar] [CrossRef]
- Ruiz, A.; Dror, E.; Handschin, C.; Furrer, R.; Perez-Schindler, J.; Bachmann, C.; Treves, S.; Zorzato, F. Over-Expression of a Retinol Dehydrogenase (SRP35/DHRS7C) in Skeletal Muscle Activates mTORC2, Enhances Glucose Metabolism and Muscle Performance. Sci. Rep. 2018, 8, 636. [Google Scholar] [CrossRef] [PubMed]
- Kowalsky, A.H.; Namkoong, S.; Mettetal, E.; Park, H.W.; Kazyken, D.; Fingar, D.C.; Lee, J.H. The GATOR2-mTORC2 Axis Mediates Sestrin2-Induced AKT Ser/Thr Kinase Activation. J. Biol. Chem. 2020, 295, 1769–1780. [Google Scholar] [CrossRef] [PubMed]
- Rion, N.; Castets, P.; Lin, S.; Enderle, L.; Reinhard, J.R.; Ruëgg, M.A. mTORC2 Affects the Maintenance of the Muscle Stem Cell Pool. Skelet. Muscle 2019, 9, 30. [Google Scholar] [CrossRef] [PubMed]
- Sanvee, G.M.; Hitzfeld, L.; Bouitbir, J.; Krähenbühl, S. mTORC2 Is an Important Target for Simvastatin-Associated Toxicity in C2C12 Cells and Mouse Skeletal Muscle–Roles of Rap1 Geranylgeranylation and Mitochondrial Dysfunction. Biochem. Pharmacol. 2021, 192, 114750. [Google Scholar] [CrossRef]
- Fry, C.S.; Drummond, M.J.; Glynn, E.L.; Dickinson, J.M.; Gundermann, D.M.; Timmerman, K.L.; Walker, D.K.; Dhanani, S.; Volpi, E.; Rasmussen, B.B. Aging Impairs Contraction-Induced Human Skeletal Muscle mTORC1 Signaling and Protein Synthesis. Skelet. Muscle 2011, 1, 11. [Google Scholar] [CrossRef]
- Sandri, M.; Barberi, L.; Bijlsma, A.Y.; Blaauw, B.; Dyar, K.A.; Milan, G.; Mammucari, C.; Meskers, C.G.M.; Pallafacchina, G.; Paoli, A.; et al. Signalling Pathways Regulating Muscle Mass in Ageing Skeletal Muscle. the Role of the IGF1-Akt-mTOR-FoxO Pathway. Biogerontology 2013, 14, 303–323. [Google Scholar] [CrossRef]
- Barns, M.; Gondro, C.; Tellam, R.L.; Radley-Crabb, H.G.; Grounds, M.D.; Shavlakadze, T. Molecular Analyses Provide Insight into Mechanisms Underlying Sarcopenia and Myofibre Denervation in Old Skeletal Muscles of Mice. Int. J. Biochem. Cell Biol. 2014, 53, 174–185. [Google Scholar] [CrossRef]
- Gaster, M.; Poulsen, P.; Handberg, A.; Schrøder, H.D.; Beck-Nielsen, H. Direct Evidence of Fiber Type-Dependent GLUT-4 Expression in Human Skeletal Muscle. Am. J. Physiol. Endocrinol. Metab. 2000, 278, E910–E916. [Google Scholar] [CrossRef]
- Petersen, K.F.; Morino, K.; Alves, T.C.; Kibbey, R.G.; Dufour, S.; Sono, S.; Yoo, P.S.; Cline, G.W.; Shulman, G.I. Effect of Aging on Muscle Mitochondrial Substrate Utilization in Humans. Proc. Natl. Acad. Sci. USA 2015, 112, 11330–11334. [Google Scholar] [CrossRef]
- Anand, A.; Nambirajan, A.; Kumar, V.; Agarwal, S.; Sharma, S.; Mohta, S.; Gopi, S.; Kaushal, K.; Gunjan, D.; Singh, N.; et al. Alterations in Autophagy and Mammalian Target of Rapamycin (mTOR) Pathways Mediate Sarcopenia in Patients with Cirrhosis. J. Clin. Exp. Hepatol. 2022, 12, 510–518. [Google Scholar] [CrossRef]
- Barnard, R.J.; Youngren, J.F.; Martin, D.A. Diet, Not Aging, Causes Skeletal Muscle Insulin Resistance. Gerontology 1995, 41, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Bian, A.; Ma, Y.; Zhou, X.; Guo, Y.; Wang, W.; Zhang, Y.; Wang, X. Association between Sarcopenia and Levels of Growth Hormone and Insulin-like Growth Factor-1 in the Elderly. BMC Musculoskelet. Disord. 2020, 21, 214. [Google Scholar] [CrossRef] [PubMed]
- Chew, J.; Tay, L.; Lim, J.P.; Leung, B.P.; Yeo, A.; Yew, S.; Ding, Y.Y.; Lim, W.S. Serum Myostatin and IGF-1 as Gender-Specific Biomarkers of Frailty and Low Muscle Mass in Community-Dwelling Older Adults. J. Nutr. Health Aging 2019, 23, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Roubenoff, R.; Parise, H.; Payette, H.A.; Abad, L.W.; D’Agostino, R.; Jacques, P.F.; Wilson, P.W.F.; Dinarello, C.A.; Harris, T.B. Cytokines, Insulin-like Growth Factor 1, Sarcopenia, and Mortality in Very Old Community-Dwelling Men and Women: The Framingham Heart Study. Am. J. Med. 2003, 115, 429–435. [Google Scholar] [CrossRef]
- Kwak, J.Y.; Hwang, H.; Kim, S.K.; Choi, J.Y.; Lee, S.M.; Bang, H.; Kwon, E.S.; Lee, K.P.; Chung, S.G.; Kwon, K.S. Prediction of Sarcopenia Using a Combination of Multiple Serum Biomarkers. Sci. Rep. 2018, 8, 8574. [Google Scholar] [CrossRef]
- Deschenes, M.R.; McCoy, R.W.; Holdren, A.N.; Eason, M.K. Gender Influences Neuromuscular Adaptations to Muscle Unloading. Eur. J. Appl. Physiol. 2009, 105, 889–897. [Google Scholar] [CrossRef]
- Phillips, B.E.; Williams, J.P.; Gustafsson, T.; Bouchard, C.; Rankinen, T.; Knudsen, S.; Smith, K.; Timmons, J.A.; Atherton, P.J. Molecular Networks of Human Muscle Adaptation to Exercise and Age. PLoS Genet. 2013, 9, e1003389. [Google Scholar] [CrossRef]
- Farnfield, M.M.; Breen, L.; Carey, K.A.; Garnham, A.; Cameron-Smith, D. Activation of MTOR Signalling in Young and Old Human Skeletal Muscle in Response to Combined Resistance Exercise and Whey Protein Ingestion. Appl. Physiol. Nutr. Metab. 2012, 37, 21–30. [Google Scholar] [CrossRef]
- Drummond, M.J.; Miyazaki, M.; Dreyer, H.C.; Pennings, B.; Dhanani, S.; Volpi, E.; Esser, K.A.; Rasmussen, B.B. Expression of Growth-Related Genes in Young and Older Human Skeletal Muscle Following an Acute Stimulation of Protein Synthesis. J. Appl. Physiol. 2009, 106, 1403–1411. [Google Scholar] [CrossRef]
- Drummond, M.J.; Dreyer, H.C.; Pennings, B.; Fry, C.S.; Dhanani, S.; Dillon, E.L.; Sheffield-Moore, M.; Volpi, E.; Rasmussen, B.B. Skeletal Muscle Protein Anabolic Response to Resistance Exercise and Essential Amino Acids Is Delayed with Aging. J. Appl. Physiol. 2008, 104, 1452–1461. [Google Scholar] [CrossRef]
- Figueiredo, V.C.; Englund, D.A.; Vechetti, I.J.; Alimov, A.; Peterson, C.A.; McCarthy, J.J. Phosphorylation of Eukaryotic Initiation Factor 4E Is Dispensable for Skeletal Muscle Hypertrophy. Am. J. Physiol. Cell Physiol. 2019, 317, C1247–C1255. [Google Scholar] [CrossRef] [PubMed]
- Rieger, F.; Grumet, M.; Edelman, G.M. N-CAM at the Vertebrate Neuromuscular Junction. J. Cell Biol. 1985, 101, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Dickson, G.; Gower, H.J.; Barton, C.H.; Prentice, H.M.; Elsom, V.L.; Moore, S.E.; Cox, R.D.; Quinn, C.; Putt, W.; Walsh, F.S. Human Muscle Neural Cell Adhesion Molecule (N-CAM): Identification of a Muscle-Specific Sequence in the Extracellular Domain. Cell 1987, 50, 1119–1130. [Google Scholar] [CrossRef]
- Markofski, M.M.; Dickinson, J.M.; Drummond, M.J.; Fry, C.S.; Fujita, S.; Gundermann, D.M.; Glynn, E.L.; Jennings, K.; Paddon-Jones, D.; Reidy, P.T.; et al. Effect of Age on Basal Muscle Protein Synthesis and mTORC1 Signaling in a Large Cohort of Young and Older Men and Women. Exp. Gerontol. 2015, 65, 1–7. [Google Scholar] [CrossRef]
- Paturi, S.; Gutta, A.K.; Katta, A.; Kakarla, S.K.; Arvapalli, R.K.; Gadde, M.K.; Nalabotu, S.K.; Rice, K.M.; Wu, M.; Blough, E. Effects of Aging and Gender on Muscle Mass and Regulation of Akt-mTOR-P70s6k Related Signaling in the F344BN Rat Model. Mech. Ageing Dev. 2010, 131, 202–209. [Google Scholar] [CrossRef]
- White, Z.; White, R.B.; McMahon, C.; Grounds, M.D.; Shavlakadze, T. High mTORC1 Signaling Is Maintained, While Protein Degradation Pathways Are Perturbed in Old Murine Skeletal Muscles in the Fasted State. Int. J. Biochem. Cell Biol. 2016, 78, 10–21. [Google Scholar] [CrossRef]
- Cuthbertson, D.J.; Babraj, J.; Leese, G.; Siervo, M. Anabolic Resistance Does Not Explain Sarcopenia in Patients with Type 2 Diabetes Mellitus, Compared with Healthy Controls, despite Reduced mTOR Pathway Activity. Clin. Nutr. 2017, 36, 1716–1719. [Google Scholar] [CrossRef]
- Kimball, S.R.; O’Malley, J.P.; Anthony, J.C.; Crozier, S.J.; Jefferson, L.S. Assessment of Biomarkers of Protein Anabolism in Skeletal Muscle during the Life Span of the Rat: Sarcopenia despite Elevated Protein Synthesis. Am. J. Physiol. Endocrinol. Metab. 2004, 287, E772–E780. [Google Scholar] [CrossRef]
- Marabita, M.; Baraldo, M.; Solagna, F.; Ceelen, J.J.M.; Sartori, R.; Nolte, H.; Nemazanyy, I.; Pyronnet, S.; Kruger, M.; Pende, M.; et al. S6K1 Is Required for Increasing Skeletal Muscle Force during Hypertrophy. Cell Rep. 2016, 17, 501–513. [Google Scholar] [CrossRef]
- Le Bacquer, O.; Combe, K.; Patrac, V.; Ingram, B.; Combaret, L.; Dardevet, D.; Montaurier, C.; Salles, J.; Giraudet, C.; Guillet, C.; et al. 4E-BP1 and 4E-BP2 Double Knockout Mice Are Protected from Aging-Associated Sarcopenia. J. Cachexia Sarcopenia Muscle 2019, 10, 696–709. [Google Scholar] [CrossRef]
- Chen, Z.; Li, L.; Wu, W.; Liu, Z.; Huang, Y.; Yang, L.; Luo, Q.; Chen, J.; Hou, Y.; Song, G. Exercise Protects Proliferative Muscle Satellite Cells against Exhaustion via the Igfbp7-Akt-mTOR Axis. Theranostics 2020, 10, 6448–6466. [Google Scholar] [CrossRef] [PubMed]
- De Sanctis, P.; Filardo, G.; Abruzzo, P.M.; Astolfi, A.; Bolotta, A.; Indio, V.; Di Martino, A.; Hofer, C.; Kern, H.; Löfler, S.; et al. Non-Coding RNAs in the Transcriptional Network That Differentiates Skeletal Muscles of Sedentary from Long-Term Endurance-and Resistance-Trained Elderly. Int. J. Mol. Sci. 2021, 22, 1539. [Google Scholar] [CrossRef] [PubMed]
- Clavel, S.; Coldefy, A.S.; Kurkdjian, E.; Salles, J.; Margaritis, I.; Derijard, B. Atrophy-Related Ubiquitin Ligases, Atrogin-1 and MuRF1 Are up-Regulated in Aged Rat Tibialis Anterior Muscle. Mech. Ageing Dev. 2006, 127, 794–801. [Google Scholar] [CrossRef] [PubMed]
- Edström, E.; Altun, M.; Hägglund, M.; Ulfhake, B. Atrogin-1/MAFbx and MuRF1 Are Downregulated in Aging-Related Loss of Skeletal Muscle. J. Gerontol. A Biol. Sci. Med. Sci. 2006, 61, 663–674. [Google Scholar] [CrossRef] [PubMed]
- DeRuisseau, K.C.; Kavazis, A.N.; Powers, S.K. Selective Downregulation of Ubiquitin Conjugation Cascade mRNA Occurs in the Senescent Rat Soleus Muscle. Exp. Gerontol. 2005, 40, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Welle, S.; Brooks, A.I.; Delehanty, J.M.; Needler, N.; Thornton, C.A. Gene Expression Profile of Aging in Human Muscle. Physiol. Genom. 2003, 14, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Whitman, S.A.; Wacker, M.J.; Richmond, S.R.; Godard, M.P. Contributions of the Ubiquitin-Proteasome Pathway and Apoptosis to Human Skeletal Muscle Wasting with Age. Pflug. Arch. Eur. J. Physiol. 2005, 450, 437–446. [Google Scholar] [CrossRef]
- Léger, B.; Derave, W.; De Bock, K.; Hespel, P.; Russell, A.P. Human Sarcopenia Reveals an Increase in SOCS-3 and Myostatin and a Reduced Efficiency of Akt Phosphorylation. Rejuvenation Res. 2008, 11, 163–175. [Google Scholar] [CrossRef]
- Gaugler, M.; Brown, A.; Merrell, E.; DiSanto-Rose, M.; Rathmacher, J.A.; Reynolds IV, T.H. PKB Signaling and Atrogene Expression in Skeletal Muscle of Aged Mice. J. Appl. Physiol. 2011, 111, 192–199. [Google Scholar] [CrossRef]
- Woo Kim, K.; Cho, H.J.; Khaliq, S.A.; Son, K.H.; Yoon, M.S. Comparative Analyses of mTOR/Akt and Muscle Atrophy-Related Signaling in Aged Respiratory and Gastrocnemius Muscles. Int. J. Mol. Sci. 2020, 21, 2862. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Sun, X.; Zhang, W.; Shi, X.; Xu, S. Di-(2-Ethyl Hexyl) Phthalate Induced Oxidative Stress Promotes Microplastics Mediated Apoptosis and Necroptosis in Mice Skeletal Muscle by Inhibiting PI3K/AKT/mTOR Pathway. Toxicology 2022, 474, 153226. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Liang, J.; Wu, L.; Zhang, H.; Lv, J.; Chen, N. Exercise-Induced Autophagy Suppresses Sarcopenia Through Akt/mTOR and Akt/FoxO3a Signal Pathways and AMPK-Mediated Mitochondrial Quality Control. Front. Physiol. 2020, 11, 583478. [Google Scholar] [CrossRef] [PubMed]
- Morisasa, M.; Yoshida, E.; Fujitani, M.; Kimura, K.; Uchida, K.; Kishida, T.; Mori, T.; Goto-Inoue, N. Fish Protein Promotes Skeletal Muscle Hypertrophy via the Akt/mTOR Signaling Pathways. J. Nutr. Sci. Vitaminol. 2022, 68, 23–31. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Q.; Zhang, Z.; Fu, R.; Zhou, T.; Long, C.; He, T.; Yang, D.; Li, Z.; Peng, S. Magnesium Supplementation Enhances mTOR Signalling to Facilitate Myogenic Differentiation and Improve Aged Muscle Performance. Bone 2021, 146, 115886. [Google Scholar] [CrossRef] [PubMed]
- Aoi, W.; Iwasa, M.; Aiso, C.; Tabata, Y.; Gotoh, Y.; Kosaka, H.; Suzuki, T. Lactococcus Cremoris Subsp. Cremoris FC-Fermented Milk Activates Protein Synthesis and Increases Skeletal Muscle Mass in Middle-Aged Mice. Biochem. Biophys. Res. Commun. 2022, 612, 176–180. [Google Scholar] [CrossRef]
- Chen, C.N.; Liao, Y.H.; Tsai, S.C.; Thompson, L.D.V. Age-Dependent Effects of Caloric Restriction on mTOR and Ubiquitin-Proteasome Pathways in Skeletal Muscles. GeroScience 2019, 41, 871–880. [Google Scholar] [CrossRef]
- Toniolo, L.; Formoso, L.; Torelli, L.; Crea, E.; Bergamo, A.; Sava, G.; Giacomello, E. Long-Term Resveratrol Treatment Improves the Capillarization in the Skeletal Muscles of Ageing C57BL/6J Mice. Int. J. Food Sci. Nutr. 2021, 72, 37–44. [Google Scholar] [CrossRef]
- Sirago, G.; Toniolo, L.; Crea, E.; Giacomello, E. A Short-Term Treatment with Resveratrol Improves the Inflammatory Conditions of Middle-Aged Mice Skeletal Muscles. Int. J. Food Sci. Nutr. 2022, 73, 630–637. [Google Scholar] [CrossRef]
- Toniolo, L.; Fusco, P.; Formoso, L.; Mazzi, A.; Canato, M.; Reggiani, C.; Giacomello, E. Resveratrol Treatment Reduces the Appearance of Tubular Aggregates and Improves the Resistance to Fatigue in Aging Mice Skeletal Muscles. Exp. Gerontol. 2018, 111, 170–179. [Google Scholar] [CrossRef]
- Barger, J.L.; Kayo, T.; Vann, J.M.; Arias, E.B.; Wang, J.; Hacker, T.A.; Wang, Y.; Raederstorff, D.; Morrow, J.D.; Leeuwenburgh, C.; et al. A Low Dose of Dietary Resveratrol Partially Mimics Caloric Restriction and Retards Aging Parameters in Mice. PLoS ONE 2008, 3, e2264. [Google Scholar] [CrossRef]
- Dutta, D.; Xu, J.; Dirain, M.L.S.; Leeuwenburgh, C. Calorie Restriction Combined with Resveratrol Induces Autophagy and Protects 26-Month-Old Rat Hearts from Doxorubicin-Induced Toxicity. Free Radic. Biol. Med. 2014, 74, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Opalach, K.; Rangaraju, S.; Madorsky, I.; Leeuwenburgh, C.; Notterpek, L. Lifelong Calorie Restriction Alleviates Age-Related Oxidative Damage in Peripheral Nerves. Rejuvenation Res. 2010, 13, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Picca, A.; Fracasso, F.; Pesce, V.; Cantatore, P.; Joseph, A.M.; Leeuwenburgh, C.; Gadaleta, M.N.; Lezza, A.M.S. Age-and Calorie Restriction-Related Changes in Rat Brain Mitochondrial DNA and TFAM Binding. Age 2013, 35, 1607–1620. [Google Scholar] [CrossRef]
- Chimienti, G.; Picca, A.; Fracasso, F.; Russo, F.; Orlando, A.; Riezzo, G.; Leeuwenburgh, C.; Pesce, V.; Lezza, A.M.S. The Age-Sensitive Efficacy of Calorie Restriction on Mitochondrial Biogenesis and Mtdna Damage in Rat Liver. Int. J. Mol. Sci. 2021, 22, 1665. [Google Scholar] [CrossRef] [PubMed]
- Gharahdaghi, N.; Rudrappa, S.; Brook, M.S.; Idris, I.; Crossland, H.; Hamrock, C.; Aziz, M.H.A.; Kadi, F.; Tarum, J.; Greenhaff, P.L.; et al. Testosterone Therapy Induces Molecular Programming Augmenting Physiological Adaptations to Resistance Exercise in Older Men. J. Cachexia Sarcopenia Muscle 2019, 10, 1276–1294. [Google Scholar] [CrossRef]
- Kim, J.E.; Kwon, E.Y.; Han, Y. A Collagen Hydrolysate Containing Tripeptides Ameliorates Sarcopenia in Middle-Aged Mice. Molecules 2022, 27, 2718. [Google Scholar] [CrossRef]
- Baek, M.O.; Cho, H.J.; Min, D.S.; Choi, C.S.; Yoon, M.S. Self-Transducible LRS-UNE-L Peptide Enhances Muscle Regeneration. J. Cachexia Sarcopenia Muscle 2022, 13, 1277–1288. [Google Scholar] [CrossRef]
- Yi, D.; Yoshikawa, M.; Sugimoto, T.; Tomoo, K.; Okada, Y.; Hashimoto, T. Effects of Maca on Muscle Hypertrophy in C2C12 Skeletal Muscle Cells. Int. J. Mol. Sci. 2022, 23, 6825. [Google Scholar] [CrossRef]
- Le Bacquer, O.; Lanchais, K.; Combe, K.; Van Den Berghe, L.; Walrand, S. Acute Rimonabant Treatment Promotes Protein Synthesis in C2C12 Myotubes through a CB1-Independent Mechanism. J. Cell. Physiol. 2021, 236, 2669–2683. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.; Kim, C.; Woo, Y.K.; Hwang, J.K. Inhibitory Effects of Chrysanthemum (Chrysanthemum Morifolium Ramat.) Extract and Its Active Compound Isochlorogenic Acid A on Sarcopenia. Prev. Nutr. Food Sci. 2021, 26, 408–416. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sirago, G.; Picca, A.; Calvani, R.; Coelho-Júnior, H.J.; Marzetti, E. Mammalian Target of Rapamycin (mTOR) Signaling at the Crossroad of Muscle Fiber Fate in Sarcopenia. Int. J. Mol. Sci. 2022, 23, 13823. https://doi.org/10.3390/ijms232213823
Sirago G, Picca A, Calvani R, Coelho-Júnior HJ, Marzetti E. Mammalian Target of Rapamycin (mTOR) Signaling at the Crossroad of Muscle Fiber Fate in Sarcopenia. International Journal of Molecular Sciences. 2022; 23(22):13823. https://doi.org/10.3390/ijms232213823
Chicago/Turabian StyleSirago, Giuseppe, Anna Picca, Riccardo Calvani, Hélio José Coelho-Júnior, and Emanuele Marzetti. 2022. "Mammalian Target of Rapamycin (mTOR) Signaling at the Crossroad of Muscle Fiber Fate in Sarcopenia" International Journal of Molecular Sciences 23, no. 22: 13823. https://doi.org/10.3390/ijms232213823
APA StyleSirago, G., Picca, A., Calvani, R., Coelho-Júnior, H. J., & Marzetti, E. (2022). Mammalian Target of Rapamycin (mTOR) Signaling at the Crossroad of Muscle Fiber Fate in Sarcopenia. International Journal of Molecular Sciences, 23(22), 13823. https://doi.org/10.3390/ijms232213823










