Influence of Selective Extraction/Isolation of Heme/Hemoglobin with Hydrophobic Imidazolium Ionic Liquids on the Precision and Accuracy of Cotinine ELISA Test
Abstract
1. Introduction
2. Results and Discussion
2.1. Optimization of Extraction Conditions (IL Kind, IL Amount, Extraction Time)
2.2. The Partitioning of the Heme/Hemoglobin in IL-Based Extraction System
2.3. Serum and Hemoglobin Enriched Serum Electrophoresis
2.4. Molecular Modeling of Interactions between IL and Heme/Hemoglobin/Cotinine
2.4.1. Quantum Mechanical Calculations
2.4.2. Molecular Dynamics Simulations
2.5. Cotinine ELISA Kit
3. Materials and Methods
3.1. Apparatus and Reagent
3.2. The Measurement of Cotinine in Human Serum
3.3. General Procedure of Heme/Hemoglobin Extraction
3.4. Electrophoresis
3.5. Molecular Modeling
3.5.1. Quantum Mechanical Calculations
3.5.2. Molecular Dynamics Simulations
3.6. Kinetics and Equilibrium of Extraction
4. Conclusions
5. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Akter, S.; Nakagawa, T.; Honda, T.; Yamamoto, S.; Kuwahara, K.; Okazaki, H.; Hu, H.; Imai, T.; Nishihara, A.; Miyamoto, T.; et al. Smoking, Smoking Cessation, and Risk of Mortality in a Japanese Working Population—Japan Epidemiology Collaboration on Occupational Health Study. Circ. J. 2018, 82, 3005–3012. [Google Scholar] [CrossRef] [PubMed]
- Samet, J.M. Tobacco smoking: The leading cause of preventable disease worldwide. Thorac. Surg. Clin. 2013, 23, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, R.; Katada, J. Effects of active smoking on postoperative outcomes in hospitalized patients undergoing elective surgery: A retrospective analysis of an administrative claims database in Japan. BMJ Open 2019, 9, e029913. [Google Scholar] [CrossRef]
- Warren, C.W.; Jones, N.R.; Eriksen, M.P.; Asma, S. Patterns of global tobacco use in young people and implications for future chronic disease burden in adults. Lancet 2006, 367, 749–753. [Google Scholar] [CrossRef]
- National Research Council. Biologic Markers of Pulmonary Toxicology; National Academy Press: Washington, DC, USA, 1989. [Google Scholar] [CrossRef]
- Benowitz, N.L.; Hukkanen, J.; Jacob, P., III. Nicotine chemistry, metabolism, kinetics and biomarkers. In Nicotine Psychopharmacology; Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2009; Volume 192, pp. 29–60. [Google Scholar] [CrossRef]
- Jaakkola, M.S.; Jaakkola, J.J.K. Assessment of exposure to environmental tobacco smoke. Eur. Respir. J. 1997, 10, 2384–2397. [Google Scholar] [CrossRef] [PubMed]
- Benowitz, N.L.; Jacob, P., III; Fong, I.; Gupta, S. Nicotine metabolic profile in man: Comparison of cigarette smoking and transdermal nicotine. J. Pharmacol. Exp. Ther. 1994, 268, 296–303. [Google Scholar]
- Etter, J.F.; Vu Duc, T.; Perneger, T.V. Saliva cotinine levels in smokers and nonsmokers. Am. J. Epidemiol. 2000, 151, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Hegaard, H.K.; Kjaergaard, H.; Moller, L.F.; Wachmann, H.; Ottesen, B. Determination of a saliva cotinine cut-off to distinguish pregnant smokers from pregnant non-smokers. Acta Obstet. Gynecol. Scand. 2007, 86, 401–406. [Google Scholar] [CrossRef]
- SRNT Subcommittee on Biochemical Verification. Biochemical verification of tobacco use and cessation. Nicotine Tob. Res. 2002, 4, 149–159. [Google Scholar] [CrossRef]
- Kim, S. Overview of Cotinine Cutoff Values for Smoking Status Classification. Int. J. Environ. Res. Public Health 2016, 13, 1236. [Google Scholar] [CrossRef]
- Avagyan, R.; Spasova, M.; Lindholm, J. Determination of Nicotine-Related Impurities in Nicotine Pouches and Tobacco-Containing Products by Liquid Chromatography–Tandem Mass Spectrometry. Separations 2021, 8, 77. [Google Scholar] [CrossRef]
- Scheidweiler, K.B.; Shakleya, D.M.; Huestis, M.A. Simultaneous quantification of nicotine, cotinine, trans-3′-hydroxycotinine, norcotinine and mecamylamine in human urine by liquid chromatography-tandem mass spectrometry. Clin. Chim. Acta 2012, 413, 978–984. [Google Scholar] [CrossRef] [PubMed]
- Chazeron, I.; Daval, S.; Ughetto, S.; Richard, D.; Nicolay, A.; Lemery, D.; Llorca, P.M.; Coudoré, F. GC-MS determined cotinine in an epidemiological study on smoking status at delivery. Pulm. Pharmacol. Ther. 2008, 21, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Baumann, F.; Regenthal, R.; Burgos-Guerrero, I.L.; Hegerl, U.; Preiss, R. Determination of nicotine and cotinine in human serum by means of LC/MS. J. Chromatogr. B 2010, 878, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Jacob, P., III; Yu, L.; Duan, M.; Ramos, L.; Yturralde, O.; Benowitz, N.L. Determination of the Nicotine Metabolites Cotinine and Trans-3′-Hydroxycotinine in Biologic fluids of Smokers and Non-Smokers using Liquid Chromatography—Tandem Mass Spectrometry: Biomarkers for Tobacco Smoke Exposure and for Phenotyping Cytochrome P450 2A6 Activity. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2011, 879, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Blount, B.C.; Wang, L. Sensitive Quantification of Nicotine in Bronchoalveolar Lavage Fluid by Acetone Precipitation Combined with Isotope-Dilution Liquid Chromatography-Tandem Mass Spectrometry. ACS Omega 2021, 6, 13962–13969. [Google Scholar] [CrossRef]
- Yasuda, M.; Ota, T.; Morikawa, A.; Mawatari, K.-I.; Fukuuchi, T.; Yamaoka, N.; Kaneko, K.; Nakagomi, K. Simultaneous determination of nicotine and cotinine in serum using high-performance liquid chromatography with fluorometric detection and postcolumn UV-photoirradiation system. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2013, 934, 41–45. [Google Scholar] [CrossRef]
- Park, S.; Lee, D.H.; Park, J.G.; Lee, Y.T.; Chung, J. A sensitive enzyme immunoassay for measuring cotinine in passive smokers. Clin. Chim. Acta 2010, 411, 1238–1242. [Google Scholar] [CrossRef]
- Baj, J.; Flieger, W.; Przygodzka, D.; Buszewicz, G.; Teresiński, G.; Pizoń, M.; Maciejewski, R.; Flieger, J. Application of HPLC-QQQ-MS/MS and New RP-HPLC-DAD System Utilizing the Chaotropic Effect for Determination of Nicotine and Its Major Metabolites Cotinine, and trans-3′-Hydroxycotinine in Human Plasma Samples. Molecules 2022, 27, 682. [Google Scholar] [CrossRef]
- Kim, I.; Darwin, W.D.; Huestis, M.A. Simultaneous determination of nicotine, cotinine, norcotinine, and trans-3′-hydroxycotinine in human oral fluid using solid phase extraction and gas chromatography-mass spectrometry. J. Chromatogr. B 2005, 814, 233–240. [Google Scholar] [CrossRef]
- Acosta, M.C.; Buchhalter, A.R.; Breland, A.B.; Hamilton, D.C.P.; Eissenberg, T. Urine cotinine as an index of smoking status in smokers during 96-hr abstinence: Comparison between gas chromatography/mass spectrometry and immunoassay test strips. Nicotine Tob. Res. 2004, 6, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Torano, J.S.; van Kan, H.J. Simultaneous determination of the tobacco smoke uptake parameters nicotine, cotinine and thiocyanate in urine, saliva and hair, using gas chromatography-mass spectrometry for characterisation of smoking status of recently exposed subjects. Analyst 2003, 128, 838–843. [Google Scholar] [CrossRef] [PubMed]
- Heinrich-Ramm, R.; Wegner, R.; Garde, A.H.; Baur, X. Cotinine excretion (tobacco smoke biomarker) of smokers and non-smokers: Comparision of GC/MS and RIA results. Int. J. Hyg. Environ. Health 2002, 205, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, H.; Inoue, R.; Yaki, K.; Saito, K. Determination of nicotine, cotinine, and related alkaloids in human urine and saliva by automated in-tube solid-phase microextraction coupled with liquid chromatography–mass spectrometry. J. Pharm. Biomed. Anal. 2009, 49, 108–114. [Google Scholar] [CrossRef]
- Gray, T.R.; Shakleya, D.M.; Huestis, M.A. Quantification of nicotine, cotinine, trans-3′-hydroxycotinine, nornicotine and norcotinine in human meconium by liquid chromatography/tandem mass spectrometry. J. Chromatogr. B 2008, 863, 107–114. [Google Scholar] [CrossRef]
- Chadwick, C.A.; Keevil, B. Measurement of cotinine in urine by liquid chromatography tandem mass spectrometry. Ann. Clin. Biochem. 2007, 44, 455–462. [Google Scholar] [CrossRef]
- Murphy, S.E.; Villalta, P.; Ho, S.W.; Weymarn, L.B. Analysis of [3′,3′-d2]-nicotine and [3′,3′-d2]-cotinine by capillary liquid chromatography–electrospray tandem mass spectrometry. J. Chromatogr. B 2007, 857, 1–8. [Google Scholar] [CrossRef][Green Version]
- Beyer, J.; Peters, F.T.; Kraemer, T.; Maurer, H.H. Detection and validated quantification of toxic alkaloids in human blood plasma—Comparison of LC-APCI-MS with LC-ESI-MS/MS. J. Mass Spectrom. 2007, 42, 621–633. [Google Scholar] [CrossRef]
- Rabbaa-Khabbaz, L.; Abi Daoud, R.; Karam-Sarkis, D. A Simple, Sensitive, and Rapid Method for the Determination of Cotinine in Urine by High-Performance Liquid Chromatography with UV Detection. J. Chromatogr. Sci. 2006, 44, 535–538. [Google Scholar] [CrossRef]
- Vin, A. Indirect ELISA. Methods Mol. Biol. 2015, 1318, 51–59. [Google Scholar]
- Lippi, G.; Cadamuro, J.; von Meyer, A.; Simundic, A.M. Practical recommendations for managing hemolyzed samples in clinical chemistry testing. Clin. Chem. Lab. Med. 2018, 56, 718–727. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Li, Z.; Jia, Y.; Zhu, B.; Cao, Z. Interference of hemolysis on the postmortem biochemical analysis of IgE by ECLIA. Int. J. Legal Med. 2021, 135, 1661–1668. [Google Scholar] [CrossRef] [PubMed]
- D’Amici, G.M.; Rinalducci, S.; Zolla, L. Depletion of hemoglobin and carbonic anhydrase from erythrocyte cytosolic samples by preparative clear native electrophoresis. Nat. Protoc. 2011, 7, 36–44. [Google Scholar] [CrossRef]
- Reimers, T.J.; McCann, J.P.; Cowan, R.G.; Concannon, P.W. Effects of storage, hemolysis, and freezing and thawing on concentrations of thyroxine, cortisol, and insulin in blood samples. Proc. Soc. Exp. Biol. Med. 1982, 170, 509–516. [Google Scholar] [CrossRef]
- Parra, M.D.; Bernal, L.J.; Cerón, J.J. Cortisol and free thyroxine determination by time-resolved fluorometry in canine serum. Can. J. Vet. Res. 2004, 68, 98–104. [Google Scholar] [PubMed]
- Koseoglu, M.; Hur, A.; Atay, A.; Cuhadar, S. Effects of hemolysis interferences on routine biochemistry parameters. Biochem. Med. 2011, 21, 79–85. [Google Scholar] [CrossRef]
- Lippi, G.; Plebani, M.; Di Somma, S.; Cervellin, G. Hemolyzed specimens: A major challenge for emergency departments and clinical laboratories. Crit. Rev. Clin. Lab. Sci. 2011, 48, 143–153. [Google Scholar] [CrossRef]
- Lippi, G.; Salvagno, G.L.; Montagnana, M.; Brocco, G.; Guidi, G.C. Influence of hemolysis on routine clinical chemistry testing. Clin. Chem. Lab. Med. 2006, 44, 311–316. [Google Scholar] [CrossRef]
- Aich, A.; Freundlich, M.; Vekilov, P.G. The free heme concentration in healthy human erythrocytes. Blood Cells Mol. Dis. 2015, 55, 402–409. [Google Scholar] [CrossRef]
- Edelstein, S.J.; Rehmar, M.J.; Olson, J.S.; Gibson, Q.H. Functional aspects of the subunit association-dissociation equilibria of hemoglobin. J. Biol. Chem. 1970, 245, 4372–4381. [Google Scholar] [CrossRef]
- Hargrove, M.S.; Whitaker, T.; Olson, J.S.; Vali, R.J.; Mathews, A.J. Quaternary structure regulates hemin dissociation from human hemoglobin. J. Biol. Chem. 1997, 272, 17385–17389. [Google Scholar] [CrossRef]
- Sachar, M.; Anderson, K.E.; Ma, X. Protoporphyrin IX: The Good, the Bad, and the Ugly. J. Pharmacol. Exp. Ther. 2016, 356, 267–275. [Google Scholar] [CrossRef]
- Donegan, R.K.; Moore, C.M.; Hanna, D.A.; Reddi, A.R. Handling heme: The mechanisms underlying the movement of heme within and between cells. Free Radic. Biol. Med. 2019, 133, 88–100. [Google Scholar] [CrossRef]
- Chou, P.P.; Kerkay, J.; Gupta, M.K. Development of a Laser Nephelometric Method for the Quantitation of Human Glycohemoglobins. Anal. Lett. 1981, 14, 1071–1087. [Google Scholar] [CrossRef]
- Frantzen, F.; Grimsrud, K.; Heggli, D.E.; Sundrehagen, E. Selective precipitation of human hemoglobin by organic solvents and metal cations. Hemoglobin 1997, 21, 155–172. [Google Scholar] [CrossRef]
- Wang, S.; Xiong, N.; Dong, X.Y.; Sun, Y. A novel nickel-chelated surfactant for affinity-based aqueous two-phase micellar extraction of histidine-rich protein. J. Chromatogr. A 2013, 1320, 118–124. [Google Scholar] [CrossRef]
- Zhang, M.; He, X.; Chen, L.; Zhang, Y. Preparation and characterization of iminodiacetic acid-functionalized magnetic nanoparticles and its selective removal of bovine hemoglobin. Nanotechnology 2011, 22, 065705. [Google Scholar] [CrossRef]
- Guo, X.; Mao, F.; Wang, W.; Yang, Y.; Bai, Z. Sulfhydryl-modified Fe3O4@SiO2 core/shell nanocomposite: Synthesis and toxicity assessment in vitro. ACS Appl. Mater. Interfaces 2015, 7, 14983–14991. [Google Scholar] [CrossRef]
- Ayyappan, S.; Mahadevan, S.; Chandramohan, P.; Srinivasan, M.P.; Philip, J.; Raj, B. Influence of Co2+ Ion concentration on the size, magnetic properties, and purity of CoFe2O4 Spinel ferrite nanoparticles. J. Phys. Chem. C 2010, 114, 6334–6341. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Yan, M.; Huang, J. Selective Removal of Hemoglobin from Blood Using Hierarchical Copper Shells Anchored to Magnetic Nanoparticles. Biomed Res. Int. 2017, 2017, 7309481. [Google Scholar] [CrossRef]
- Schröder, C. Proteins in Ionic Liquids: Current Status of Experiments and Simulations. Top. Curr. Chem. 2017, 375, 25. [Google Scholar] [CrossRef]
- Bento, R.M.F.; Almeida, C.A.S.; Neves, M.C.; Tavares, A.P.M.; Freire, M.G. Advances Achieved by Ionic-Liquid-Based Materials as Alternative Supports and Purification Platforms for Proteins and Enzymes. Nanomaterials 2021, 11, 2542. [Google Scholar] [CrossRef]
- Cheng, D.H.; Chen, X.W.; Shu, Y.; Wang, J.H. Selective extraction/isolation of hemoglobin with ionic liquid 1-butyl-3-trimethylsilylimidazolium hexafluorophosphate (BtmsimPF6). Talanta 2008, 75, 1270–1278. [Google Scholar] [CrossRef]
- Zeng, Q.; Li, Z.; Wang, Y.B. Substituent effects on gas-phase homolytic Fe–N bond energies of m-G-C6H4NHFe(CO)2(η5-C5H5) and m-G-C6H4N(COMe)Fe(CO)2(η5-C5H5) studied using density functional theory methods. J. Phys. Org. Chem. 2018, 31, e3782. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Shevchenco, A.; Wilm, M.; Vorm, O.; Mann, M. Mass Spectrometric Sequencing of Proteins from Silver-Stained Polyacrylamide Gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef]
- Rappe, A.K.; Casewit, C.J.; Colwell, K.S.; Goddard, W.A., III; Skiff, W.M. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J. Am. Chem. Soc. 1992, 114, 10024–10035. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785. [Google Scholar] [CrossRef]
- Hehre, W.J.; Radom, L.; Schleyer, P.V.R.; Pople, J. AB INITIO Molecular Orbital Theory; John Wiley: New York, NY, USA, 1986. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09, Revision E.01; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, A.D., Jr. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone ϕ, ψ and Side-Chain χ1 and χ2 Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed]
- Parrinello, M.; Rahman, A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J. Appl. Phys. 1981, 52, 7182. [Google Scholar] [CrossRef]
- Hockney, R.W. The potential calculation and some applications. Methods Comput. Phys. 1970, 9, 135–211. [Google Scholar]
- Hess, B.; Bekker, H.; Berendsen, H.; Fraaije, J. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald: An N⋅log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926. [Google Scholar] [CrossRef]
- Kirkwood, J.G. Statistical Mechanics of Fluid Mixtures. J. Chem. Phys. 1935, 3, 300. [Google Scholar] [CrossRef]
- Bennett, C.H. Efficient Estimation of Free Energy Differences from Monte Carlo Data. J. Comput. Phys. 1976, 22, 245–268. [Google Scholar] [CrossRef]
- Kazmi, S.; Awan, Z.; Hashmi, S. Simulation Study of Ionic Liquid Utilization for Desulfurization of Model Gasoline. Iran. J. Chem. Chem. Eng. IJCCE 2019, 38, 209–221. [Google Scholar] [CrossRef]
- Sharifi, A.; Barazandeh, M.; Saeed Abaee, M.; Mirzaei, M. K2CO3/H2O in [omim][BF4] Ionic Liquid: A Green Medium for Efficient Room-Temperature Synthesis of N-Substituted 1,4-Benzoxazin-3-ones. J. Heterocycl. Chem. 2012, 49, 933–938. [Google Scholar] [CrossRef]
- Sharifi, A.; Barazandeh, M.; Saeed Abaee, M.; Mirzaei, M. [Omim][BF4], agreen and recyclable ionic liquid medium for the one-pot chemoselective synthesis of benzoxazinones. Tetrahedron Lett. 2010, 51, 1852–1855. [Google Scholar] [CrossRef]
- Wan, H.; Huang, D.Y.; Cai, Y.; Guan, G.F. Extraction of phenolic compounds with [omim]BF4 ionic liquid. J. Chem. Eng. Chin. Univ. 2008, 22, 162–165. (In Chinese) [Google Scholar]

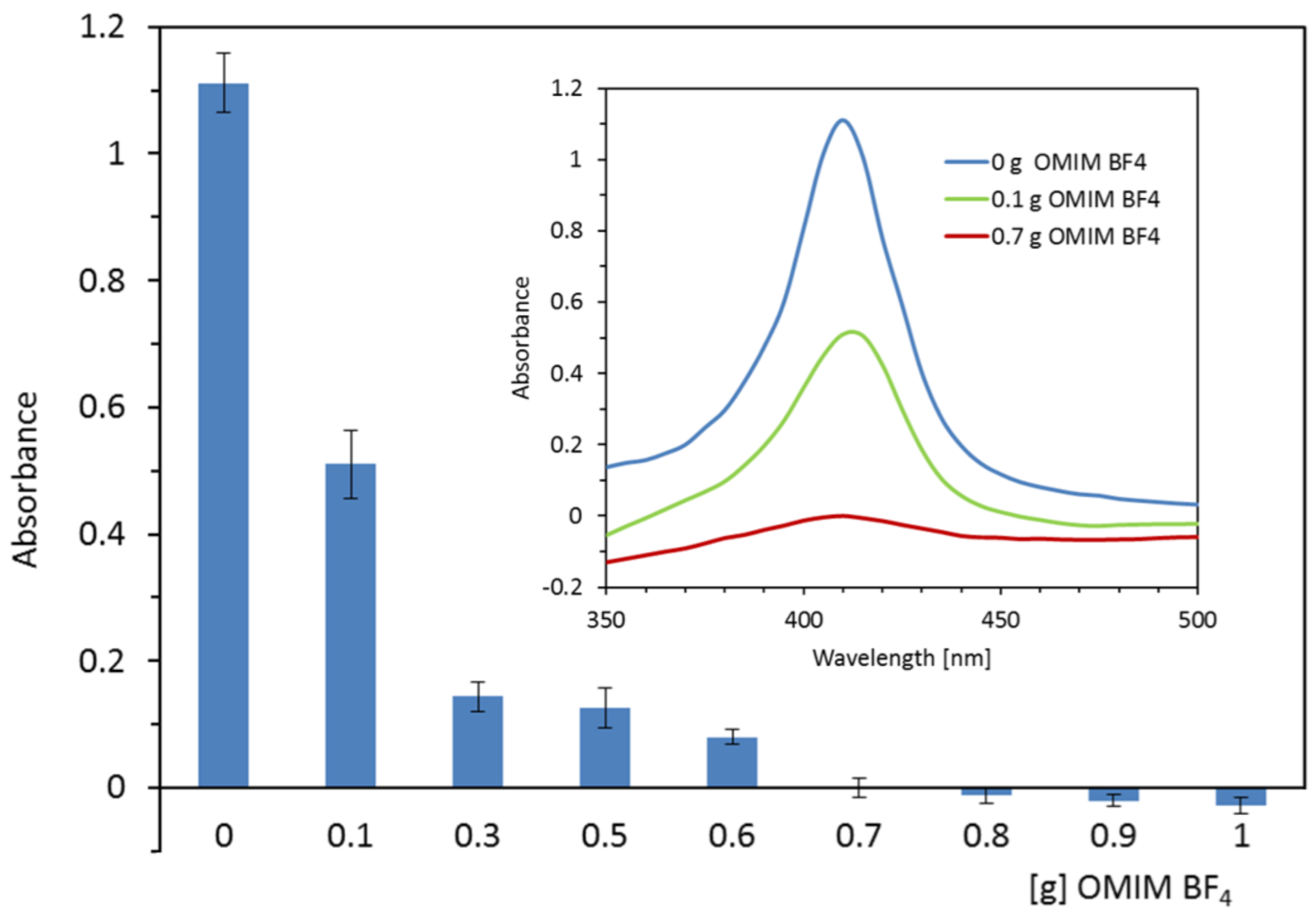


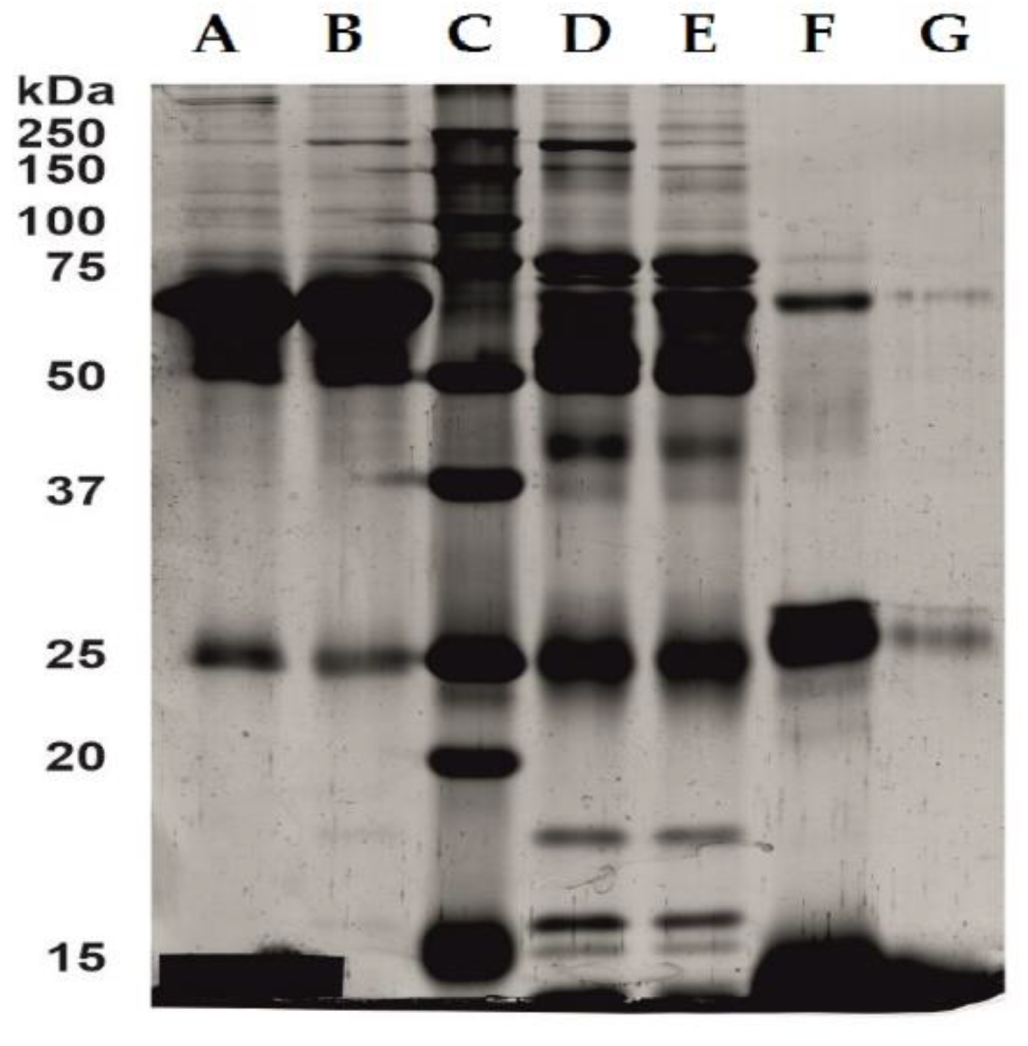
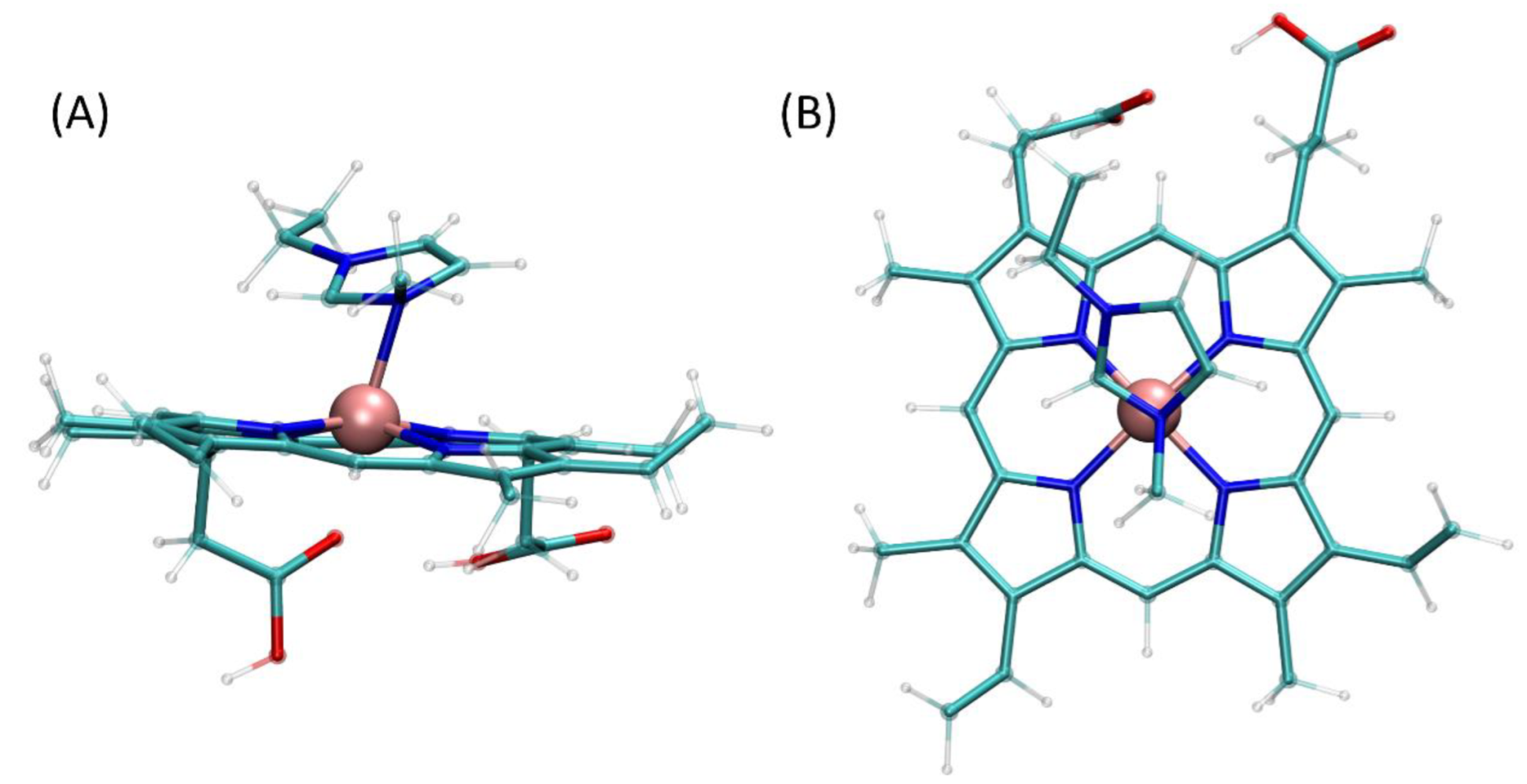


| Mode of Measurements | Parameter | Hemoglobin Concentration in Serum Samples | |||
|---|---|---|---|---|---|
| 0 mg mL−1 | 0.05 mg mL−1 | 0.1 mg mL−1 | 0.2 mg mL−1 | ||
| Measurements with the cotinine ELISA Kit without removing heme/hemoglobin | Absorbance | 0.562 0.652 0.517 | 0.560 0.519 0.445 | 0.465 0.521 0.580 | 0.345 0.442 0.396 |
| Concentration [ng mL−1] | 21.904 19.950 22.971 | 21.955 22.917 24.867 | 24.314 22.891 21.496 | 28.042 24.953 26.314 | |
| Concentration mean value [ng mL−1] | 21.608 | 23.246 | 22.900 | 26.436 | |
| SD | 1.532 | 1.484 | 1.409 | 1.548 | |
| CV% | 7.090 | 6.382 | 6.153 | 5.856 | |
| Relative error [%] | 8.04 | 16.23 | 14.5 | 32.18 | |
| Measurements with the cotinine ELISA Kit after heme/hemoglobin extraction | Absorbance | - | 0.625 0.546 0.493 | 0.830 0.803 0.759 | 0.625 0.642 0.684 |
| Concentration [ng mL−1] | - | 20.523 22.288 23.588 | 16.464 16.964 17.793 | 20.515 20.145 19.287 | |
| Concentration mean value [ng mL−1] | - | 22.133 | 17.074 | 19.982 | |
| SD | - | 1.538 | 0.671 | 0.630 | |
| CV% | - | 6.951 | 3.932 | 3.153 | |
| Relative error [%] | - | 10.66 | 14.6 | −0.09 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flieger, J.; Tatarczak-Michalewska, M.; Flieger, W.; Baj, J.; Buszewicz, G.; Teresiński, G.; Maciejewski, R.; Wawrzykowski, J.; Przygodzka, D.; Lutsyk, V.; et al. Influence of Selective Extraction/Isolation of Heme/Hemoglobin with Hydrophobic Imidazolium Ionic Liquids on the Precision and Accuracy of Cotinine ELISA Test. Int. J. Mol. Sci. 2022, 23, 13692. https://doi.org/10.3390/ijms232213692
Flieger J, Tatarczak-Michalewska M, Flieger W, Baj J, Buszewicz G, Teresiński G, Maciejewski R, Wawrzykowski J, Przygodzka D, Lutsyk V, et al. Influence of Selective Extraction/Isolation of Heme/Hemoglobin with Hydrophobic Imidazolium Ionic Liquids on the Precision and Accuracy of Cotinine ELISA Test. International Journal of Molecular Sciences. 2022; 23(22):13692. https://doi.org/10.3390/ijms232213692
Chicago/Turabian StyleFlieger, Jolanta, Małgorzata Tatarczak-Michalewska, Wojciech Flieger, Jacek Baj, Grzegorz Buszewicz, Grzegorz Teresiński, Ryszard Maciejewski, Jacek Wawrzykowski, Dominika Przygodzka, Valery Lutsyk, and et al. 2022. "Influence of Selective Extraction/Isolation of Heme/Hemoglobin with Hydrophobic Imidazolium Ionic Liquids on the Precision and Accuracy of Cotinine ELISA Test" International Journal of Molecular Sciences 23, no. 22: 13692. https://doi.org/10.3390/ijms232213692
APA StyleFlieger, J., Tatarczak-Michalewska, M., Flieger, W., Baj, J., Buszewicz, G., Teresiński, G., Maciejewski, R., Wawrzykowski, J., Przygodzka, D., Lutsyk, V., & Płaziński, W. (2022). Influence of Selective Extraction/Isolation of Heme/Hemoglobin with Hydrophobic Imidazolium Ionic Liquids on the Precision and Accuracy of Cotinine ELISA Test. International Journal of Molecular Sciences, 23(22), 13692. https://doi.org/10.3390/ijms232213692







