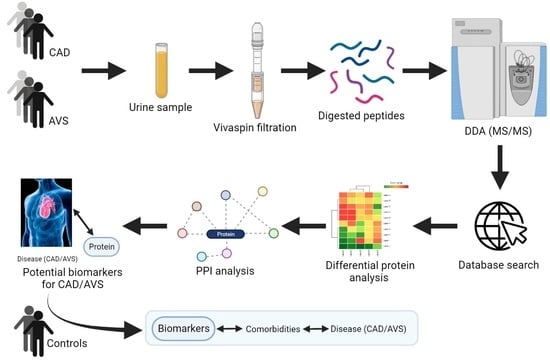
Coronary Artery Disease and Aortic Valve Stenosis: A Urine Proteomics Study
Abstract
1. Introduction
2. Results
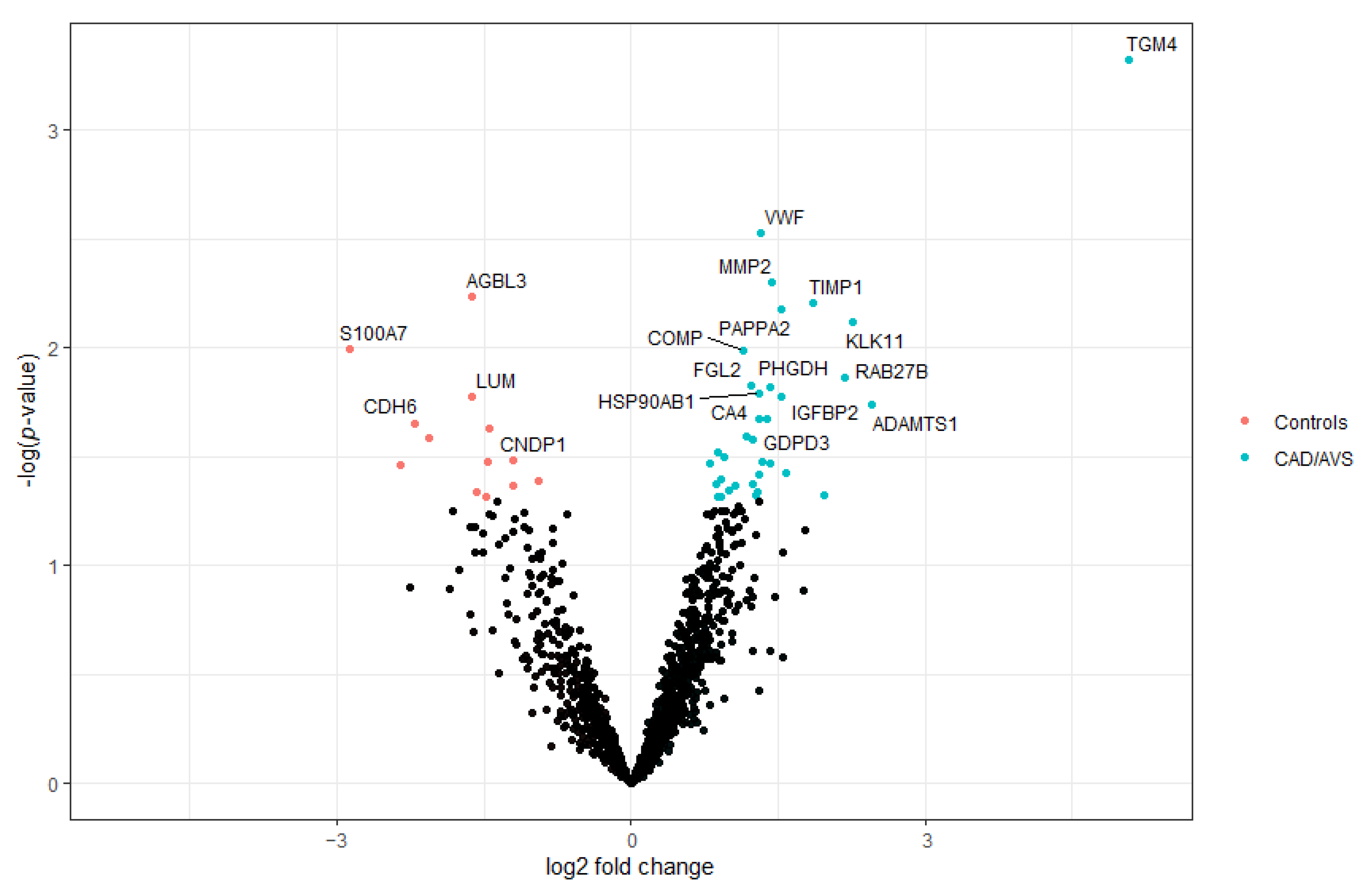
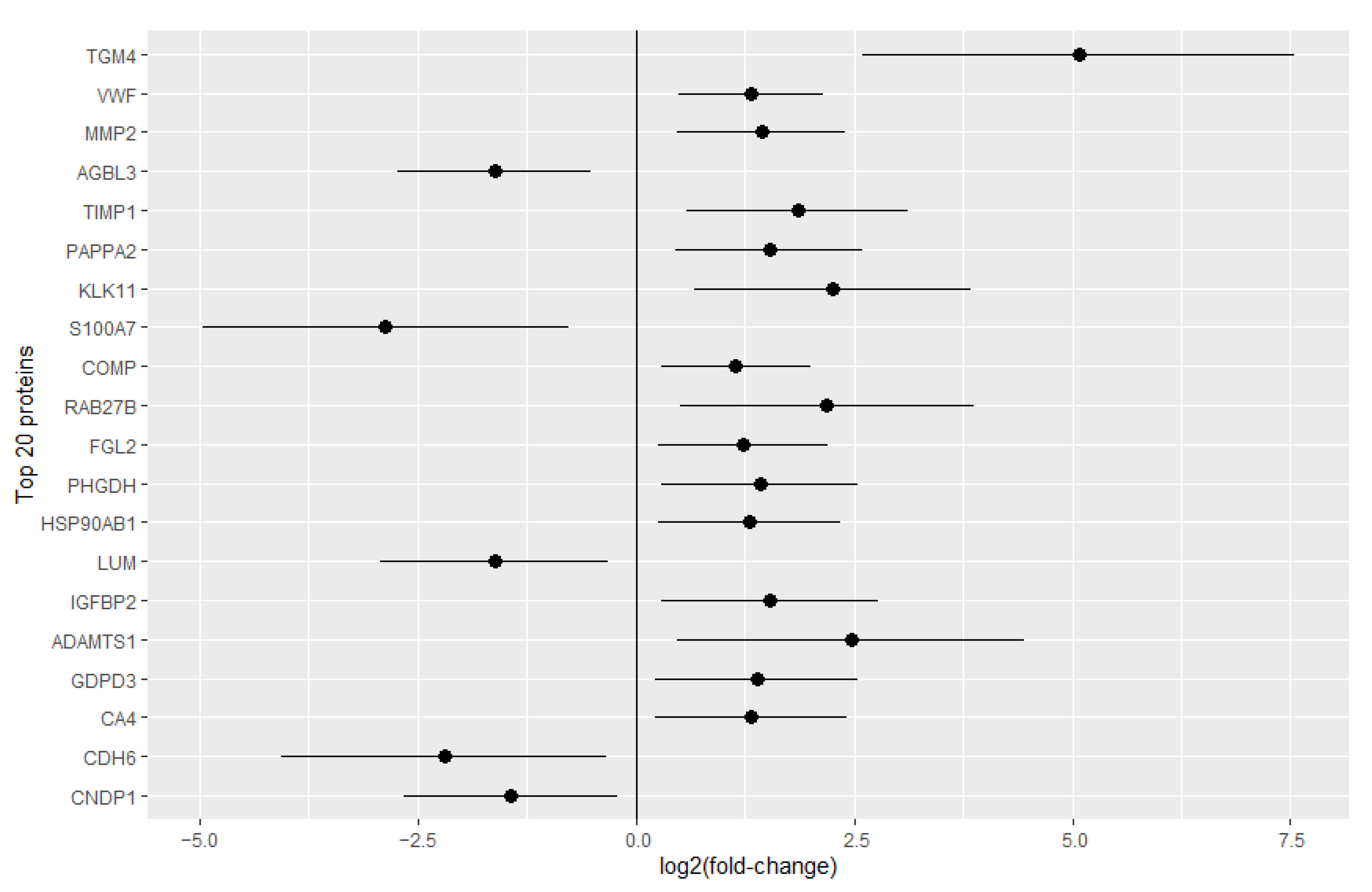
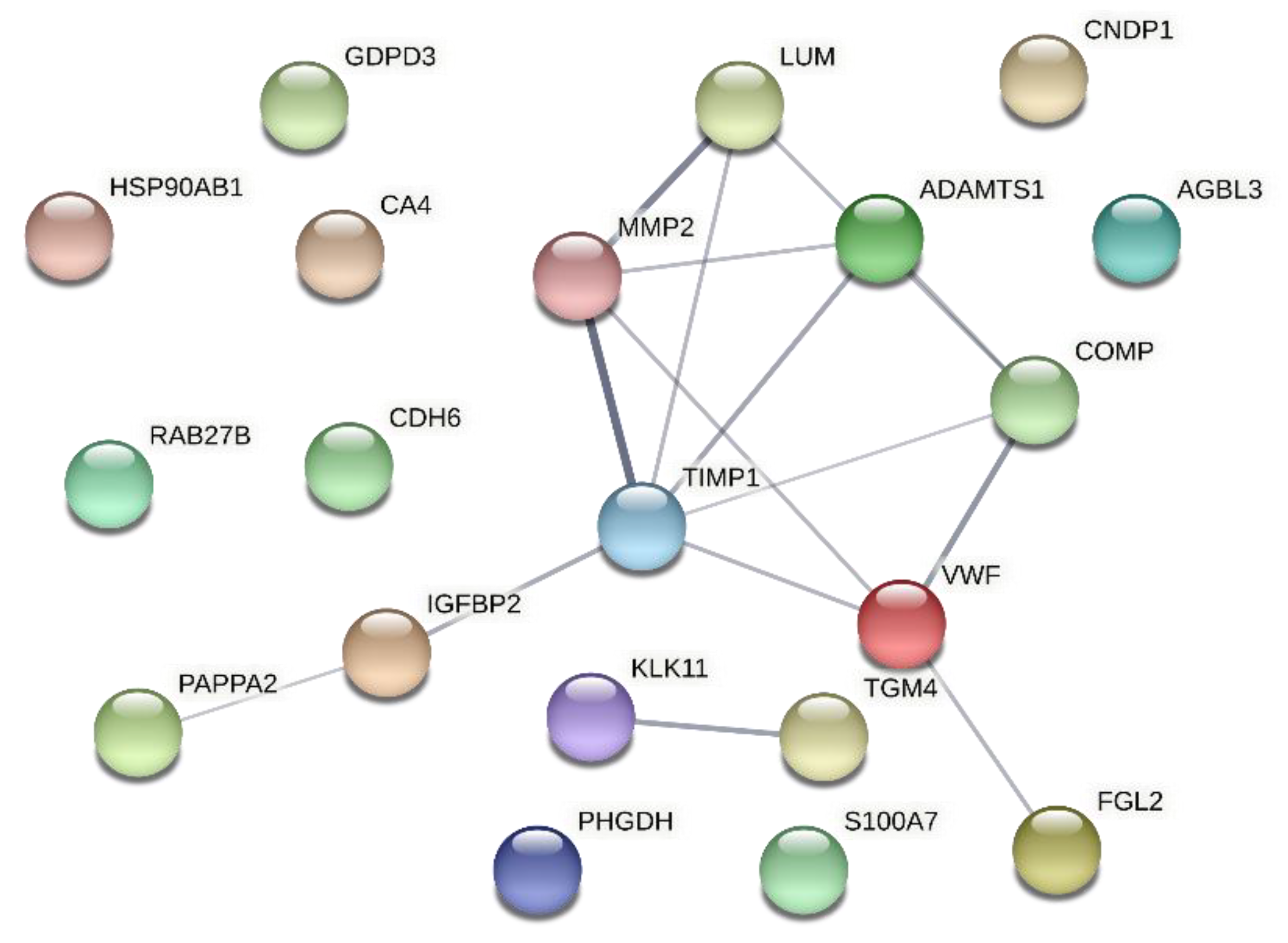
2.1. Identification of Potential CAD/AVS Biomarkers through Urine Proteomics
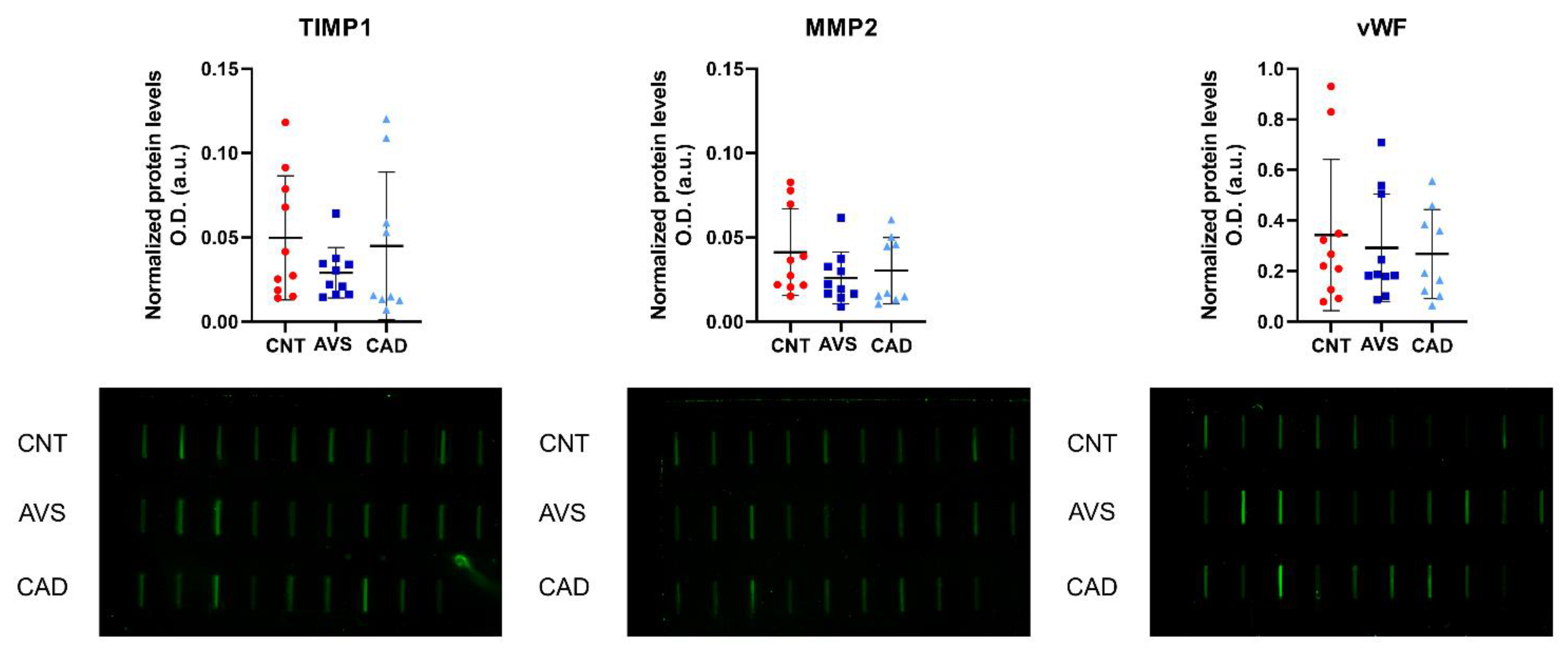
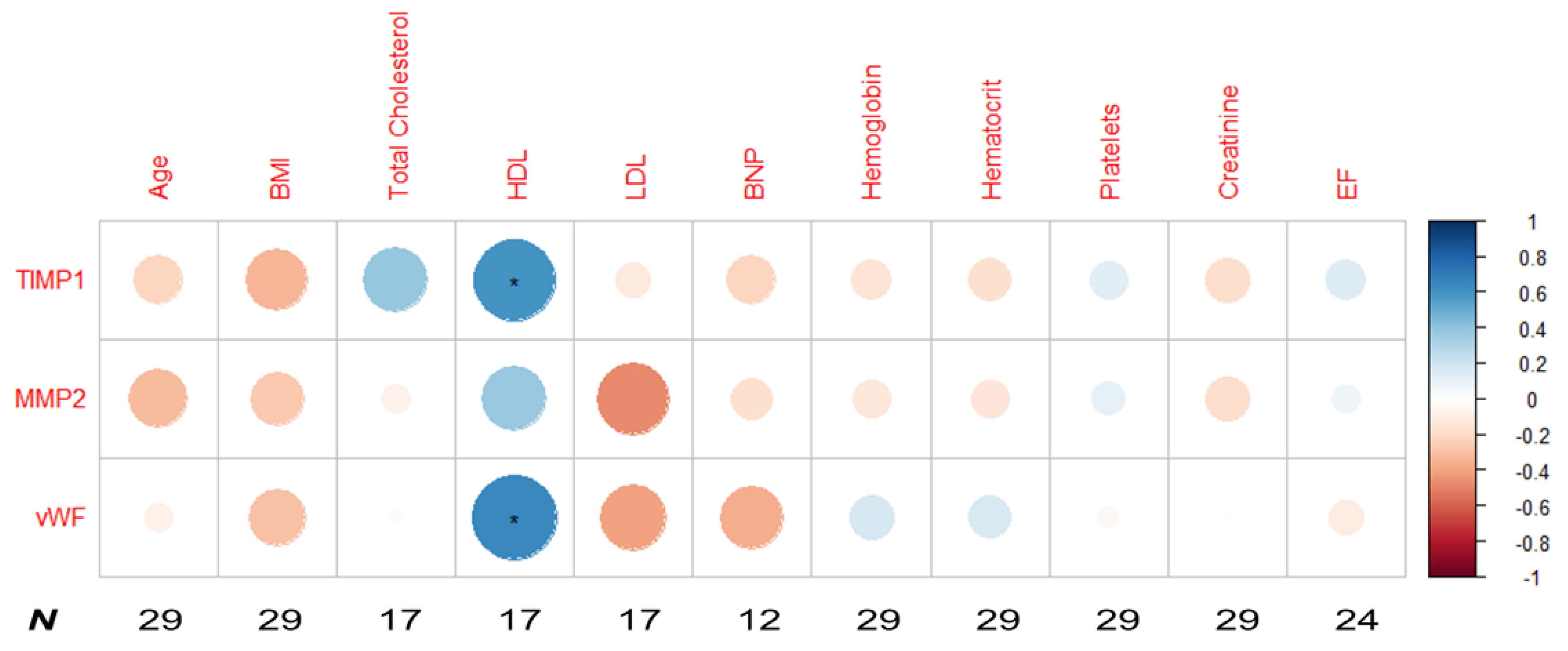
2.2. Slot Blot Relative Quantification of the Putative Biomarkers of Coronary Artery Disease/Aortic Valve Stenosis
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Preparation
4.2. LC-MS/MS Analysis
4.3. Data Analysis
4.4. Bioinformatics Analysis
4.5. Slot Blot Relative Quantification of Urine Proteins
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mendis, S.; Puska, P.; Norrving, B. Global Atlas on Cardiovascular Disease Prevention and Control; WHO: Geneva, Switzerland; World Heart Federation: Geneva, Switzerland; World Stroke Organ: Geneva, Switzerland, 2011. [Google Scholar]
- Brown, J.C.; Gerhardt, T.E.; Kwon, E. Risk Factors for Coronary Artery Disease; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- GBD 2017 Causes of Death Collaborators. Global, Regional, and National Age-Sex-Specific Mortality for 282 Causes of Death in 195 Countries and Territories, 1980–2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1736. [Google Scholar] [CrossRef]
- el Sabbagh, A.; Nishimura, R.A. Clinical Conundrum of Coronary Artery Disease and Aortic Valve Stenosis. J. Am. Heart Assoc. 2017, 6, e005593. [Google Scholar] [CrossRef] [PubMed]
- Ambrose, J.A.; Singh, M. Pathophysiology of Coronary Artery Disease Leading to Acute Coronary Syndromes. F1000Prime Rep. 2015, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Hanson, M.A.; Fareed, M.T.; Argenio, S.L.; Agunwamba, A.O.; Hanson, T.R. Coronary Artery Disease. Prim. Care 2013, 40, 1. [Google Scholar] [CrossRef] [PubMed]
- Mohler, E.R., 3rd; Gannon, F.; Reynolds, C.; Zimmerman, R.; Keane, M.G.; Kaplan, F.S. Bone Formation and Inflammation in Cardiac Valves. Circulation 2001, 103, 1522. [Google Scholar] [CrossRef]
- Dweck, M.R.; Boon, N.A.; Newby, D.E. Calcific Aortic Stenosis: A Disease of the Valve and the Myocardium. J. Am. Coll. Cardiol. 2012, 60, 1854. [Google Scholar] [CrossRef]
- Wissler, R.W. Update on the Pathogenesis of Atherosclerosis. Am. J. Med. 1991, 91, S3. [Google Scholar] [CrossRef]
- Lindman, B.R.; Clavel, M.-A.; Mathieu, P.; Iung, B.; Lancellotti, P.; Otto, C.M.; Pibarot, P. Calcific Aortic Stenosis. Nat. Rev. Dis. Prim. 2016, 2, 16006. [Google Scholar] [CrossRef]
- O’Brien, K.D.; Reichenbach, D.D.; Marcovina, S.M.; Kuusisto, J.; Alpers, C.E.; Otto, C.M. Apolipoproteins B, (a), and E Accumulate in the Morphologically Early Lesion of “degenerative” Valvular Aortic Stenosis. Arterioscler. Thromb. Vasc. Biol. 1996, 16, 523. [Google Scholar] [CrossRef]
- Regmi, M.; Siccardi, M.A. Coronary Artery Disease Prevention. [Updated 2021 Aug 11]; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Rogers, F.J. Aortic Stenosis: New Thoughts on a Cardiac Disease of Older People. J. Osteopath. Med. 2013, 113, 820. [Google Scholar] [CrossRef][Green Version]
- Ross, R. Atherosclerosis—An Inflammatory Disease. N. Engl. J. Med. 1999, 340, 115. [Google Scholar] [CrossRef] [PubMed]
- Grech, E.D. Pathophysiology and Investigation of Coronary Artery Disease. BMJ 2003, 326, 1027. [Google Scholar] [CrossRef] [PubMed]
- Sverdlov, A.L.; Ngo, D.T.; Chapman, M.J.; Ali, O.A.; Chirkov, Y.Y.; Horowitz, J.D. Pathogenesis of Aortic Stenosis: Not Just a Matter of Wear and Tear. Am. J. Cardiovasc. Dis. 2011, 1, 185. [Google Scholar] [PubMed]
- Moore, K.J.; Koplev, S.; Fisher, E.A.; Tabas, I.; Björkegren, J.L.M.; Doran, A.C.; Kovacic, J.C. Macrophage Trafficking, Inflammatory Resolution, and Genomics in Atherosclerosis: JACC Macrophage in CVD Series (Part 2). J. Am. Coll. Cardiol. 2018, 72, 2181. [Google Scholar] [CrossRef]
- Shah, R.V.; Desai, N.R.; O’gara, P.T. Asymptomatic Severe Aortic Stenosis: Silence of the Lambs? Circulation 2010, 122, 1734–1739. [Google Scholar] [CrossRef]
- Vick, G.W., 3rd. The Gold Standard for Noninvasive Imaging in Coronary Heart Disease: Magnetic Resonance Imaging. Curr. Opin. Cardiol. 2009, 24, 567. [Google Scholar] [CrossRef]
- Saikrishnan, N.; Kumar, G.; Sawaya, F.J.; Lerakis, S.; Yoganathan, A.P. Accurate Assessment of Aortic Stenosis. Circulation 2014, 129, 244. [Google Scholar] [CrossRef]
- Cassar, A.; Holmes, D.R.J.; Rihal, C.S.; Gersh, B.J. Chronic Coronary Artery Disease: Diagnosis and Management. Mayo Clin. Proc. 2009, 84, 1130. [Google Scholar] [CrossRef]
- Joseph, J.; Naqvi, S.Y.; Giri, J.; Goldberg, S. Aortic Stenosis: Pathophysiology, Diagnosis, and Therapy. Am. J. Med. 2017, 130, 253. [Google Scholar] [CrossRef]
- Rusnak, J.; Fastner, C.; Behnes, M.; Mashayekhi, K.; Borggrefe, M.; Akin, I. Biomarkers in Stable Coronary Artery Disease. Curr. Pharm. Biotechnol. 2017, 18, 456. [Google Scholar] [CrossRef]
- McCarthy, C.P.; McEvoy, J.W.; Januzzi, J.L.J. Biomarkers in Stable Coronary Artery Disease. Am. Heart J. 2018, 196, 82. [Google Scholar] [CrossRef] [PubMed]
- Redfors, B.; Furer, A.; Lindman, B.R.; Burkhoff, D.; Marquis-Gravel, G.; Francese, D.P.; Ben-Yehuda, O.; Pibarot, P.; Gillam, L.D.; Leon, M.B.; et al. Biomarkers in Aortic Stenosis: A Systematic Review. Struct. Heart 2017, 1, 18. [Google Scholar] [CrossRef]
- Collet, J.-P.; Thiele, H.; Barbato, E.; Barthélémy, O.; Bauersachs, J.; Bhatt, D.L.; Dendale, P.; Dorobantu, M.; Edvardsen, T.; Folliguet, T.; et al. 2020 ESC Guidelines for the Management of Acute Coronary Syndromes in Patients Presenting without Persistent ST-Segment Elevation. Eur. Heart J. 2021, 42, 1289. [Google Scholar] [CrossRef] [PubMed]
- Knuuti, J.; Wijns, W.; Saraste, A.; Capodanno, D.; Barbato, E.; Funck-Brentano, C.; Prescott, E.; Storey, R.F.; Deaton, C.; Cuisset, T.; et al. 2019 ESC Guidelines for the Diagnosis and Management of Chronic Coronary Syndromes. Eur. Heart J. 2020, 41, 407. [Google Scholar] [CrossRef] [PubMed]
- van Holten, T.C.; Waanders, L.F.; de Groot, P.G.; Vissers, J.; Hoefer, I.E.; Pasterkamp, G.; Prins, M.W.J.; Roest, M. Circulating Biomarkers for Predicting Cardiovascular Disease Risk; a Systematic Review and Comprehensive Overview of Meta-Analyses. PLoS ONE 2013, 8, e62080. [Google Scholar] [CrossRef] [PubMed]
- Vahanian, A.; Beyersdorf, F.; Praz, F.; Milojevic, M.; Baldus, S.; Bauersachs, J.; Capodanno, D.; Conradi, L.; De Bonis, M.; De Paulis, R.; et al. 2021 ESC/EACTS Guidelines for the Management of Valvular Heart Disease. Eur. Heart J. 2022, 43, 561. [Google Scholar] [CrossRef] [PubMed]
- Basak, T.; Tanwar, V.S.; Bhardwaj, G.; Bhardwaj, N.; Ahmad, S.; Garg, G.; Sreenivas, V.; Karthikeyan, G.; Seth, S.; Sengupta, S. Plasma Proteomic Analysis of Stable Coronary Artery Disease Indicates Impairment of Reverse Cholesterol Pathway. Sci. Rep. 2016, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Ferrannini, E.; Manca, M.L.; Ferrannini, G.; Andreotti, F.; Andreini, D.; Latini, R.; Magnoni, M.; Williams, S.A.; Maseri, A.; Maggioni, A.P. Differential Proteomics of Cardiovascular Risk and Coronary Artery Disease in Humans. Front. Cardiovasc. Med. 2021, 8, 790289. [Google Scholar] [CrossRef]
- Ljungberg, J.; Janiec, M.; Bergdahl, I.A.; Holmgren, A.; Hultdin, J.; Johansson, B.; Näslund, U.; Siegbahn, A.; Fall, T.; Söderberg, S. Proteomic Biomarkers for Incident Aortic Stenosis Requiring Valvular Replacement. Circulation 2018, 138, 590. [Google Scholar] [CrossRef]
- Hortin, G.L.; Sviridov, D. The Dynamic Range Problem in the Analysis of the Plasma Proteome. J. Proteomics 2010, 73, 629. [Google Scholar] [CrossRef]
- Decramer, S.; de Peredo, A.G.; Breuil, B.; Mischak, H.; Monsarrat, B.; Bascands, J.-L.; Schanstra, J.P. Urine in Clinical Proteomics. Mol. Cell. Proteom. 2008, 7, 1850. [Google Scholar] [CrossRef] [PubMed]
- Snell-Bergeon, J.K.; Maahs, D.M.; Ogden, L.G.; Kinney, G.L.; Hokanson, J.E.; Schiffer, E.; Rewers, M.; Mischak, H. Evaluation of Urinary Biomarkers for Coronary Artery Disease, Diabetes, and Diabetic Kidney Disease. Diabetes Technol. Ther. 2009, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Iwata, A.; Futami, M.; Yamashita, M.; Imaizumi, S.; Kuwano, T.; Ike, A.; Sugihara, M.; Nishikawa, H.; Zhang, B.; et al. Impact of von Willebrand Factor on Coronary Plaque Burden in Coronary Artery Disease Patients Treated with Statins. Medicine 2018, 97, e0589. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Wang, X.; Peng, X.; Feng, S.; Zhao, J.; Liao, L.; Zhang, Y.; Hou, Y.; Liu, J. Prognostic Value of Plasma von Willebrand Factor Levels in Major Adverse Cardiovascular Events: A Systematic Review and Meta-Analysis. BMC Cardiovasc. Disord. 2020, 20, 72. [Google Scholar] [CrossRef]
- Yan, B.; Wang, Q.; Du, W.; Zhai, S.; Gou, C.; Hu, T.; Xia, L.; Ruan, C.; Zhao, Y. Elevated Plasma von Willebrand Factor Antigen and Activity Levels Are Associated With the Severity of Coronary Stenosis. Clin. Appl. Thromb. 2020, 26, 1076029619900552. [Google Scholar] [CrossRef]
- van Belle, E.; Rauch, A.; Vincent, F.; Robin, E.; Kibler, M.; Labreuche, J.; Jeanpierre, E.; Levade, M.; Hurt, C.; Rousse, N.; et al. Von Willebrand Factor Multimers during Transcatheter Aortic-Valve Replacement. N. Engl. J. Med. 2016, 375, 335. [Google Scholar] [CrossRef]
- Kaden, J.J.; Vocke, D.C.; Fischer, C.S.; Grobholz, R.; Brueckmann, M.; Vahl, C.F.; Hagl, S.; Haase, K.K.; Dempfle, C.E.; Borggrefe, M. Expression and Activity of Matrix Metalloproteinase-2 in Calcific Aortic Stenosis. Z. Kardiol. 2004, 93, 124. [Google Scholar] [CrossRef]
- Galis, Z.S.; Khatri, J.J. Matrix Metalloproteinases in Vascular Remodeling and Atherogenesis. Circ. Res. 2002, 90, 251. [Google Scholar] [CrossRef]
- Orbe, J.; Fernandez, L.; Rodríguez, J.A.; Rábago, G.; Belzunce, M.; Monasterio, A.; Roncal, C.; Páramo, J.A. Different Expression of MMPs/TIMP-1 in Human Atherosclerotic Lesions. Relation to Plaque Features and Vascular Bed. Atherosclerosis 2003, 170, 269. [Google Scholar] [CrossRef]
- Fitzsimmons, P.J.; Forough, R.; Lawrence, M.E.; Gantt, D.S.; Rajab, M.H.; Kim, H.; Weylie, B.; Spiekerman, A.M.; Dehmer, G.J. Urinary Levels of Matrix Metalloproteinase 9 and 2 and Tissue Inhibitor of Matrix Metalloproteinase in Patients with Coronary Artery Disease. Atherosclerosis 2007, 194, 196. [Google Scholar] [CrossRef]
- Sundström, J.; Evans, J.C.; Benjamin, E.J.; Levy, D.; Larson, M.G.; Sawyer, D.B.; Siwik, D.A.; Colucci, W.S.; Wilson, P.W.F.; Vasan, R.S. Relations of Plasma Total TIMP-1 Levels to Cardiovascular Risk Factors and Echocardiographic Measures: The Framingham Heart Study. Eur. Heart J. 2004, 25, 1509. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.W.; Chen, J.; Ling, M.; Fu, X.; Blevins, T.; Parsons, S.; Le, J.; Harris, J.; Martin, T.R.; Konkle, B.A.; et al. High-Density Lipoprotein Modulates Thrombosis by Preventing von Willebrand Factor Self-Association and Subsequent Platelet Adhesion. Blood 2016, 127, 637. [Google Scholar] [CrossRef] [PubMed]
- Leutert, M.; Rodríguez-Mias, R.A.; Fukuda, N.K.; Villén, J. R2-P2 Rapid-Robotic Phosphoproteomics Enables Multidimensional Cell Signaling Studies. Mol. Syst. Biol. 2019, 15, e9021. [Google Scholar] [CrossRef] [PubMed]
- Türker, C.; Akal, F.; Joho, D.; Panse, C.; Barkow-Oesterreicher, S.; Rehrauer, H.; Schlapbach, R. B-Fabric: The Swiss Army Knife for Life Sciences. In Proceedings of the EDBT ’10: 13th International Conference on Extending Database Technology, Lausanne, Switzerland, 22–26 March 2010; Volume 717, p. 720. [Google Scholar]
- Cox, J.; Mann, M. MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008, 26, 1367. [Google Scholar] [CrossRef] [PubMed]
- Wolski, C.P.W.; Grossmann, J. SRMService—R-Package to Report Quantitative Mass Spectrometry Data. Available online: https://github.com/protViz/SRMService (accessed on 1 March 2022).
| Discovery Cohort | Testing Cohort | |||||||
|---|---|---|---|---|---|---|---|---|
| (CAD/AVS + Controls) (N = 23) | CAD/AVS (N = 12) | Controls (N = 11) | p-Value | (CAD/AVS + Controls) (N = 29) | CAD/AVS (N = 19) | Controls (N = 10) | p-Value | |
| Sex (man) | 12 + 11 | 67% | 82% | n.s. | 19 + 10 | 100% | 50% | <0.001 |
| Age (years) | 12 + 5 | 70 (65–75) | 63 (59–76) | n.s. | 19 + 10 | 78 (74–81) | 69 (63–75) | <0.01 |
| Body Mass Index | 11 + 5 | 29 (25–30) | 30 (24–35) | n.s. | 19 + 10 | 27 (4) | 29 (9) | n.s. |
| Total cholesterol (mg/dL) | 3 + 5 | 142 (122–194) | 161 (107–179) | n.s. | 7 + 10 | 164 (139–189) | 142 (129–161) | n.s. |
| HDL (mg/dL) | 3 + 5 | 35 (34–40) | 46 (39–46) | n.s. | 7 + 10 | 55 (40–56) | 46 (42–60) | <0.05 |
| LDL (mg/dL) | 3 + 5 | 78 (65–128) | 77 (65–101) | n.s. | 7 + 10 | 85 (80–112) | 67.5 (58–79) | <0.05 |
| Hemoglobin (g/dL) | 12 + 5 | 14 (13–15) | 14 (13–15) | n.s. | 19 + 10 | 14 (13–15) | 12 (11–13) | <0.05 |
| Hematocrit (%) | 12 + 5 | 41 (40–44) | 42 (37–44) | n.s. | 19 + 10 | 42 (39–43) | 37 (33–38) | <0.01 |
| Platelets (thousands/mL) | 12 + 5 | 237 (198–246) | 277 (235–280) | n.s. | 19 + 10 | 207 (181–228) | 270 (214–280) | <0.01 |
| Creatinine (mg/dL) | 12 + 5 | 0.8 (0.7–1) | 0.87 (0.8–1) | n.s. | 19 + 10 | 0.95 (0.9–1.1) | 0.22 (0.7–1) | n.s. |
| Ejection fraction (%) | 7 + 5 | 36 (34–54) | 63 (62–65) | <0.001 | 15 + 9 | 49 (40–62) | 63 (62–64) | <0.001 |
| Hypertension | 12 + 11 | 83% | 45% | n.s. | 19 + 10 | 95% | 90% | n.s. |
| Dyslipidemia | 12 + 11 | 75% | 45% | n.s. | 19 + 10 | 79% | 100% | n.s. |
| Ex-smoker or smoker | 12 + 5 | 50% | 60% | n.s. | 19 + 10 | 68% | 40% | n.s. |
| Diabetes mellitus | 12 + 11 | 42% | 27% | n.s. | 19 + 10 | 58% | 60% | n.s. |
| Angina | 10 + 11 | 50% | 0% | n.s. | 19 + 10 | 74% | 0% | n.s. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perpétuo, L.; Barros, A.S.; Dalsuco, J.; Nogueira-Ferreira, R.; Resende-Gonçalves, P.; Falcão-Pires, I.; Ferreira, R.; Leite-Moreira, A.; Trindade, F.; Vitorino, R. Coronary Artery Disease and Aortic Valve Stenosis: A Urine Proteomics Study. Int. J. Mol. Sci. 2022, 23, 13579. https://doi.org/10.3390/ijms232113579
Perpétuo L, Barros AS, Dalsuco J, Nogueira-Ferreira R, Resende-Gonçalves P, Falcão-Pires I, Ferreira R, Leite-Moreira A, Trindade F, Vitorino R. Coronary Artery Disease and Aortic Valve Stenosis: A Urine Proteomics Study. International Journal of Molecular Sciences. 2022; 23(21):13579. https://doi.org/10.3390/ijms232113579
Chicago/Turabian StylePerpétuo, Luís, António S. Barros, Jéssica Dalsuco, Rita Nogueira-Ferreira, Pedro Resende-Gonçalves, Inês Falcão-Pires, Rita Ferreira, Adelino Leite-Moreira, Fábio Trindade, and Rui Vitorino. 2022. "Coronary Artery Disease and Aortic Valve Stenosis: A Urine Proteomics Study" International Journal of Molecular Sciences 23, no. 21: 13579. https://doi.org/10.3390/ijms232113579
APA StylePerpétuo, L., Barros, A. S., Dalsuco, J., Nogueira-Ferreira, R., Resende-Gonçalves, P., Falcão-Pires, I., Ferreira, R., Leite-Moreira, A., Trindade, F., & Vitorino, R. (2022). Coronary Artery Disease and Aortic Valve Stenosis: A Urine Proteomics Study. International Journal of Molecular Sciences, 23(21), 13579. https://doi.org/10.3390/ijms232113579