EGFR T751_I759delinsN Mutation in Exon19 Detected by NGS but Not by Real-Time PCR in a Heavily-Treated Patient with NSCLC
Abstract
1. Introduction
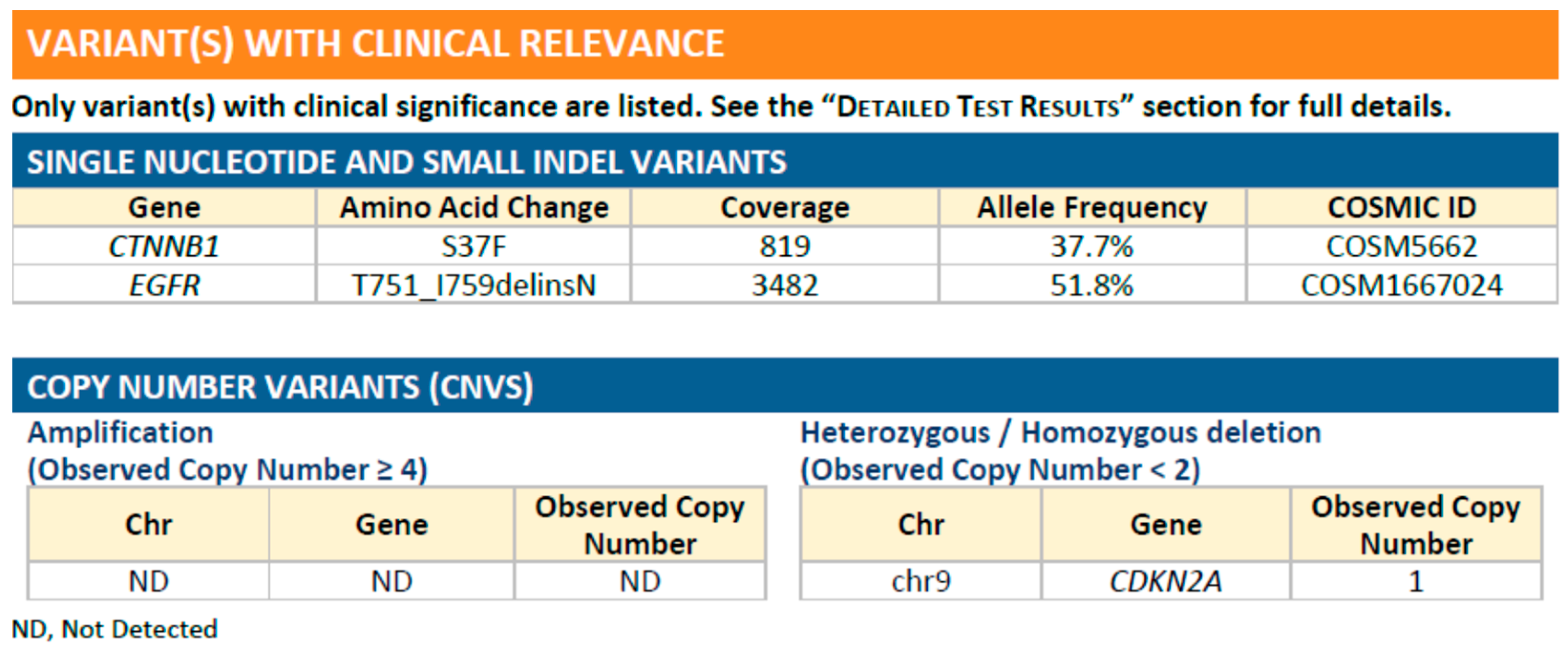
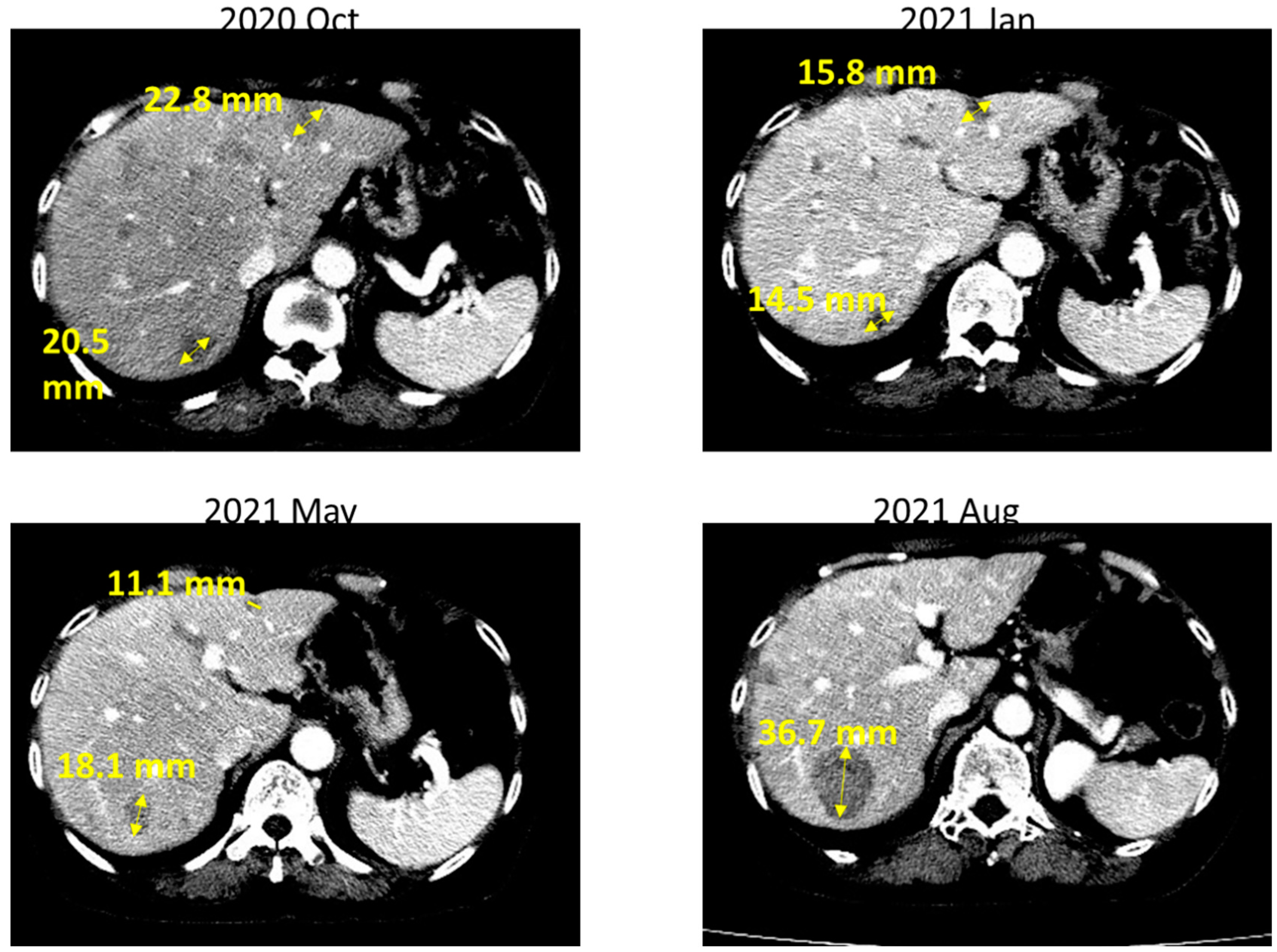
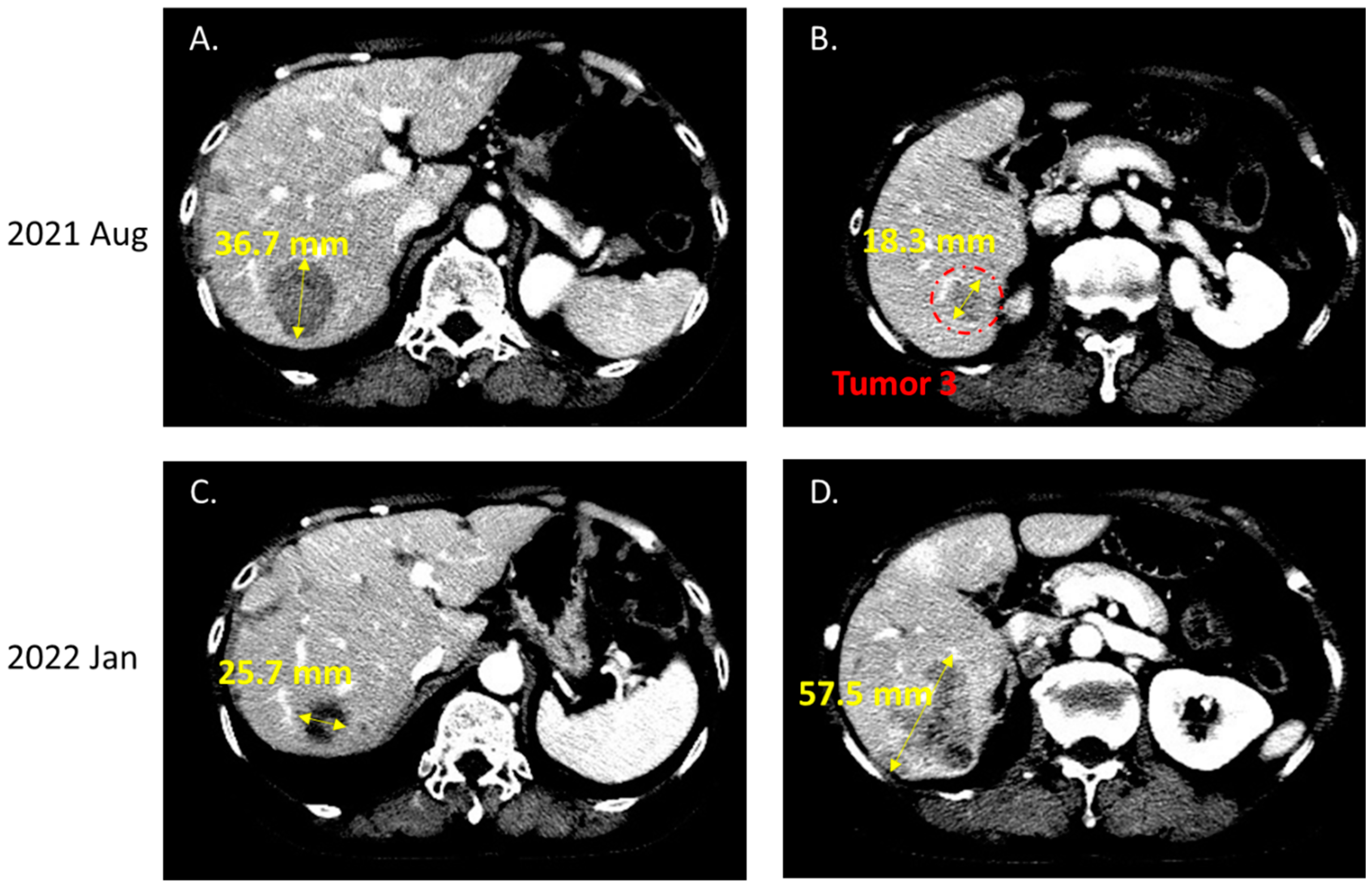
2. Case Report
3. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Melosky, B.; Kambartel, K.; Hantschel, M.; Bennetts, M.; Nickens, D.J.; Brinkmann, J.; Kayser, A.; Moran, M.; Cappuzzo, F. Worldwide Prevalence of Epidermal Growth Factor Receptor Mutations in Non-Small Cell Lung Cancer: A Meta-Analysis. Mol. Diagn. Ther. 2022, 26, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Gao, Q.; Jiang, X.; Zhan, Z.; Yan, Q.; Li, Z.; Huang, C. Comparison of Clinicopathological Features and Prognosis between ALK Rearrangements and EGFR Mutations in Surgically Resected Early-stage Lung Adenocarcinoma. J. Cancer 2019, 10, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Lindeman, N.I.; Cagle, P.T.; Beasley, M.B.; Chitale, D.A.; Dacic, S.; Giaccone, G.; Jenkins, R.B.; Kwiatkowski, D.J.; Saldivar, J.S.; Squire, J.; et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: Guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J. Thorac. Oncol. 2013, 8, 823–859. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, E.; Sugita, M.; Furukawa, K.; Takahashi, H.; Uchida, O.; Kawaguchi, Y.; Ohira, T.; Matsubayashi, J.; Ikeda, N.; Hirsch, F.R.; et al. Frequency and significance of epidermal growth factor receptor mutations detected by PCR methods in patients with non-small cell lung cancer. Oncol. Lett. 2019, 17, 5125–5131. [Google Scholar] [CrossRef]
- Bruno, R.; Fontanini, G. Next Generation Sequencing for Gene Fusion Analysis in Lung Cancer: A Literature Review. Diagnostics 2020, 10, 521. [Google Scholar] [CrossRef]
- Shen, C.; Chiang, C.; Luo, Y.; Huang, H.; Chiu, C. FP12.05 The Intrinsic Limitation and Clinical Impact of Mutant Allele-Specific Real-Time PCR-Based EGFR Mutation Assay in NSCLC. J. Thorac. Oncol. 2021, 16, S966. [Google Scholar] [CrossRef]
- Fang, Y.F.; Liu, P.C. Afatinib and osimertinib in lung adenocarcinoma harbored EGFR T751_I759delinsS mutation: A case report. Thorac. Cancer 2021, 12, 3429–3432. [Google Scholar] [CrossRef]
- Lettig, L.; Sahnane, N.; Pepe, F.; Cerutti, R.; Albeni, C.; Franzi, F.; Veronesi, G.; Ogliari, F.; Pastore, A.; Tuzi, A.; et al. EGFR T790M detection rate in lung adenocarcinomas at baseline using droplet digital PCR and validation by ultra-deep next generation sequencing. Transl. Lung Cancer Res. 2019, 8, 584–592. [Google Scholar] [CrossRef]
- Wu, C.E.; Ng, C.T.; Tan, K.T. Transient Response of Olaparib on Pulmonary Artery Sarcoma Harboring Multiple Homologous Recombinant Repair Gene Alterations. J. Pers. Med. 2021, 11, 357. [Google Scholar] [CrossRef]
- Tfayli, A.H.; Fakhri, G.B.; Al Assaad, M.S. Prevalence of the epidermal growth factor receptor mutations in lung adenocarcinoma patients from the Middle East region. Ann. Thorac. Med. 2019, 14, 173–178. [Google Scholar] [CrossRef]
- Chen, J.; Yang, H.; Teo, A.S.M.; Amer, L.B.; Sherbaf, F.G.; Tan, C.Q.; Alvarez, J.J.S.; Lu, B.; Lim, J.Q.; Takano, A.; et al. Genomic landscape of lung adenocarcinoma in East Asians. Nat. Genet. 2020, 52, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Li, B.; Lin, D.; Zhou, Q.; Huang, D. Advances in Targeted Therapy and Immunotherapy for Non-small Cell Lung Cancer Based on Accurate Molecular Typing. Front. Pharmacol. 2019, 10, 230. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.I.; Chiang, C.L.; Shiao, T.H.; Luo, Y.H.; Chao, H.S.; Huang, H.C.; Chiu, C.H. Real-world evidence of the intrinsic limitations of PCR-based EGFR mutation assay in non-small cell lung cancer. Sci. Rep. 2022, 12, 13566. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, H.; d’Arienzo, P.D.; Yousaf, N.; Cui, W.; Popat, S. Inadequacy of PCR genotyping in advanced non-small cell lung cancer: EGFR L747_A755delinsSS exon 19 deletion is not detected by the real-time PCR Idylla EGFR mutation test but is detected by ctDNA next generation sequencing and responds to osimertinib. Eur. J. Cancer 2022, 166, 38–40. [Google Scholar] [CrossRef]
- Letovanec, I.; Finn, S.; Zygoura, P.; Smyth, P.; Soltermann, A.; Bubendorf, L.; Speel, E.J.; Marchetti, A.; Nonaka, D.; Monkhorst, K.; et al. Evaluation of NGS and RT-PCR Methods for ALK Rearrangement in European NSCLC Patients: Results from the European Thoracic Oncology Platform Lungscape Project. J. Thorac. Oncol. 2018, 13, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Bates, J.A.; Taylor, E.J. Scorpion ARMS primers for SNP real-time PCR detection and quantification of Pyrenophora teres. Mol. Plant. Pathol. 2001, 2, 275–280. [Google Scholar] [CrossRef]
- Shi, Y.; Au, J.S.; Thongprasert, S.; Srinivasan, S.; Tsai, C.M.; Khoa, M.T.; Heeroma, K.; Itoh, Y.; Cornelio, G.; Yang, P.C. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J. Thorac. Oncol. 2014, 9, 154–162. [Google Scholar] [CrossRef]
- Siggillino, A.; Ulivi, P.; Pasini, L.; Reda, M.S.; Chiadini, E.; Tofanetti, F.R.; Baglivo, S.; Metro, G.; Crino, L.; Delmonte, A.; et al. Detection of EGFR Mutations in Plasma Cell-Free Tumor DNA of TKI-Treated Advanced-NSCLC Patients by Three Methodologies: Scorpion-ARMS, PNAClamp, and Digital PCR. Diagnostics 2020, 10, 1062. [Google Scholar] [CrossRef]
- Kimura, H.; Ohira, T.; Uchida, O.; Matsubayashi, J.; Shimizu, S.; Nagao, T.; Ikeda, N.; Nishio, K. Analytical performance of the cobas EGFR mutation assay for Japanese non-small-cell lung cancer. Lung Cancer 2014, 83, 329–333. [Google Scholar] [CrossRef]
- Xu, C.W.; Lei, L.; Wang, W.X.; Lin, L.; Zhu, Y.C.; Wang, H.; Miao, L.Y.; Wang, L.P.; Zhuang, W.; Fang, M.Y.; et al. Molecular Characteristics and Clinical Outcomes of EGFR Exon 19 C-Helix Deletion in Non-Small Cell Lung Cancer and Response to EGFR TKIs. Transl. Oncol. 2020, 13, 100791. [Google Scholar] [CrossRef]
- Paz-Ares, L.; Tan, E.H.; O’Byrne, K.; Zhang, L.; Hirsh, V.; Boyer, M.; Yang, J.C.; Mok, T.; Lee, K.H.; Lu, S.; et al. Afatinib versus gefitinib in patients with EGFR mutation-positive advanced non-small-cell lung cancer: Overall survival data from the phase IIb LUX-Lung 7 trial. Ann. Oncol. 2017, 28, 270–277. [Google Scholar] [CrossRef]
- Yang, J.C.-H.; Wu, Y.-L.; Schuler, M.; Sebastian, M.; Popat, S.; Yamamoto, N.; Zhou, C.; Hu, C.-P.; O’Byrne, K.; Feng, J.; et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): Analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol. 2015, 16, 141–151. [Google Scholar] [CrossRef]
- Wu, C.E.; Chang, C.F.; Huang, C.Y.; Yang, C.T.; Kuo, C.S.; Hsu, P.C.; Chang, J.W. Comparison of Different Tyrosine Kinase Inhibitors for Treatment of Poor Performance Status Patients with EGFR-Mutated Lung Adenocarcinoma. Cancers 2022, 14, 674. [Google Scholar] [CrossRef]
- Chang, J.W.; Huang, C.Y.; Fang, Y.F.; Chang, C.F.; Yang, C.T.; Kuo, C.S.; Hsu, P.C.; Wu, C.E. Risk Stratification Using a Novel Nomogram for 2190 EGFR-Mutant NSCLC Patients Receiving the First or Second Generation EGFR-TKI. Cancers 2022, 14, 977. [Google Scholar] [CrossRef]
- Tan, D.S.; Yom, S.S.; Tsao, M.S.; Pass, H.I.; Kelly, K.; Peled, N.; Yung, R.C.; Wistuba, I.I.; Yatabe, Y.; Unger, M.; et al. The International Association for the Study of Lung Cancer Consensus Statement on Optimizing Management of EGFR Mutation-Positive Non-Small Cell Lung Cancer: Status in 2016. J. Thorac. Oncol. 2016, 11, 946–963. [Google Scholar] [CrossRef]
- Chen, C.H.; Chang, J.W.; Chang, C.F.; Huang, C.Y.; Yang, C.T.; Kuo, C.S.; Fang, Y.F.; Hsu, P.C.; Wu, C.E. Real-world Afatinib Outcomes in Advanced Non-small Cell Lung Cancer Harboring EGFR Mutations. Anticancer Res. 2022, 42, 2145–2157. [Google Scholar] [CrossRef]
- Chang, J.W.; Huang, C.Y.; Fang, Y.F.; Chang, C.F.; Yang, C.T.; Kuo, C.S.; Hsu, P.C.; Wu, C.E. Epidermal growth factor receptor tyrosine kinase inhibitors for de novo T790M mutation: A retrospective study of 44 patients. Thorac. Cancer 2022, 13, 1888–1897. [Google Scholar] [CrossRef]
- Yu, H.A.; Arcila, M.E.; Rekhtman, N.; Sima, C.S.; Zakowski, M.F.; Pao, W.; Kris, M.G.; Miller, V.A.; Ladanyi, M.; Riely, G.J. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin. Cancer Res. 2013, 19, 2240–2247. [Google Scholar] [CrossRef]
- Buder, A.; Hochmair, M.J.; Filipits, M. The Allele Frequency of EGFR Mutations Predicts Survival in Advanced EGFR T790M-Positive Non-small Cell Lung Cancer Patients Treated with Osimertinib. Target. Oncol. 2021, 16, 77–84. [Google Scholar] [CrossRef]
- Wang, Y.; He, Y.; Tian, P.; Wang, W.; Wang, K.; Chuai, S.; Li, Y.; Zhao, S.; Wang, Y.; Li, W. Low T790M relative allele frequency indicates concurrent resistance mechanisms and poor responsiveness to osimertinib. Transl. Lung Cancer Res. 2020, 9, 1952–1962. [Google Scholar] [CrossRef]
- Lim, S.M.; Syn, N.L.; Cho, B.C.; Soo, R.A. Acquired resistance to EGFR targeted therapy in non-small cell lung cancer: Mechanisms and therapeutic strategies. Cancer Treat. Rev. 2018, 65, 1–10. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, Z.-T.; Chan, T.-M.; Wu, C.-E. EGFR T751_I759delinsN Mutation in Exon19 Detected by NGS but Not by Real-Time PCR in a Heavily-Treated Patient with NSCLC. Int. J. Mol. Sci. 2022, 23, 13451. https://doi.org/10.3390/ijms232113451
Chang Z-T, Chan T-M, Wu C-E. EGFR T751_I759delinsN Mutation in Exon19 Detected by NGS but Not by Real-Time PCR in a Heavily-Treated Patient with NSCLC. International Journal of Molecular Sciences. 2022; 23(21):13451. https://doi.org/10.3390/ijms232113451
Chicago/Turabian StyleChang, Zi-Ting, Tien-Ming Chan, and Chiao-En Wu. 2022. "EGFR T751_I759delinsN Mutation in Exon19 Detected by NGS but Not by Real-Time PCR in a Heavily-Treated Patient with NSCLC" International Journal of Molecular Sciences 23, no. 21: 13451. https://doi.org/10.3390/ijms232113451
APA StyleChang, Z.-T., Chan, T.-M., & Wu, C.-E. (2022). EGFR T751_I759delinsN Mutation in Exon19 Detected by NGS but Not by Real-Time PCR in a Heavily-Treated Patient with NSCLC. International Journal of Molecular Sciences, 23(21), 13451. https://doi.org/10.3390/ijms232113451




