Abstract
Fruit size is an important fruit quality trait that influences the production and commodity values of loquats (Eriobotrya japonica Lindl.). The Small Auxin Upregulated RNA (SAUR) gene family has proven to play a vital role in the fruit development of many plant species. However, it has not been comprehensively studied in a genome-wide manner in loquats, and its role in regulating fruit size remains unknown. In this study, we identified 95 EjSAUR genes in the loquat genome. Tandem duplication and segmental duplication contributed to the expansion of this gene family in loquats. Phylogenetic analysis grouped the SAURs from Arabidopsis, rice, and loquat into nine clusters. By analyzing the transcriptome profiles in different tissues and at different fruit developmental stages and comparing two sister lines with contrasting fruit sizes, as well as by functional predictions, a candidate gene (EjSAUR22) highly expressed in expanding fruits was selected for further functional investigation. A combination of Indoleacetic acid (IAA) treatment and virus-induced gene silencing revealed that EjSAUR22 was not only responsive to auxin, but also played a role in regulating cell size and fruit expansion. The findings from our study provide a solid foundation for understanding the molecular mechanisms controlling fruit size in loquats, and also provide potential targets for manipulation of fruit size to accelerate loquat breeding.
1. Introduction
Auxin plays a key role in regulating the growth and development of plants, which is achieved through activities such as controlling the plasma membrane H+-ATPase, or the regulation of plant genes [1,2]. The auxin-responsive genes in plants were classified into many gene families, including the Auxin/Indoleacetic Acid (Aux/IAA), Gretchen Hagen 3 (GH3), Small Auxin-Up RNA (SAUR), Auxin Response Factor (ARF), and Glutathione S-transferase (GST) families [3,4,5,6]. Among the three early auxin-responsive gene families (Aux/IAA, GH3, and SAUR), the SAUR genes can quickly respond to auxin stimuli within minutes, indicating that their transcription is greatly affected by auxin [7].
The first SAUR gene was identified in soybeans in their elongating hypocotyls, and was found to be rapidly induced by auxin [8]. In the past two decades, the SAUR gene families have been analyzed in many plant species, such as Arabidopsis [4], rice [9], sorghum [10], maize [3], citrus [11], watermelon [12], and apple [13]. It has been reported that many members of the SAURs family were derived from tandem duplications and segmental duplications, resulting in functional redundancy in some SAUR paralogues [14]. Despite the availability of these studies, few studies reported the functional characterization of the SAUR genes.
Various functions of SAURs have been reported from studies in Arabidopsis. The overexpression of AtSAUR32 contributes to a hookless phenotype in Arabidopsis, which can be rescued by exogenous auxin [15]. AtSAUR63 plays a role in cell elongation, and its overexpression can lead to the elongation of several tissues, such as the hypocotyls, petals, and stamen filaments [16]. The overexpression of AtSAUR36 and AtSAUR49 promotes leaf senescence [17,18]. The AtSAUR19 subfamily was reported to play a role in promoting cell expansion [19]. In addition, studies have shown that many SAURs participate in the synthesis and transport of auxin, and some members may play a positive role in fruit expansion [20,21].
Cultivated loquat (Eriobotrya japonica Lindl.) is a fruit tree crop in Rosaceae. Fruit quality traits greatly influence its production and commodity values, which makes them an important aspect of loquat research, for example, fruit weight/size [22,23,24]. Research has shown that cell number and cell size are two key factors determining the fruit size, while auxin is capable of promoting cell division and elongation [25]. Therefore, the levels and signal transduction of auxin are closely related to fruit size [26]. Despite the availability of several reference genomes for loquats [24,27,28], currently, no studies have been performed to analyze the SAUR gene family systematically and comprehensively in loquats. This has limited our understanding of the role of SAURs in regulating fruit weight/size in loquats. In this study, we performed a genome-wide analysis of the SAUR gene family in loquats and identified a candidate SAUR associated with cell expansion and fruit size. The findings from our study provide valuable information for fruit size breeding in loquats and lay a solid foundation for understanding the molecular mechanisms controlling fruit size in loquats.
2. Results
2.1. Identification and Annotation of the SAUR Family
Based on the reference genome of ‘Seventh Star’ [24], a total of 95 EjSAUR genes were identified from the loquat genome (Table 1). The EjSAURs were named EjSAUR1-EjSAUR95 according to their locations (chromosome one-seventeen, top to bottom). The predicted peptide lengths ranged from 90 to 226 amino acids, the predicted molecular mass ranged from 9.98 to 25.48 kDa, while the theoretical PI ranged from 4.92 to 10.14 (Table 1). The prediction of subcellular localization revealed that the majority of the EjSAURs are localized in the mitochondria (43) and nucleus (31) (Table 1).

Table 1.
Summary of identified Small Auxin-up RNA (SAUR) gene family in loquat.
2.2. Phylogenetic Relationships, Gene Structure, and Conserved Motifs
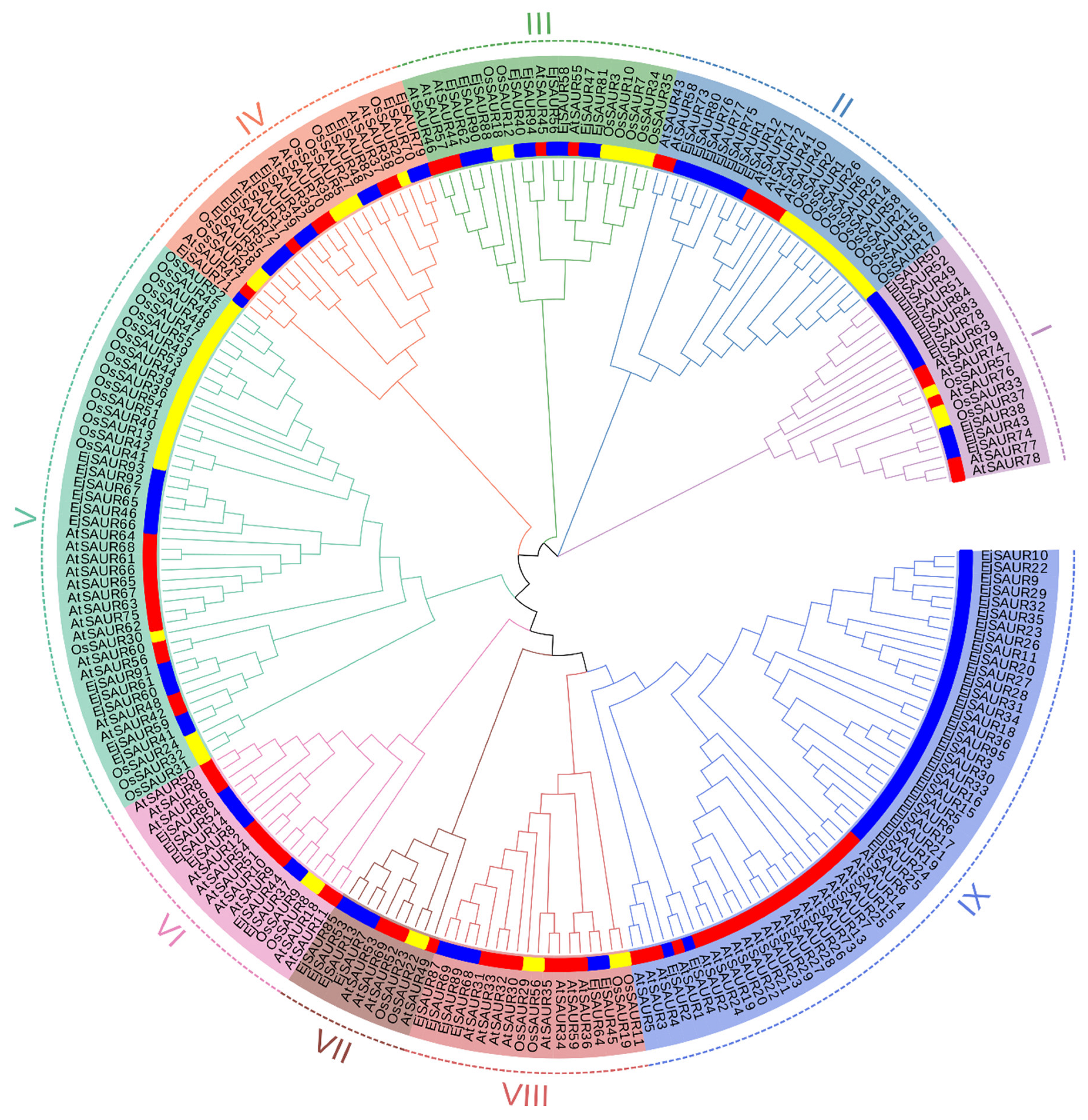
To investigate the phylogenetic relationships among the EjSAURs as well as the SAURs from Arabidopsis and rice, a phylogenetic tree was constructed based on their protein sequences using the neighbor-joining method in MEGA11 (Figure 1 and Table S1). The EjSAURs together with other SAURs were assigned to nine different clusters. Clusters IX and V contained the largest numbers of SAURs (53 and 45). Cluster VII contained the smallest numbers of SAURs, including two OsSAURs, four EjSAURs, and four AtSAURs. The majority of the EjSAURs were assigned to Cluster IX, similar with Arabidopsis, while most of the SAURs from rice were assigned to Clusters II and V. Interestingly, Cluster IX only contained EjSAURs and AtSAURs, which suggested that loquats and Arabidopsis (both dicots) may have retained the most duplication events of the SAUR gene families during evolution.
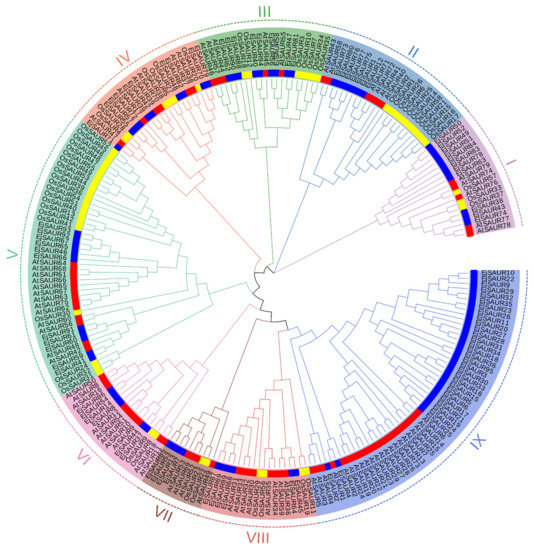
Figure 1.
Unrooted neighbor-joining phylogenetic tree of small auxin-up RNAs (SAURs) in loquats, Arabidopsis, and rice. The phylogenetic tree was constructed using SAUR protein sequences from Arabidopsis (red), rice (yellow), and loquats (blue) using the neighbor-joining method in MEGA11. A total of 1000 bootstraps were used. The genes belonging to different clusters (I–IX) are highlighted in different colors.
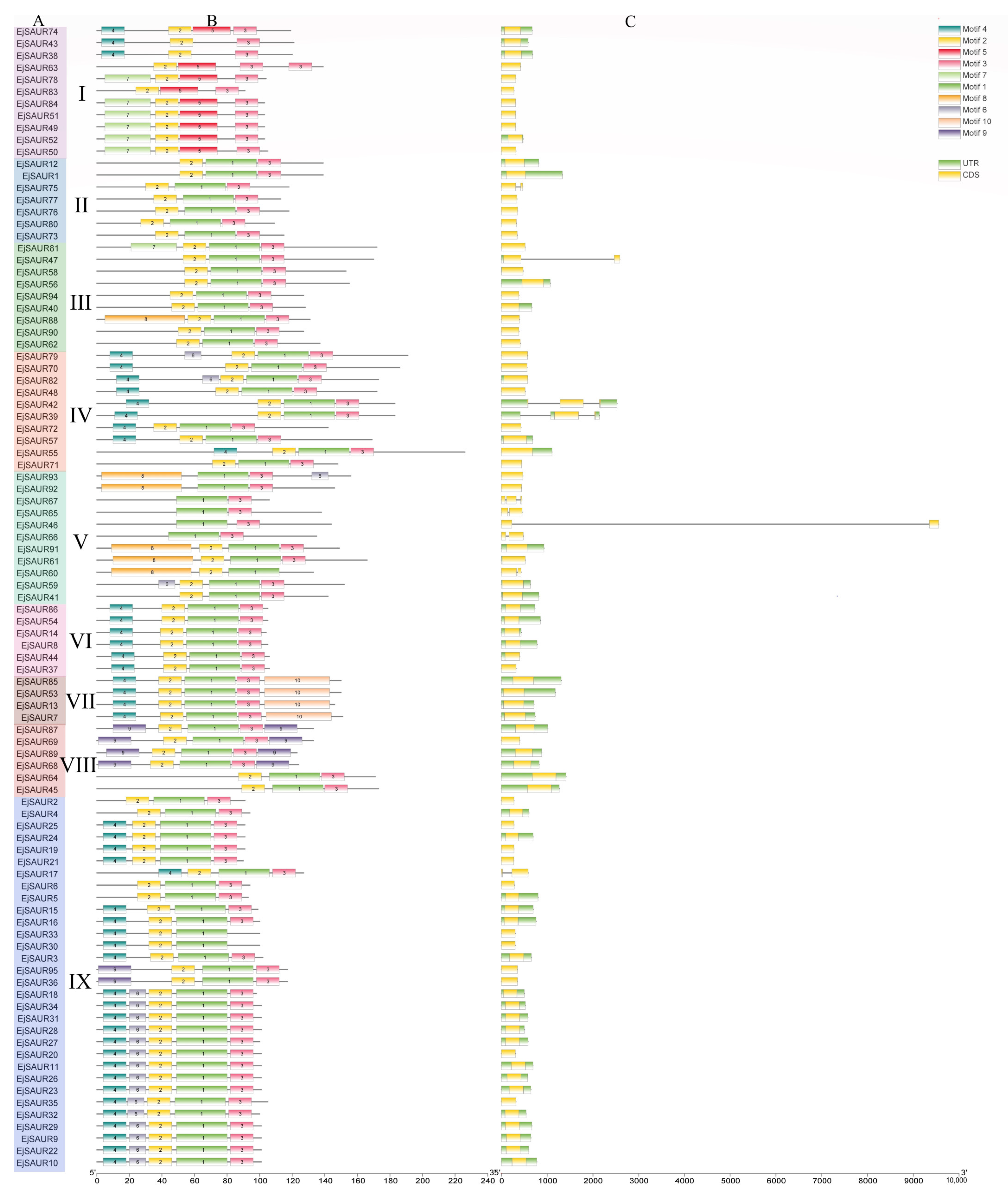
The exon–intron structures of EjSAURs are shown in Figure 2. Most members of the SAUR gene families contained no intron. A total of 85 EjSAURs did not contain an intron, which is consistent with the cases reported in other plant species. Among the remaining 10 EjSAURs, seven contained one intron, while the other three contained two introns. Interestingly, most members of Clusters VI, VII, VIII, and IX contained UTRs. Most members of Clusters VI and VII contained long 5′ UTRs, while most members of Cluster VIII contained long 3′ UTRs, which might be related with their mRNA stability, translation efficiency, and gene expressions [29,30,31]. In total, 10 different motifs were identified and shown in Figure 2, while motifs one, two, and three were the most conservative/common motifs of EjSAURs, as 76 EjSAURs contained these three motifs. The EjSAUR members assigned to the same clusters tend to have similar motifs, implying that they may play similar functions.
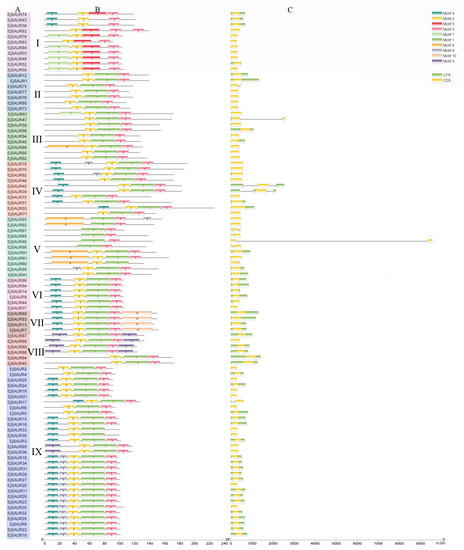
Figure 2.
Conserved motifs and exon–intron structures of the predicted EjSAUR proteins. (A) EjSAURs of different clusters (I–IX). (B) Each motif is represented by a colored box. (C) Exon–intron structures of EjSAUR proteins. The exons and introns are represented by boxes and gray lines, respectively.
2.3. Cis Elements in the Promoters of EjSAURs
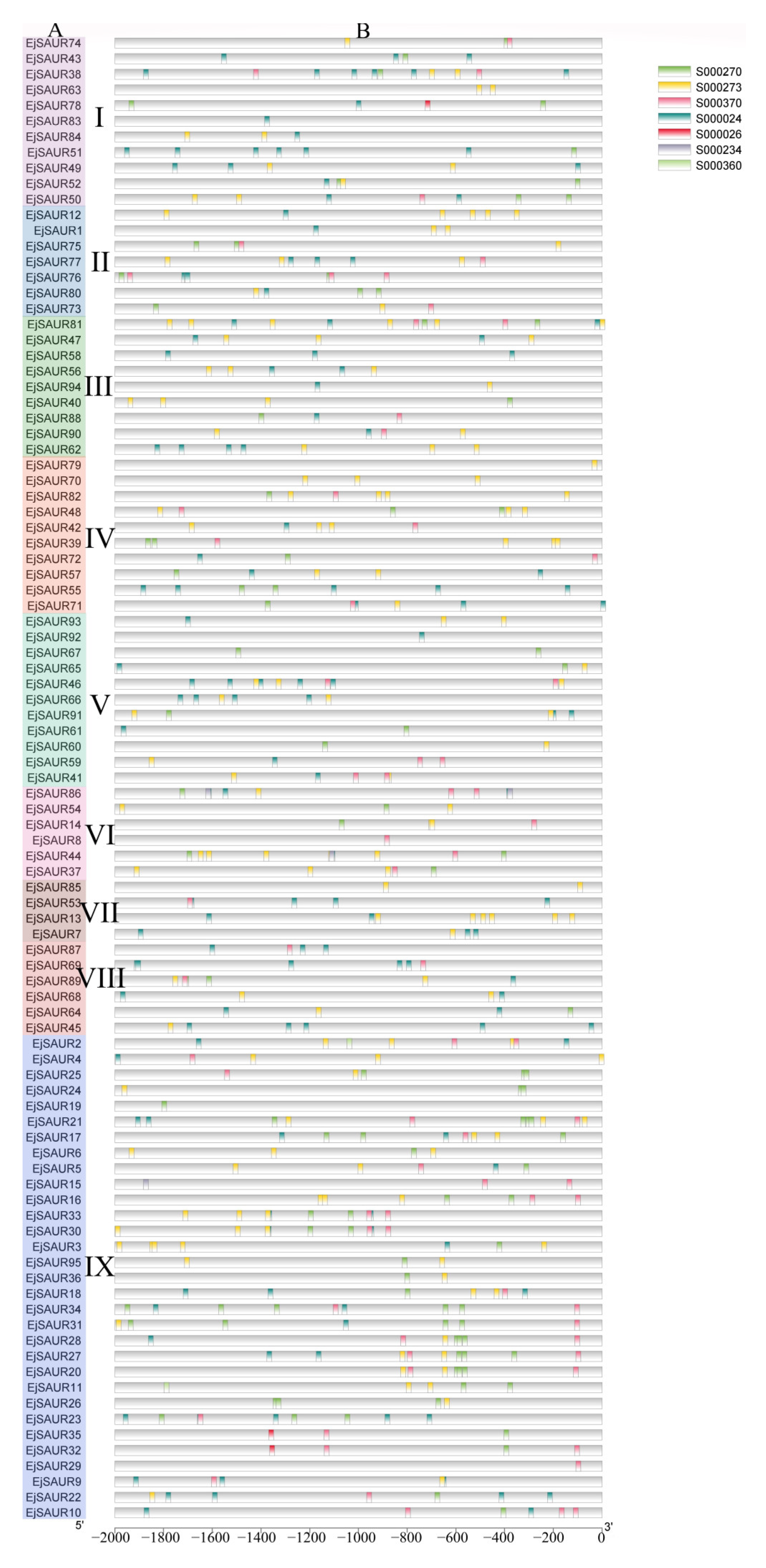
To elucidate the possible regulatory mechanisms under exogenous auxin stimulations, putative auxin-responsive cis-elements were searched in the 2000 bp promoter regions upstream of the transcription start site of the EjSAUR genes. In reference to a study in maize [3], the NEW PLACE website was used to search for seven cis-elements, including TGA-box (S000234), ARF binding (S000270), Dof protein binding (S000273), NDE element (S000360 and S000370), ASF-1 binding (S000024), and AuxRE (S000026). The results showed that all 95 EjSAURs contain at least one of the elements in their promoter regions (Figure 3). Except for AuxRE, TGA-box, and the NDE element (S000360), the remaining four cis-elements seemed universal in many EjSAURs. The presence of these identified cis elements suggests that these EjSAURs are potentially responsive to auxin stimuli and may play a role in plant hormone signaling.
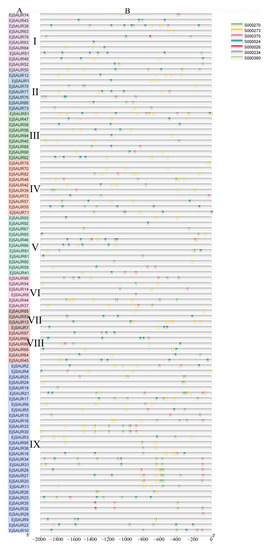
Figure 3.
Distribution of major auxin-responsive cis-elements in the promoter regions of the EjSAUR genes. (A) EjSAURs of different clusters (I–IX). (B) Seven putative cis-elements are represented by different colors as indicated in the figure.
2.4. Chromosomal Locations, Gene Duplication, and Synteny Analysis
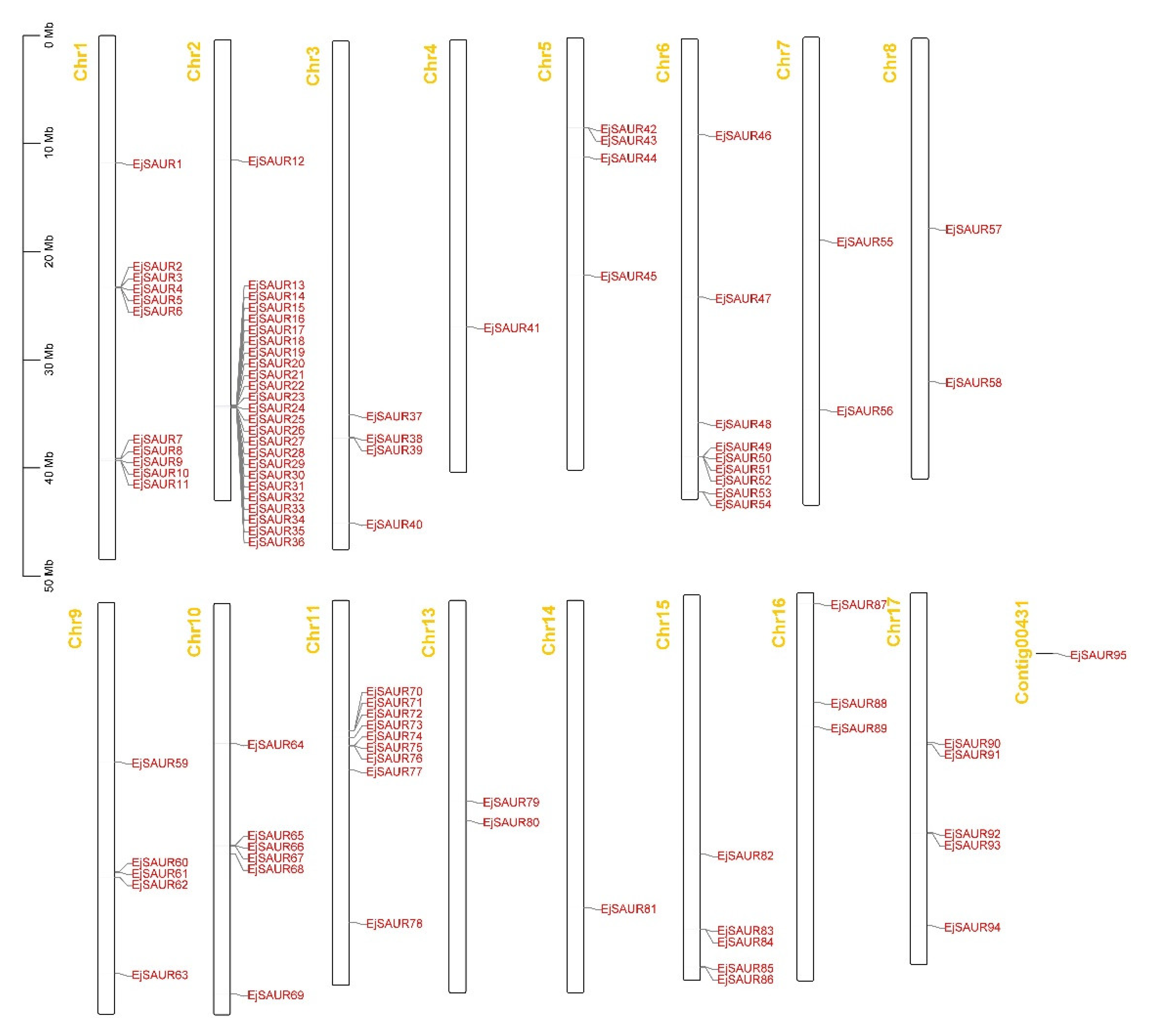
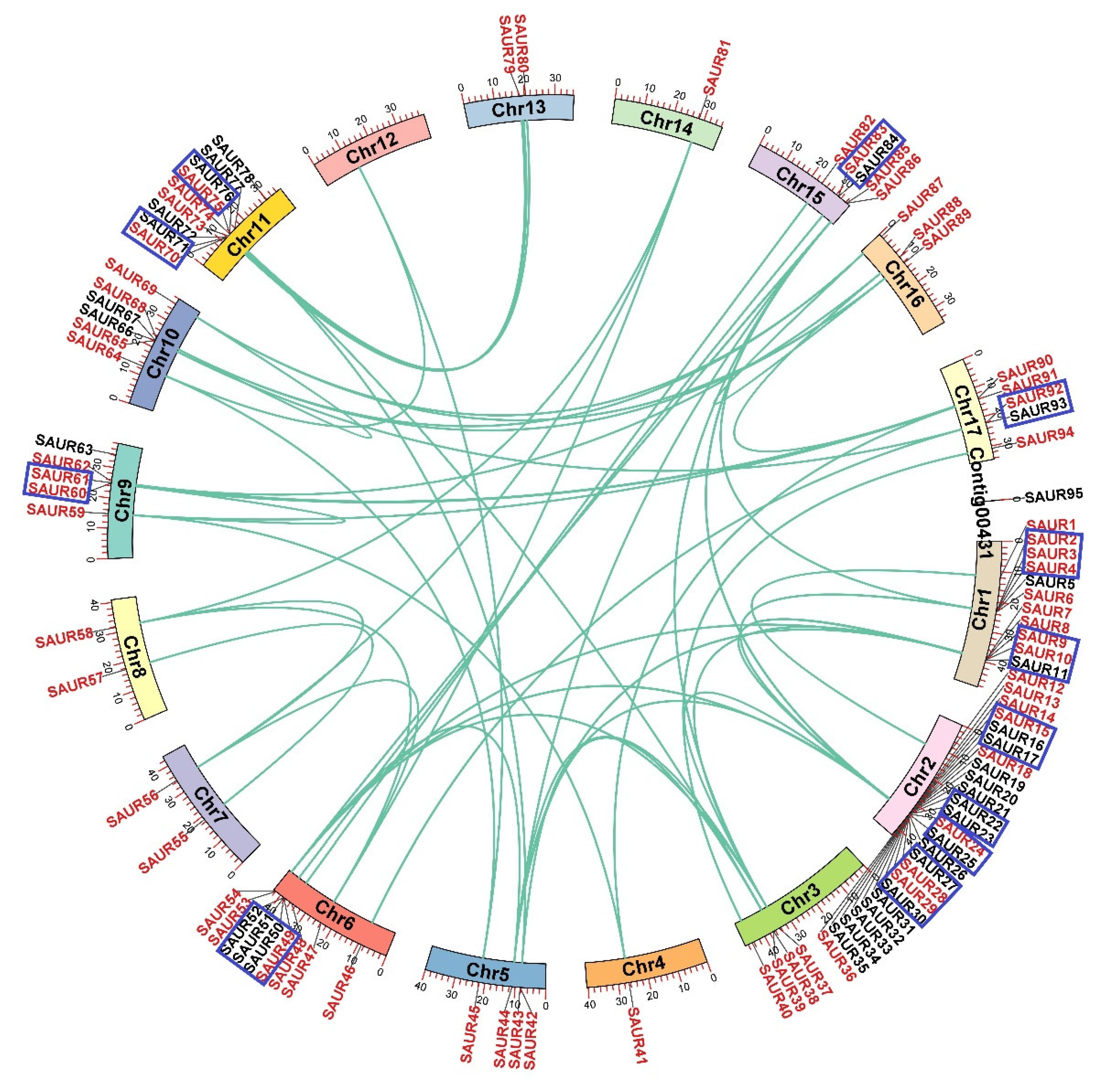
The chromosomal locations of the 95 EjSAURs were plotted to a map (Figure 4). Obviously, they were non-evenly distributed throughout the 17 chromosomes. Surprisingly and interestingly, chromosome two contained a large cluster of EjSAURs (24 members). In contrast, some chromosomes contained only one EjSAUR, such as chromosomes four and fourteen. All tandem and segmental duplicated EjSAURs were plotted to a circos map (Figure 5). Further analysis revealed that between one and four tandem=duplication events of EjSAURs were observed on chromosomes one, two, six, nine, eleven, fifteen, and seventeen (Figure 5). As expected, chromosome two contained four tandem-duplication events, which may explain the large number of EjSAURs on this chromosome. In addition, a total of 61 segmental duplication pairs were identified by MCScanX [32], which may be related to the whole-genome duplication events during the evolution. Therefore, both tandem duplication and segmental duplication contributed to the expansion of EjSAURs in loquats.
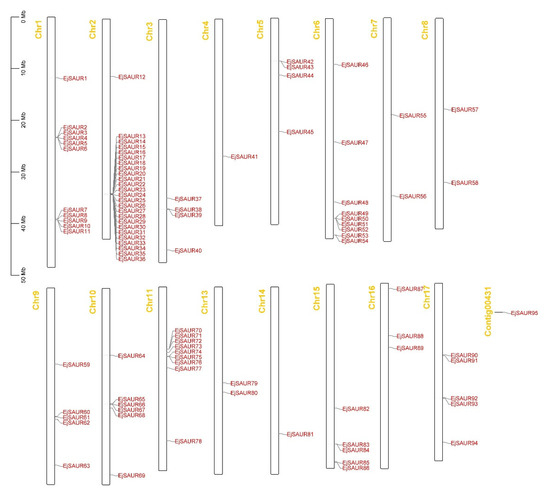
Figure 4.
Gene locations of EjSAURs. The chromosome number is indicated at the top of each chromosome.
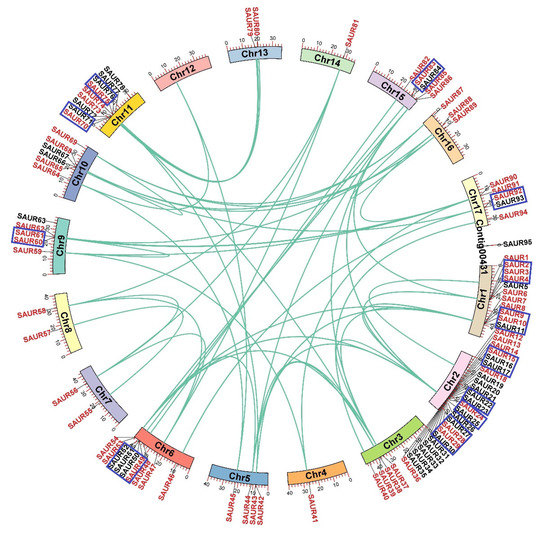
Figure 5.
Tandem and segmental duplications of SAURs in loquats. The genes with a blue rectangule are tandem duplicated genes. The genes with a red font and connected with green lines are segmental duplicated genes.
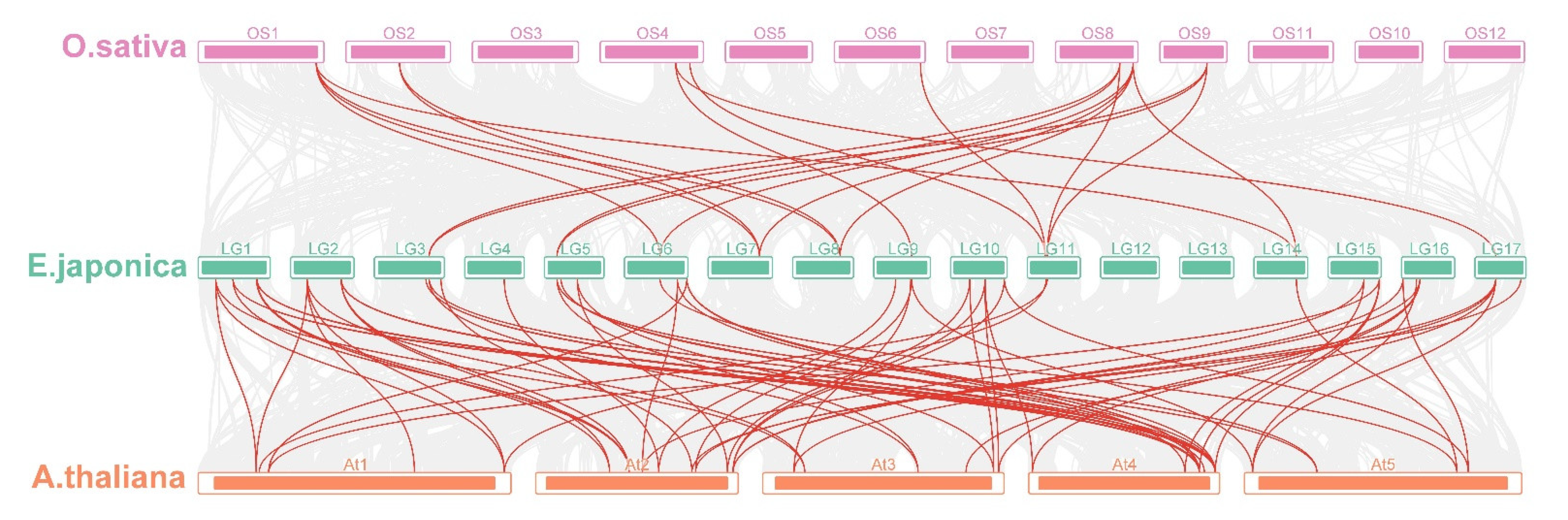
The number of nonsynonymous substitutions per nonsynonymous site (Ka), the number of synonymous substitutions per synonymous site (Ks), and the Ka/Ks values were calculated for all duplicated gene pairs of EjSAURs (Table S2). The results show that almost all pairs had Ka/Ks < 1, implying that these genes were under purifying selection. However, the Ka/Ks values of the segmentally duplicated EjSAUR49/EjSAUR83 (Ka/Ks = 1.30) and tandemly duplicated EjSAUR49/EjSAUR50 (Ka/Ks = 1.53) were >1, suggesting that they were under positive selection and their functions may be differentiated or even that new functions could be evolved. By visualizing the SAURs with syntenic relationships between species, we found that the number of collinear pairs between loquats and Arabidopsis was much more than that between loquats and rice (Figure 6). A total of 45 EjSAURs had a syntenic relationship with the SAURs from Arabidopsis (85 collinear pairs), whereas only 12 EjSAURs had a syntenic relationship with the SAURs from rice (22 collinear pairs). There were nine EjSAURs (EjSAUR38/39/42/43/47/62/74/81/90) with collinear pairs in both Arabidopsis and rice, implying that they may share the same ancestral genes and have similar functions (Figure 6 and Table S2).
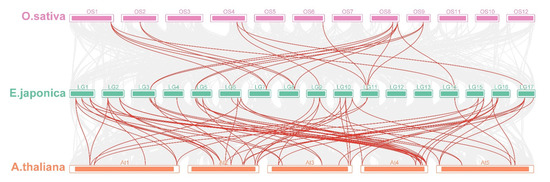
Figure 6.
Multicollinearity analysis of SAURs from loquats, Arabidopsis, and rice. The red lines connect SAUR genes with a collinearity relationship. The grey lines connect other genes with a collinearity relationship.
2.5. Expression Profiles of EjSAURs in Different Tissues
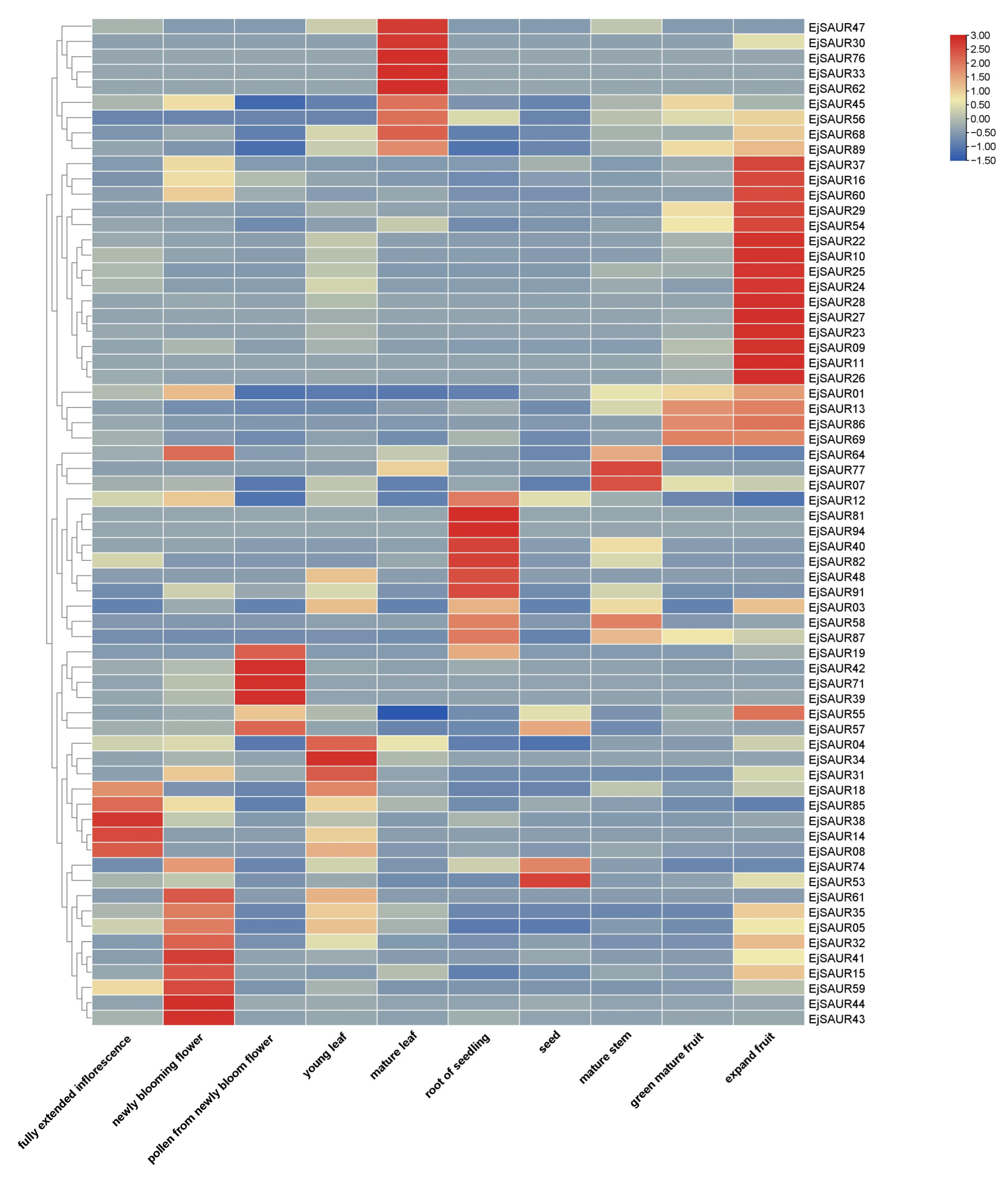
To obtain a global view on the expression patterns of the EjSAURs in various organs and tissues, we re-analyzed a total of 10 transcriptomes of the inflorescence, flower, pollen, young leaf, mature leaf, root, seed, stem, expanding fruit, and green-mature fruit of a cultivar ‘Jiefangzhong’. An expression heatmap was constructed (Figure 7). The results show that many EjSAURs maintain high expression levels in the flower, mature leaf, root, and expanding fruit, implying that these EjSAURs play an important role in the growth and development of loquats, including fruit development. Interestingly, there were 14 EjSAURs (mostly from Cluster IX), such as EjSAUR22, EjSAUR26, and EjSAUR29, with very high expression levels only in expanding fruits, suggesting a vital role in fruit development. In addition, a minor proportion of EjSAURs were not expressed at all in any of the tissues, implying that they may not play important roles in the growth and development of loquats.
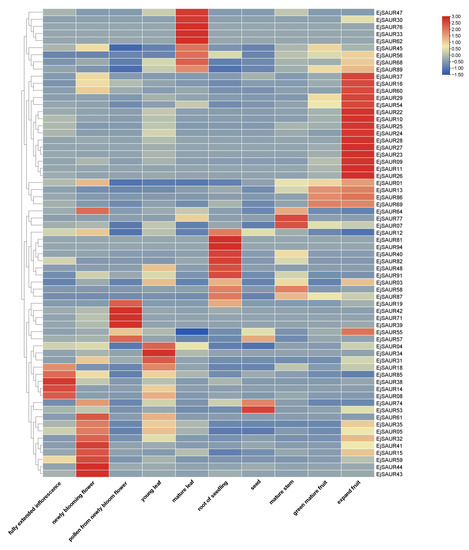
Figure 7.
Tissue-specific expressions of SAUR genes in loquats. The transcripts per million (TPM) values were used for heatmap construction using TBtools.
2.6. Observation of Fruit Development and Expression Patterns of Three EjSAURs
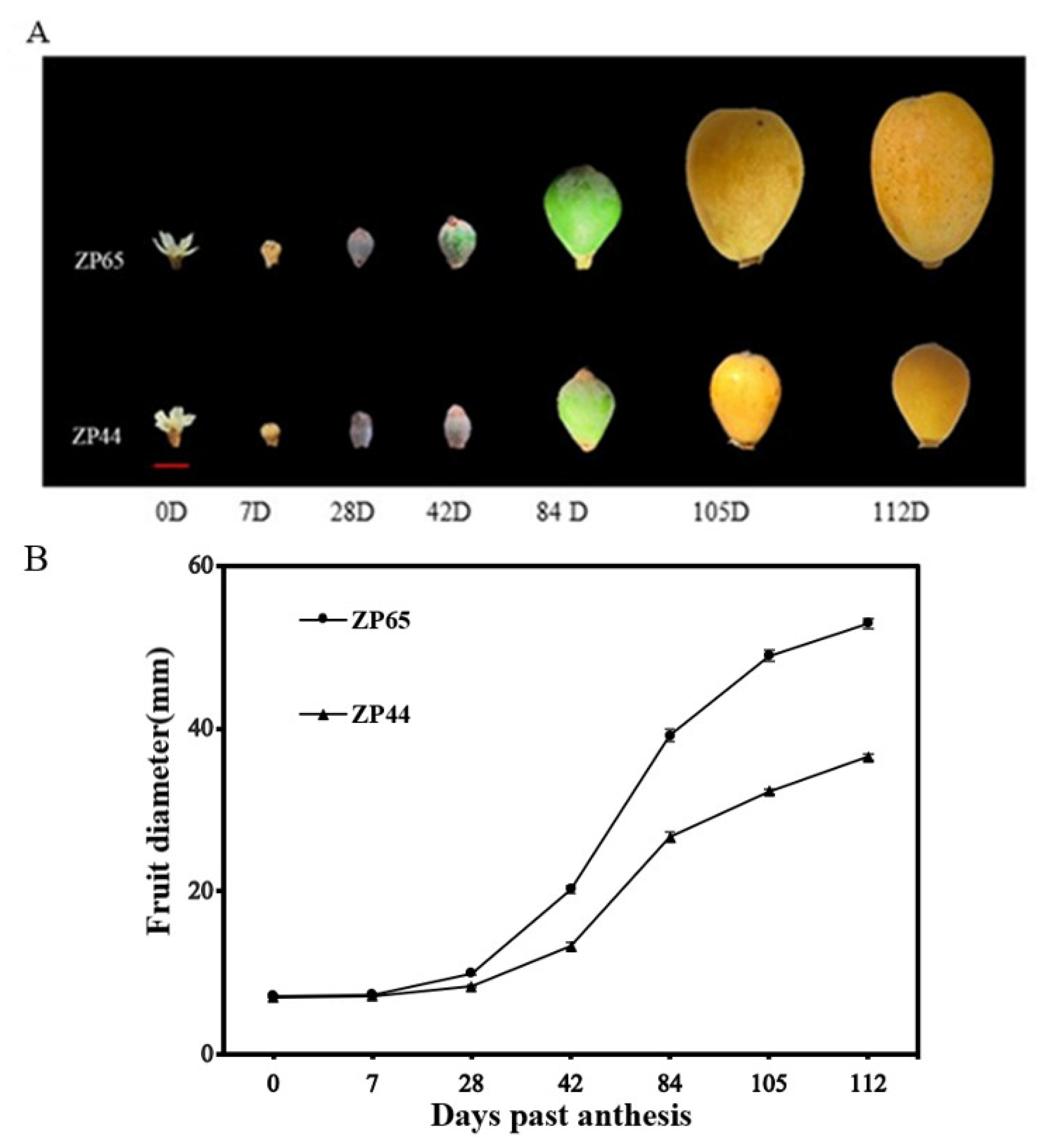
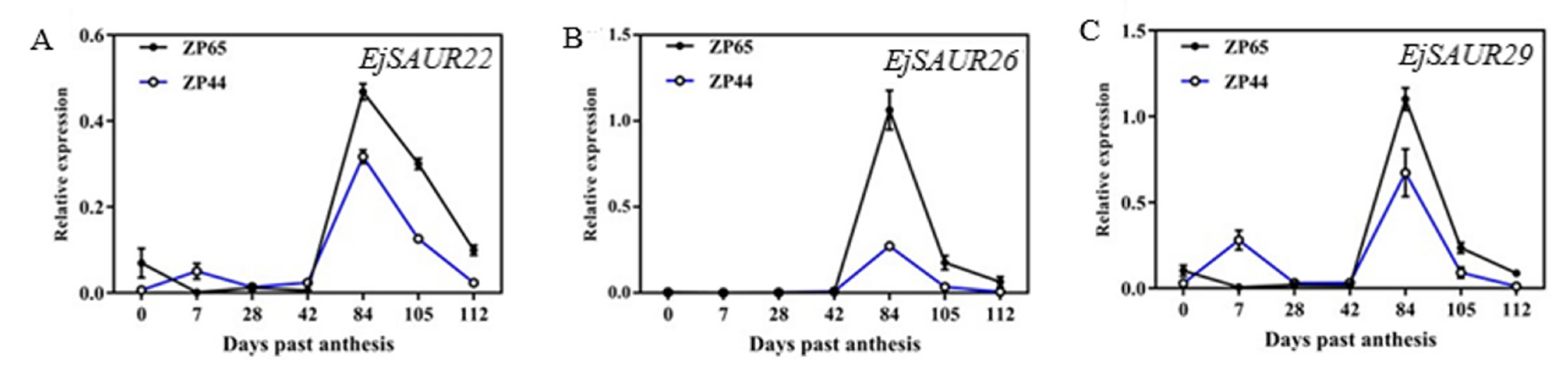
The availability of two sister lines (ZP44 and ZP65) with contrasting fruit size performances enabled us to perform a detailed study of their fruit growth changes and evaluate the expression patterns of three EjSAURs (EjSAUR22, EjSAUR26, and EjSAUR29) across these fruit developmental stages, which were randomly selected out of the fourteen EjSAURs displaying high expression levels at the fruit-expanding stage. In total, seven stages were selected for fruit growth observations, including 0 day past anthesis (DPA), 7 DPA, 28 DPA, 42 DPA, 84 DPA, 105 DPA, and 112 DPA (Figure 8A). The results showed that the fruit size followed an ‘S’ curve pattern (Figure 8B). At earlier stages, the fruits grew relatively slowly, and no big difference was observed between the fruit sizes of ZP44 and ZP65. At 28-84 DPA, the fruits of both ZP44 and ZP65 expanded quickly, but ZP65 expanded at a much higher rate compared with that of ZP44. At around 105 DPA, the fruit sizes of both ZP44 and ZP65 started to plateau.
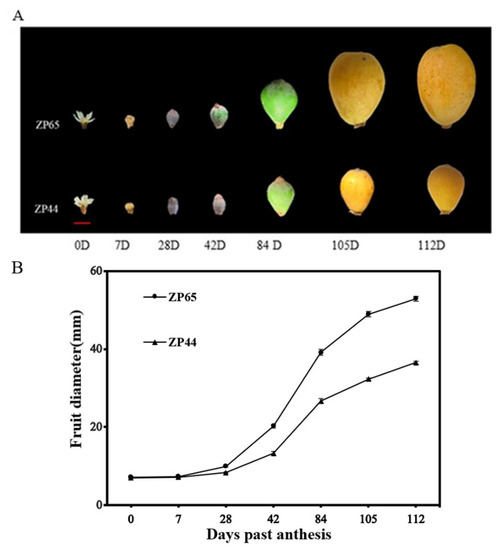
Figure 8.
The fruit growth patterns of ZP44 and ZP65. (A) The receptacles and fruits at seven developmental stages. (B) Changes of fruit diameters along the seven developmental stages. 15 fruits at each stage for each line were measured. The error bars indicate standard errors.
The expression patterns of EjSAUR22, EjSAUR26, and EjSAUR29 were investigated across the above seven developmental stages in ZP44 and ZP65. The three genes showed similar expression patterns in both ZP44 and ZP65: they maintained low expression levels at 0–42 DPA, started increasing after 42 DPA, reached to a peak at 84 DPA, and decreased thereafter (Figure 9A–C). However, the expressions of these three EjSAURs were all considerably higher in ZP65 (large-fruited) than that in ZP44 (small-fruited).
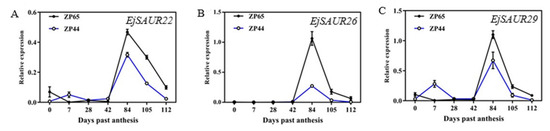
Figure 9.
The expression patterns of three EjSAURs at different fruit developmental stages. (A–C) The expression patterns of EjSAUR22 (A), EjSAUR26 (B), and EjSAUR29 (C). The error bars indicate standard errors.
2.7. EjSAUR22, EjSAUR26, and EjSAUR29 Responses to IAA treatment
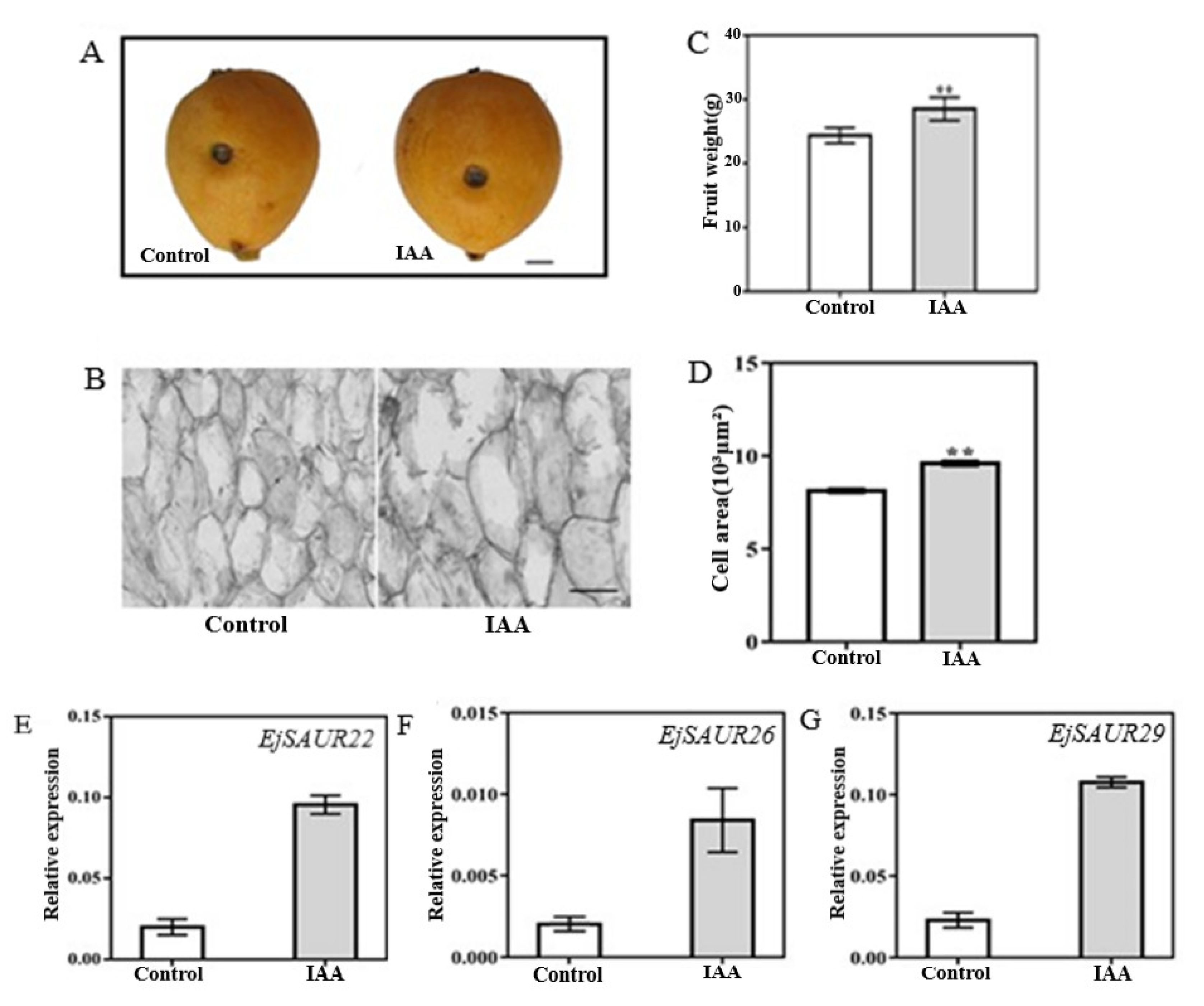
To investigate whether the above three EjSAURs were responsive to IAA, we injected IAA solution (10−7 M) into the fruits of the cultivar ‘Zaozhong No. 6′ at 63 DPA [23]. An exogenous injection of IAA into the fruits at this early expanding stage of ‘Zaozhong No. 6′ showed that the fruits (after reaching maturity at 116 DPA) were significantly larger (p < 0.05) than those of the control, and similar results were observed for fruit weight and cell area (Figure 10A–D). Importantly, at 14 days after treatment (DAT), the expressions of EjSAUR22, EjSAUR26, and EjSAUR29 were all up-regulated in the IAA-treatment group compared with those in the control group, suggesting that they were all auxin-responsive (Figure 10E–G).
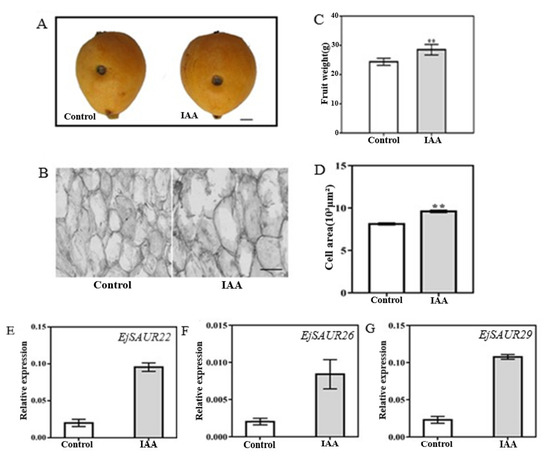
Figure 10.
The influence of the exogenous IAA treatment on fruit growth and the expressions of three EjSAURs. (A) Photos of mature fruits from the control and IAA-treatment groups. The wounds indicate the positions where IAA was injected into. The bar represents 1 cm. (B) Microscopic observation of cell size in fruits from the control and IAA-treatment group. The bar represents 100 μm. (C) Comparison of fruit weights. More than 15 fruits were measured. ‘**’ indicates p-value < 0.01. (D) Comparison of cell areas. ‘**’ indicates p-value < 0.01. (E) The expressions of EjSAUR22, EjSAUR26, and EjSAUR29. The error bars indicate standard errors.
2.8. VIGS Support EjSAUR22′s Role in Cell Expansion and Fruit Size
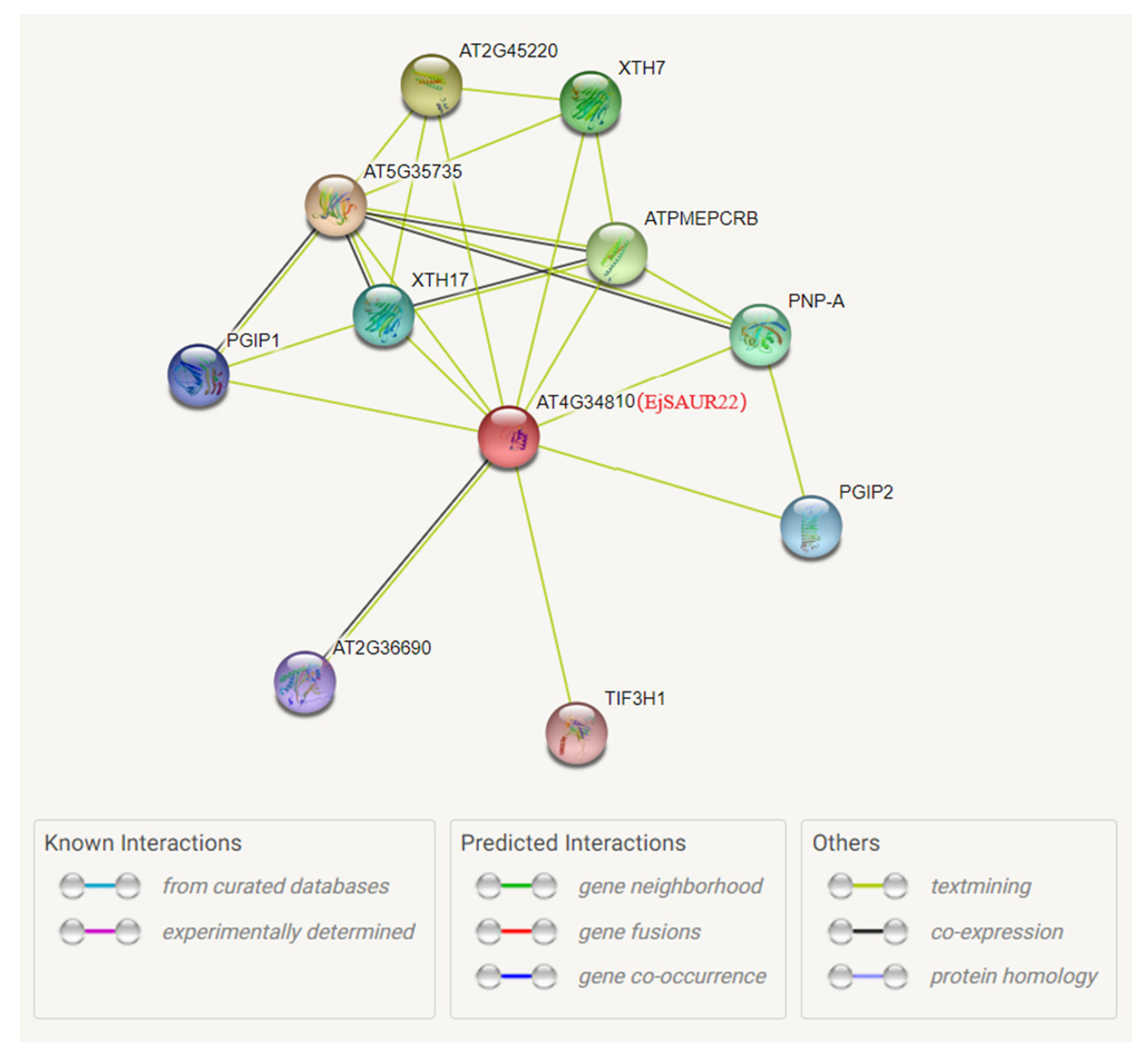
Among the above three EjSAURs, EjSAUR22 showed a syntenic relationship with the gene AT4G34810 in Arabidopsis. Therefore, we further investigated its potential functions by analyzing the protein–protein interaction network of its collinear gene (AT4G34810) in Arabidopsis using the STRING database (Figure 11). Among the top interacting proteins with AT4G34810 were AT5G35735, an auxin-responsive family protein that may act as a catecholamine-responsive trans-membrane electron transporter, and AT4G02330, which may act in the modification of cell walls via demethylesterification of cell wall pectin. This analysis suggested that EjSAUR22 may play a role in fruit expansion by regulating cell size. Subsequently, we further investigated the function of EjSAUR22.
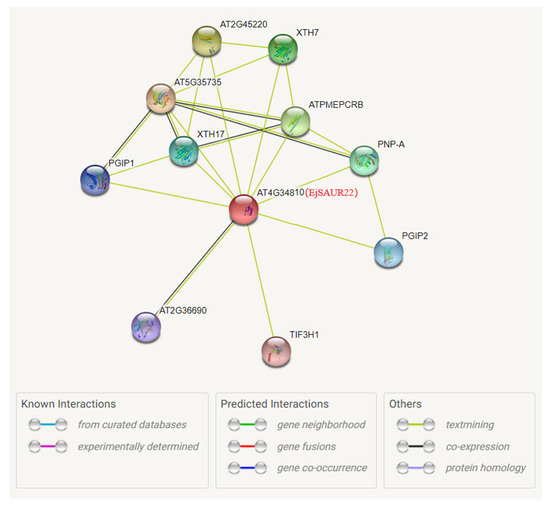
Figure 11.
The predicted protein–protein interaction network of AT4G34810. The AT4G34810 collinear with EjSAUR22 was used to investigate the potential interacted proteins using the STRING tool.
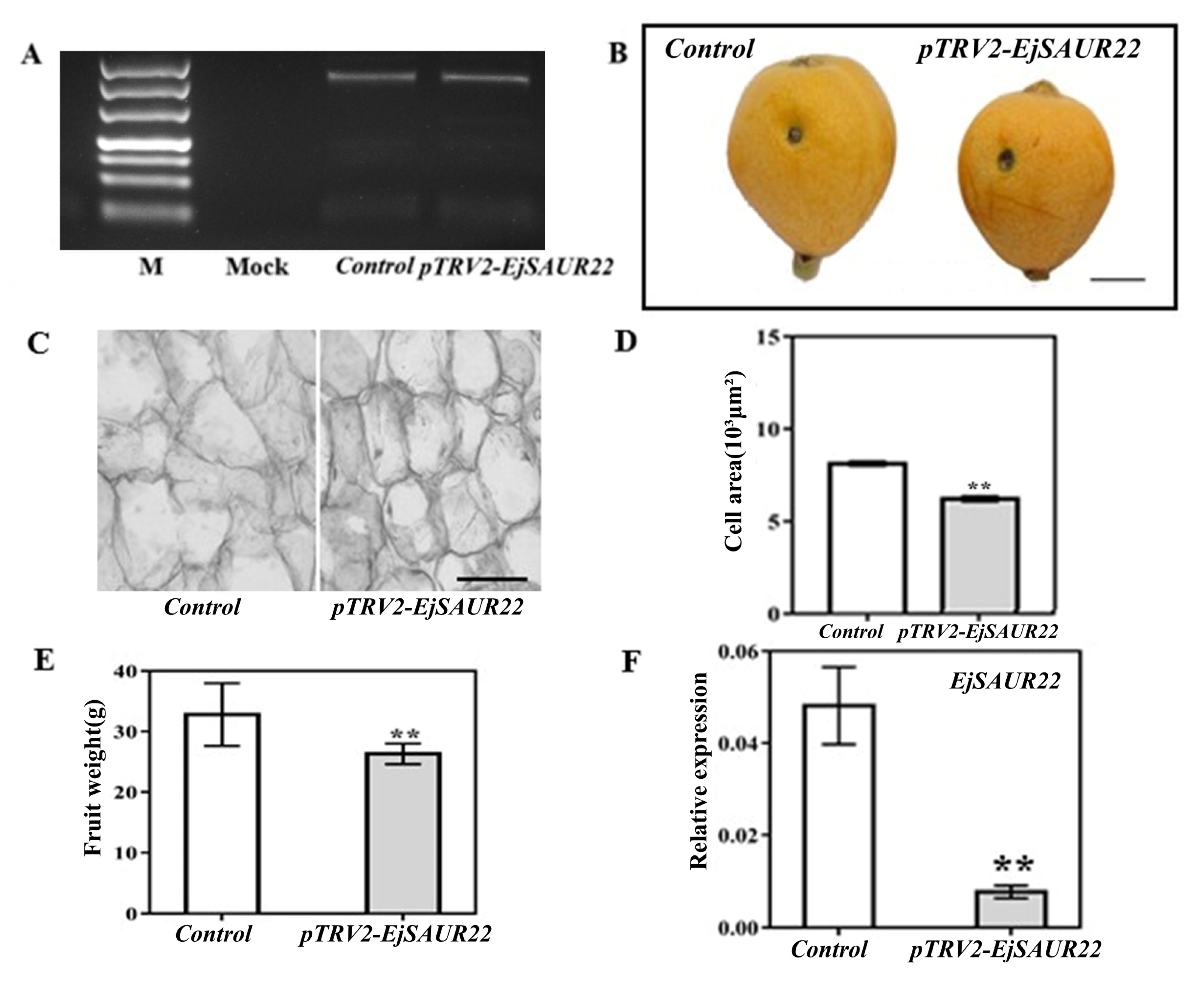
For validation of the function of EjSAUR22, virus induced gene silencing (VIGS) was performed at the early fruit-expanding stage (63 DPA) in fruits of ‘Zaozhong No. 6′ (Figure 12). The amplification of the coat-protein-coding sequence of TRV2 confirmed its presence in both TRV1+TRV2-empty (control) and TRV1+TRV2-EjSAUR (treatment) fruits, while there was no amplification in the mock fruits (no vector injection) (Figure 12A). After reaching maturity (116 DPA), fruits from the treatment group were found to be significantly smaller than those from the control group (Figure 12B,C, p < 0.01). Further histological observations revealed that the cells in the fruits of the treatment group were smaller than those in the control group (Figure 12D). As expected, at 14 DAT, the expression of EjSAUR22 was significantly reduced (p < 0.05) in the treatment group compared with that in the control group.
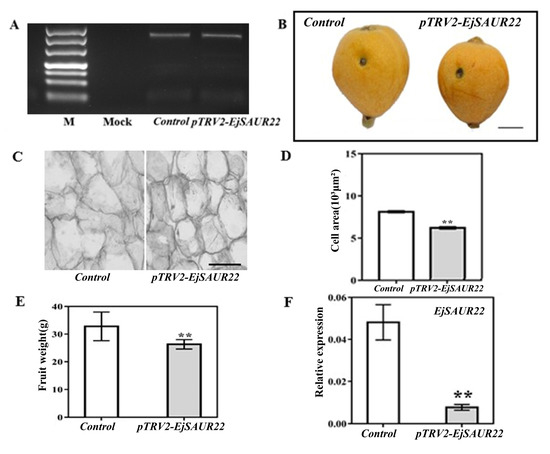
Figure 12.
The influence of VIGS on fruit growth and expressions of EjSAUR22. (A) The detection of the coat-protein-coding sequence (cDNA) of TRV2. (B) Photos of mature fruits from the control and TRV2-EjSAUR22 groups. The bar represents 1 cm. (C) Microscopic observation of cell sizes in fruits from the control and TRV2-EjSAUR22 group. The bar represents 100 μm. (D) Comparison of cell areas. A total of 15 samples were used. (E) Comparison of fruit weights. More than 15 fruits were measured. (F) The expressions of EjSAUR22. The error bars indicate standard errors. ‘**’ indicates p-value < 0.01.
Collectively, our results revealed that EjSAUR22, EjSAUR26, and EjSAUR29 were responsive to IAA treatment, and EjSAUR22 may play an important role in the fruit development of loquats by influencing the cell size and facilitating fruit expansion.
3. Discussion
Fruit quality traits are directly associated with consumer satisfaction and economic returns of fruit trees. Therefore, they have been gaining popularity in the research area. Fruit size is one of the important traits that influence the first impressions from consumers. The SAUR gene family regulated by auxin has proven to play a vital role in fruit development of many plant species [14]. However, this gene family has not been studied in-depth in loquats. Moreover, whether it is associated with fruit size or what members regulate fruit size remain understudied. In this study, we carried out a comprehensive analysis of the SAUR gene family in loquats. By assessing their transcriptional profiles in different tissues and at different fruit developmental stages comparing two sister lines with contrasting fruit sizes, a candidate EjSAUR gene was selected and further investigated. A combination of the IAA treatment experiment and functional validation using VIGS proved its role in regulating cell size and fruit expansion. These results not only provide genomic and genetic resources for fruit size breeding in loquats, but also lay a foundation for understanding the molecular mechanisms of auxin signaling and fruit expansion in loquats.
Previously, many other studies on the SAUR gene family revealed between 60–140 members in each plant species [14]. Similarly in the current study, a total of 95 EjSAURs were identified in the reference genome of ‘Seventh Star’. This number is relatively larger than that reported in Arabidopsis (72) [4], rice [9], and maize (79) [3]. It suggests that the EjSAUR family in loquats has experienced expansion during the evolutionary history. This is supported by the tandem duplication and segmental duplication events identified in the current study, which may be associated with the whole genome duplication in loquats [24]. Interestingly, chromosome two contained the largest number of EjSAURs compared with other chromosomes in loquats and harbored multiple tandem-duplication events, which may imply the special role of chromosome two in the growth and development of loquats. In accordance with the cases in many plants such as Arabidopsis, rice, and apples, the majority of the EjSAURs contain no introns. This seems to be a common feature of the SAUR gene family [25]. In accordance with studies in rice [9] and poplar [33], the EjSAURs showed tissue- or organ-specific gene expressions, indicating that their functions have probably diverged. The EjSAURs from this study were classified into nine clusters. In loquat, most EjSAURs in Cluster IX showed high expressions in expanding fruits, suggesting their role in fruit development. On the basis of collinearity analysis (Figure 6), we found that loquat shared more ortholog pairs of SAUR genes with Arabidopsis (85 pairs) compared with rice (22 pairs), which implied that the two dicots may share some duplication events. Among these collinear pairs, the protein–protein interaction network of the Arabidopsis gene AT4G34810 collinear with EjSAUR22 was used to predict the potential function of EjSAUR22. Due to a potential role of EjSAUR22 in regulating cell size based on this prediction, we selected it for VIGS assay, which supported its function in cell expansion and fruit expansion. Although it would be ideal to obtain transgenic loquat plants to investigate the functions of the EjSAURs, the genetic transformation system is still not well established in loquats. Furthermore, the long juvenile phase would also make this time-consuming.
Currently, fruit size breeding in loquats is still in its infancy, especially compared with several major fruit trees, such as apple and peach. The first reference genome of loquats was released in 2020 [27]. Therefore, the research at molecular and genomics level has just been initiated. In comparison, numerous approaches have already been widely applied in fruit size breeding in apples and peachs, including quantitative trait locus (QTL) mapping, marker-assisted breeding, genome-wide association study (GWAS), and genomic selection [34,35,36]. We believe the future direction of fruit size breeding in loquats will be integrating these advanced techniques with multi-omics, including genomics, transcriptomics, metabolomics, and proteomics, as well as with advanced phenotyping methods.
4. Materials and Methods
4.1. Plant Materials
All loquat trees (13-year-old) were grown in the Eriobotrya Germplasm Resource Preservation Garden (South China Agricultural University, Guangzhou, China) under regular management conditions. Two sister lines (ZP44 and ZP65) with contrasting fruit size performances were used for growth observations and gene expression assays at 0 days past anthesis (DPA), 7 DPA, 28 DPA, 42 DPA, 84 DPA, 105 DPA, and 112 DPA. For each analysis, 15 fruits at each stage for each line were used. Tissues close to the ovule or along the equatorial plane of the fruits were used as materials.
The cultivar ‘Zaozhong No. 6′ was used for IAA treatment. A total of 30 inflorescences at similar developmental stages and with similar sizes were selected. IAA was dissolved in 0.1% ethanol to obtain an IAA solution (10−7 M) for treatment, while the control used 0.1% ethanol solution. At the early fruit-expanding stage (63 DPA) in reference to our previous report [23], an Injex-30 injector (Thesera, Daegu, Korea) was used for injecting the solutions into the fruits along the equatorial plane. After reaching maturity at 116 DPA, fruits were collected for weight/size measurement and histological analyses. The fruit flesh tissues close to the ovule and along the equatorial plane were used. At 14 days after treatment (DAT), gene expression assays were performed.
4.2. Identification of the SAUR Gene Family
Among the three available reference genomes of E. japonica [24,27,28], the ‘Seventh Star’ genome [27] with the highest BUSCO score (99.1%, embryophyta_odb10) of the annotated gene models was used for genome-wide mining of SAUR genes. All SAUR proteins from Arabidopsis (AtSAURs) were searched and downloaded from the TAIR database (https://www.arabidopsis.org/ (accessed on 6 April 2021)). Two strategies were applied for identifying the SAUR genes in loquats. Firstly, the hidden Markov model (HMM) of the Auxin-inducible domain (PF02519) was downloaded from the Pfam database (https://www.ebi.ac.uk/interpro/ (accessed on 6 April 2021)) and used as a query to search against the protein sequences of the ‘Seventh Star’ with HMMER v3.3.1 from Sean R. Eddy and the HMMER development team (Cambridge, MA, USA) [37] under an e-value cutoff of 1 × 10−5. Secondly, the protein sequences of AtSAURs were used as the query to compare against the loquat proteins with BLASTP under an e-value cutoff 1 × 10−5 and identity >40%. Subsequently, the protein sequences of the above-identified genes were subjected to a further analysis using the Pfam database, the NCBI Conserved Domain tool, and the SMART database (https://smart.embl.de/ (accessed on 12 April 2021)) to confirm the presence of the SAUR domain. The ProtParam tool (https://web.expasy.org/protparam/ (accessed on 12 April 2021)) was applied to predict the physicochemical parameters, such as length, molecular weight, and isoelectric point of the identified SAUR proteins. The CELLO v2.5 server, developed by Chin-Sheng Yu and the team (Hsinchu, China), was used to investigate the subcellular localization [38].
4.3. Phylogenetic Analysis
A phylogenetic analysis was performed using the SAUR genes from Arabidopsis, rice, and loquats. The SAUR proteins from rice (OsSAURs) were obtained from a previous study [39]. The protein sequences were aligned using MAFFT [40]. MEGA11 was used for phylogenetic tree construction with the neighbor-joining (NJ) method and 1000 bootstraps, and other parameters were as default [41].
4.4. Chromosomal Locations, Gene Duplication, and Protein–-Protein Interaction Network
The physical locations of the identified SAUR genes on the 17 chromosomes of the loquat genome were visualized using TBtools [42]. Synteny analysis was performed using MCScanX [32] in TBtools for the SAUR genes between Arabidopsis & loquats and rice & loquats, as well as within loquats. Multi-collinearity analysis was performed and visualized using TBtools based on the results of the synteny analysis. Gene tandem-duplication events and segmental duplication events were also catalogued from the output of MCScanX. The nonsynonymous (Ka)/synonymous (Ks) analysis was performed using the Simple Ka/Ks Calculator within TBtools on the genes associated with tandem or segmental duplication events. The orthologs of EjSAURs from Arabidopsis were used to construct the protein–protein interaction networks with STRING (https://string-db.org/cgi/input.pl (accessed on 28 April 2021)).
4.5. Analysis of Gene Structure, Conserved Motifs, and cis-Elements of EjSAURs
The gene structure information was extracted from the GFF3 file of the ‘Seventh Star’ reference genome, which was visualized using TBtools. Conserved motifs were identified using MEME v5.4.1 (https://meme-suite.org/meme/ (accessed on 17 April 2021)). The 2 Kb sequences upstream of the EjSAURs were extracted and submitted to the New PLACE website (https://www.dna.affrc.go.jp/PLACE/?action=newplace (accessed on 17 April 2021)) for searching auxin-responsive cis-elements in their promoters.
4.6. RNA Extraction and qRT-PCR
RNA samples were extracted using the EASYspin Plus plant RNA extraction kit (Aidlab, Beijing, China). The first-strand cDNA was synthesized using the PrimeScriptTM RT reagent kit (TaKaRa, Kusatsu, Japan). qRT-PCR was performed following our previous report [43]. Primers for qRT-PCR were designed using BatchPrimer3 (v1.0) from Frank M You and the development team (Davis, CA, USA) [44]. All primer sequences are provided in Table S3. EjRPL18 was used as the reference gene [45].
4.7. Virus Induced Gene Silencing
The coding sequences of EjSAUR22 were cloned into the TRV2 vector to perform virus-induced gene silencing (VIGS) in the fruits of the cultivar ‘Zaozhong No. 6′. The VIGS experiment was carried out following the same method described previously [43], except that the Agrobacterium tumefaciens strain GV3101 was used. The TRV2-empty, TRV2-EjSAUR22, and TRV1 vectors were introduced into the Agrobacterium tumefaciens strain GV3101. At the early fruit-expanding stage (63 DPA), in reference to our previous report [23], an Injex-30 infector was used to inject the TRV1+TRV2:EjSAUR22 mixed Agrobacterium cells into the fruit near the equator. The TRV1+TRV2:Empty mixed Agrobacterium cells were used as a control. At 7 DAT, samples were collected to detect the presence and levels of the TRV2 virus. At 14 DAT, gene expression assays were performed.
4.8. RNA-Seq Analysis
To investigate the tissue-specific expressions of SAURs, our previously published transcriptome data from the cultivar ‘Jiefangzhong’ [24], covering tissues including the inflorescence, flower, pollen, young leaf, mature leaf, root, seed, stem, young fruit, expanding fruit, and mature fruit were re-analyzed using the ‘Seventh Star’ reference genome following the same method described previously [22]. The gene-expression heatmap was constructed using transcripts per million (TPM) values and TBtools.
5. Conclusions
Collectively, we performed a comprehensive analysis of the EjSAUR gene family in loquats, including investigating their physio-chemical features, evolutionary relationships, chromosomal locations, gene structures, cis-regulatory elements, and syntenic relationships. Through mining RNA-seq data, we obtained a group of EjSAURs highly expressed in expanding fruits that may play vital roles in the development loquat fruits. The results from the current study provide a theoretical foundation for future exploration of the features and functions of more EjSAUR genes. The EjSAUR22 associated with cell expansion and fruit size could facilitate a deeper understanding of auxin signaling in the fruit development of loquats. Moreover, it may serve as a potential target for the manipulation of fruit size and accelerated breeding in loquats.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms232113271/s1.
Author Contributions
X.Y., Z.P., and Z.L. designed and supervised the project. W.L. performed bioinformatics data analysis under supervision by Z.P. and D.P., X.G. performed experiments with help from W.S., C.Z., J.L., S.L. and Z.L. secured plant materials. Z.P., W.L., and X.G. wrote the original manuscript draft. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Key R&D Program of China (2019YFD1000200) and the National Natural Science Foundation of China (31901973).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hoffmann, M.; Hentrich, M.; Pollmann, S. Auxin-Oxylipin Crosstalk: Relationship of Antagonists. J. Integr. Plant Biol. 2011, 53, 429–445. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Hayashi, K.; Kinoshita, T. Auxin Activates the Plasma Membrane H+-ATPase by Phosphorylation during Hypocotyl Elongation in Arabidopsis. Plant Physiol. 2012, 159, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Hao, X.; Cao, J. Small Auxin Upregulated RNA (SAUR) Gene Family in Maize: Identification, Evolution, and Its Phylogenetic Comparison with Arabidopsis, Rice, and Sorghum. J. Integr. Plant Biol. 2014, 56, 133–150. [Google Scholar] [CrossRef]
- Hagen, G.; Guilfoyle, T. Auxin-Responsive Gene Expression: Genes, Promoters and Regulatory Factors. Plant Mol. Biol. 2002, 49, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Guilfoyle, T.J.; Hagen, G. Auxin Response Factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.P.; Nourizadeh, S.D.; Peer, W.A.; Xu, J.; Bandyopadhyay, A.; Murphy, A.S.; Goldsbrough, P.B. Arabidopsis AtGSTF2 Is Regulated by Ethylene and Auxin, and Encodes a Glutathione S-Transferase That Interacts with Flavonoids. Plant J. 2003, 36, 433–442. [Google Scholar] [CrossRef]
- Franco, A.R.; Gee, M.A.; Guilfoyle, T.J. Induction and Superinduction of Auxin-Responsive MRNAs with Auxin and Protein Synthesis Inhibitors. J. Biol. Chem. 1990, 265, 15845–15849. [Google Scholar] [CrossRef]
- McClure, B.A.; Guilfoyle, T. Characterization of a Class of Small Auxin-Inducible Soybean Polyadenylated RNAs. Plant Mol. Biol. 1987, 9, 611–623. [Google Scholar] [CrossRef]
- Jain, M.; Tyagi, A.K.; Khurana, J.P. Genome-Wide Analysis, Evolutionary Expansion, and Expression of Early Auxin-Responsive SAUR Gene Family in Rice (Oryza sativa). Genomics 2006, 88, 360–371. [Google Scholar] [CrossRef]
- Wang, S.; Bai, Y.; Shen, C.; Wu, Y.; Zhang, S.; Jiang, D.; Guilfoyle, T.J.; Chen, M.; Qi, Y. Auxin-Related Gene Families in Abiotic Stress Response in Sorghum Bicolor. Funct. Integr. Genom. 2010, 10, 533–546. [Google Scholar] [CrossRef]
- Xie, R.; Dong, C.; Ma, Y.; Deng, L.; He, S.; Yi, S.; Lv, Q.; Zheng, Y. Comprehensive Analysis of SAUR Gene Family in Citrus and Its Transcriptional Correlation with Fruitlet Drop from Abscission Zone A. Funct. Integr. Genom. 2015, 15, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Huang, X.; Bao, Y.; Wang, B.; Zeng, H.; Cheng, W.; Tang, M.; Li, Y.; Ren, J.; Sun, Y. Genome-Wide Identification of SAUR Genes in Watermelon (Citrullus lanatus). Physiol. Mol. Biol. Plants 2017, 23, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Lu, S.; Xie, M.; Wu, M.; Ding, S.; Khaliq, A.; Ma, Z.; Mao, J.; Chen, B. Identification and Expression Analysis of the Small Auxin-up RNA (SAUR) Gene Family in Apple by Inducing of Auxin. Gene 2020, 750, 144725. [Google Scholar] [CrossRef] [PubMed]
- Stortenbeker, N.; Bemer, M. The SAUR Gene Family: The Plant’s Toolbox for Adaptation of Growth and Development. J. Exp. Bot. 2019, 70, 17–27. [Google Scholar] [CrossRef]
- Park, J.-E.; Kim, Y.-S.; Yoon, H.-K.; Park, C.-M. Functional Characterization of a Small Auxin-up RNA Gene in Apical Hook Development in Arabidopsis. Plant Sci. 2007, 172, 150–157. [Google Scholar] [CrossRef]
- Chae, K.; Isaacs, C.G.; Reeves, P.H.; Maloney, G.S.; Muday, G.K.; Nagpal, P.; Reed, J.W. Arabidopsis SMALL AUXIN UP RNA63 Promotes Hypocotyl and Stamen Filament Elongation. Plant J. 2012, 71, 684–697. [Google Scholar] [CrossRef]
- Hou, K.; Wu, W.; Gan, S.S. SAUR36, a Small Auxin up RNA Gene, Is Involved in the Promotion of Leaf Senescence in Arabidopsis. Plant Physiol. 2013, 161, 1002–1009. [Google Scholar] [CrossRef]
- Wen, Z.; Mei, Y.; Zhou, J.; Cui, Y.; Wang, D.; Wang, N.N. SAUR49 Can Positively Regulate Leaf Senescence by Suppressing SSPP in Arabidopsis. Plant Cell Physiol. 2020, 61, 644–658. [Google Scholar] [CrossRef]
- Spartz, A.K.; Lee, S.H.; Wenger, J.P.; Gonzalez, N.; Itoh, H.; Inzé, D.; Peer, W.A.; Murphy, A.S.; Overvoorde, P.J.; Gray, W.M. The SAUR19 Subfamily of SMALL AUXIN UP RNA Genes Promote Cell Expansion. Plant J. 2012, 70, 978–990. [Google Scholar] [CrossRef]
- Kant, S.; Bi, Y.M.; Zhu, T.; Rothstein, S.J. SAUR39, a Small Auxin-up RNA Gene, Acts as a Negative Regulator of Auxin Synthesis and Transport in Rice. Plant Physiol. 2009, 151, 691–701. [Google Scholar] [CrossRef]
- Li, M.; Chen, R.; Gu, H.; Cheng, D.; Guo, X.; Shi, C.; Li, L.; Xu, G.; Gu, S.; Wu, Z.; et al. Grape Small Auxin Upregulated RNA (SAUR) 041 Is a Candidate Regulator of Berry Size in Grape. Int. J. Mol. Sci. 2021, 22, 1818. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Zhao, C.; Li, S.; Guo, Y.; Xu, H.; Hu, G.; Liu, Z.; Chen, X.; Chen, J.; Lin, S.; et al. Integration of Genomics, Transcriptomics and Metabolomics Identifies Candidate Loci Underlying Fruit Weight in Loquat. Hortic. Res. 2022, 9, uhac037. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Zhu, Y.; Zhang, L.; Yang, X.; Gao, Y.; Lin, S. The Cellular Physiology of Loquat (Eriobotrya japonica Lindl.) Fruit with a Focus on How Cell Division and Cell Expansion Processes Contribute to Pome Morphogenesis. Sci. Hortic. 2017, 224, 142–149. [Google Scholar] [CrossRef]
- Su, W.; Jing, Y.; Lin, S.; Yue, Z.; Yang, X.; Xu, J.; Wu, J.; Zhang, Z.; Xia, R.; Zhu, J.; et al. Polyploidy Underlies Co-Option and Diversification of Biosynthetic Triterpene Pathways in the Apple Tribe. Proc. Natl. Acad. Sci. USA 2021, 118, e2101767118. [Google Scholar] [CrossRef]
- Pattison, R.J.; Csukasi, F.; Catalá, C. Mechanisms Regulating Auxin Action during Fruit Development. Physiol. Plant 2014, 151, 62–72. [Google Scholar] [CrossRef]
- Sundberg, E.; Østergaard, L. Distinct and Dynamic Auxin Activities during Reproductive Development. Cold Spring Harbor. Perspect. Biol. 2009, 1, a001628. [Google Scholar] [CrossRef]
- Jiang, S.; An, H.; Xu, F.; Zhang, X. Chromosome-Level Genome Assembly and Annotation of the Loquat (Eriobotrya japonica) Genome. Gigascience 2020, 9, giaa015. [Google Scholar] [CrossRef]
- Wang, Y. A Draft Genome, Resequencing, and Metabolomes Reveal the Genetic Background and Molecular Basis of the Nutritional and Medicinal Properties of Loquat (Eriobotrya japonica (Thunb.) Lindl). Hortic. Res. 2021, 8, 231. [Google Scholar] [CrossRef]
- Navarro, E.; Mallén, A.; Hueso, M. Dynamic Variations of 3′UTR Length Reprogram the MRNA Regulatory Landscape. Biomedicines 2021, 9, 1560. [Google Scholar] [CrossRef]
- Aguilar-Hernández, V.; Guzmán, P. Spliceosomal Introns in the 5′ Untranslated Region of Plant BTL RING-H2 Ubiquitin Ligases Are Evolutionary Conserved and Required for Gene Expression. BMC Plant Biol. 2013, 13, 179. [Google Scholar] [CrossRef]
- Kamo, K.; Kim, A.Y.; Park, S.H.; Joung, Y.H. The 5′UTR-Intron of the Gladiolus Polyubiquitin Promoter GUBQ1 Enhances Translation Efficiency in Gladiolus and Arabidopsis. BMC Plant Biol. 2012, 12, 79. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Yan, H.; Luo, S.; Pan, F.; Wang, Y.; Xiang, Y. Genome-Wide Analysis of Poplar SAUR Gene Family and Expression Profiles under Cold, Polyethylene Glycol and Indole-3-Acetic Acid Treatments. Plant Physiol. Biochem. 2018, 128, 50–65. [Google Scholar] [CrossRef]
- Muranty, H.; Troggio, M.; Sadok, I.B.; Rifaï, M.A.; Auwerkerken, A.; Banchi, E.; Velasco, R.; Stevanato, P.; Van De Weg, W.E.; Di Guardo, M. Accuracy and Responses of Genomic Selection on Key Traits in Apple Breeding. Hortic. Res. 2015, 2, 15060. [Google Scholar] [CrossRef] [PubMed]
- Laurens, F.; Aranzana, M.J.; Arus, P.; Bassi, D.; Bink, M.; Bonany, J.; Caprera, A.; Corelli-Grappadelli, L.; Costes, E.; Durel, C.E. An Integrated Approach for Increasing Breeding Efficiency in Apple and Peach in Europe. Hortic. Res. 2018, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cao, K.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Zhao, P.; Guo, J.; Ding, T.; Guan, L. Genomic Analyses of an Extensive Collection of Wild and Cultivated Accessions Provide New Insights into Peach Breeding History. Genome biology 2019, 20, 36. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.S.; Lin, C.J.; Hwang, J.K. Predicting Subcellular Localization of Proteins for Gram-Negative Bacteria by Support Vector Machines Based on n-Peptide Compositions. Protein. Sci. 2004, 13, 1402–1406. [Google Scholar] [CrossRef]
- Zhang, H.; Yu, Z.; Yao, X.; Chen, J.; Chen, X.; Zhou, H.; Lou, Y.; Ming, F.; Jin, Y. Genome-Wide Identification and Characterization of Small Auxin-up RNA (SAUR) Gene Family in Plants: Evolution and Expression Profiles during Normal Growth and Stress Response. BMC Plant Biol. 2021, 21, 4. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Shao, Z.; Wang, M.; Gan, X.; Yang, X.; Lin, S. EjBZR1 Represses Fruit Enlargement by Binding to the EjCYP90 Promoter in Loquat. Hortic. Res. 2021, 8, 152. [Google Scholar] [CrossRef] [PubMed]
- You, F.M.; Huo, N.; Gu, Y.Q.; Luo, M.C.; Ma, Y.; Hane, D.; Lazo, G.R.; Dvorak, J.; Anderson, O.D. BatchPrimer3: A High Throughput Web Application for PCR and Sequencing Primer Design. BMC Bioinform. 2008, 9, 253. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Yuan, Y.; Zhang, L.; Jiang, Y.; Gan, X.; Bai, Y.; Peng, J.; Wu, J.; Liu, Y.; Lin, S. Selection of the Optimal Reference Genes for Expression Analyses in Different Materials of Eriobotrya Japonica. Plant Methods 2019, 15, 7. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).